SUMMARY
The capability of “Candidatus Accumulibacter” to use nitrate as an electron acceptor for phosphorus uptake was investigated using two activated sludge communities. The two communities were enriched in Accumulibacter clade IA and clade IIA, respectively. By performing a series of batch experiments, we found that clade IA was able to couple nitrate reduction with phosphorus uptake, but clade IIA could not. These results agree with a previously proposed hypothesis that different populations of Accumulibacter have different nitrate reduction capabilities, and they will help to understand the ecological roles that these two clades provide.
Keywords: Enhanced biological phosphorus removal, “Candidatus Accumulibacter phosphatis”, denitrification
Enhanced biological phosphorus removal (EBPR) activated sludge has been applied for decades to economically reduce wastewater effluent phosphorus (P) levels. Using molecular techniques, as yet uncultured organisms related to “Candidatus Accumulibacter phosphatis” (hereafter referred to as Accumulibacter) are frequently identified as the major polyphosphate accumulating organisms (PAOs) in lab-scale EBPR systems fed with acetate or propionate (Hesselmann et al., 1999; Crocetti et al., 2000) and some full-scale EBPR facilities (Zilles et al., 2002; Kong et al., 2004; He et al., 2008). A rich diversity of Accumulibacter can be organized into two main Types (I and II), using the polyphosphate kinase gene (ppk1) as a genetic marker. Each Type is comprised of several distinct clades (He et al., 2007; Peterson et al., 2008). Lab-scale acetate-fed reactors studied to date have exclusively contained clades IA, IIA, and IID (Peterson et al., 2008; Wilmes et al., 2008). Accumulibacter has been reported to be able to use nitrate as an electron acceptor to achieve anoxic P-uptake, based on work conducted on activated sludge communities (Dabert et al., 2001; Zeng et al., 2003; Kong et al., 2004). However, a metagenomic analysis of clade IIA-enriched EBPR sludges indicated a lack of any clearly identifiable respiratory nitrate reductase homolog in Accumulibacter (Garcia Martin et al., 2006). Recently, Carvalho and colleagues (2007) reported varying nitrate reduction capabilities in two lab-scale reactors fed with acetate and propionate, respectively, and containing two morphologically distinct Accumulibacter populations. Further work determined that Accumulibacter populations established in a lab-scale bioreactor under nitrite reducing conditions could not reduce nitrate (Guisasola et al., 2009). The differences in nitrate reduction activities suggested finer phenotypic and ecological differences among members of the Accumulibacter lineage. Therefore, we hypothesized that clade IA can use nitrate as electron acceptor for anoxic P-uptake, while clade IIA cannot.
Two lab-scale acetate-fed EPBR sequencing batch reactors (R1 and R2), with good and stable P removal performance were operated in parallel under essentially identical conditions. R1 was inoculated from a local EBPR wastewater treatment plant, and R2 was inoculated from R1. Detailed operational conditions were described elsewhere (He et al., 2006). Briefly, the influent contained (mg/L total feed): CH3COONa•3H2O (425); casamino acids (31), yeast extract (8.6), NaH2PO4•H2O (62.3) and other mineral salts to achieve the chemical oxygen demand (COD) to P ratio of 14. The pH was controlled at 7.0–7.5 and allyl-thiourea was added to the reactors at 4 mg/L to inhibit nitrification (Hooper and Terry, 1973). As a result, both reactors had not been exposed to nitrate prior to this study.
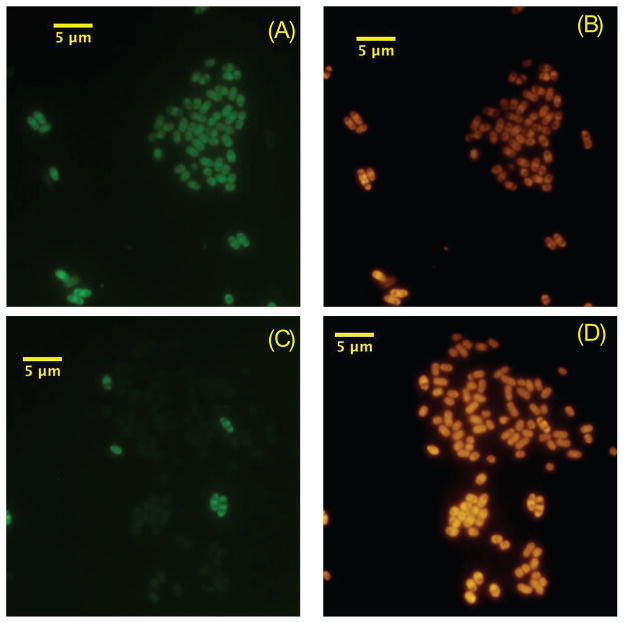
The relative abundance of Accumulibacter was estimated by 16S rRNA-targeted fluorescent in situ hybridization (FISH) with the PAOMIX probes (PAO 462, PAO 651 and PAO 846) (Crocetti et al., 2000) and counterstaining total cells with 1 μg/ml of 4′6-diamidino-2-phenylindole (DAPI). The slides were viewed and a total of 10 images per sample were captured using a Zeiss Axioplan 2 epifluorescent microscope (Carl Zeiss, Thornwood, N.Y.) equipped with an Olympus DP70 color camera (Center Valley, PA). The images were analyzed using DP Controller v2.2.1 (Olympus, Center Valley, PA). To quantify the % Accumulibacter of total DAPI-stained cells, the images for each sample were manually counted. Both reactors were enriched (>50% of total DAPI-stained cells) in Accumulibacter. Originally, Accumulibacter clades were defined based on the ppk genes (He et al., 2007). Later, the 16S rRNA sequence of clade IIA was identified from its finished genome, which also contains the clade IIA ppk (JGI img/m: http://img.jgi.doe.gov/cgi-bin/m/main.cgi). Therefore, in addition, two probes (Acc-I-444 and Acc-II-444) (Table 1) were designed to differentiate between these two clades in our reactors. They target the same site in the 16S rRNA, and are different from each other by two adjacent nucleotides. Based on an in silico prediction of formamide denaturation profiles according to Yilmaz and Noguera (2007), the melting formamide concentrations are approximately 35% and 42% for Acc-I-444 and Acc-II-444, respectively. Accordingly, experiments were performed with 35% formamide, to maximize specificity while gaining sufficient signal intensity from hybridized cells. When applied independently, a non-labeled competitor probe targeting the other Type was added to eliminate cross hybridization. Figure 1 shows the FISH images hybridized by the two probes and the PAOMIX on the same sample.
Table 1.
Probes designed to distinguish Accumulibacter clade IA from IIA
| Probe | Sequence (5′-3′) | Target1 | ΔG°overall’ (kcal/mol)2 | Formamide |
|---|---|---|---|---|
| Acc-I-444 | CCCAAGCAATTTCTTCCCC | Clade IA and other Type I clades | −13.7 | 35% |
| Acc-II-444 | CCCGTGCAATTTCTTCCCC | Clade IIA, IIC, and IID | −14.4 | 35% |
We emphasize that the probes are suitable for use in well-characterized lab-scale bioreactors only. Probe Acc-I-444 also targets some (but not all) members of other Type I clades. Probe Acc-II-444 also targets some (but not all) members of clades IIC and IID. Clades were defined previously (He et al., 2007; Peterson et al., 2008). However, in reactors included in this study, Accumulibacter IA and IIA were the only clades detected using qPCR. Therefore, the two probes could be used to distinguish these two clades in our systems.
Thermodynamic probe affinity calculated as described elsewhere (Yilmaz and Noguera, 2004). Design value was set to ≤− 13 kcal/mol (Yilmaz et al., 2006).
Figure 1.
FISH images of sludge from reactor enriched in Accumulibacter clade IIA (R2). Images in panels (A) and (B) were collected in the same field, and panels (C) and (D) were collected in the same field. Cells in panels (B) and (D) were hybridized with PAOMIX probes and labeled cells are therefore members of the Accumulibacter lineage (Crocetti et al., 2000). Labeled cells (green) in panel (A) are hybridized with Acc-II-444, and labeled cells (green) in panel (C) are hybridized with Acc-I-444. Comparison of panels (C) and (D) demonstrates the heterogeneity within the Accumulibacter community, since many cells identified as Accumulibacter by PAOMIX in (D) are not hybridized with Acc-I-444 in (C).
To determine the relative abundance of Accumulibacter clades IA and IIA accurately, quantitative real-time PCR (qPCR) with Accumulibacter clade-specific primers targeting ppk1 was performed as described (He et al., 2007). The distribution patterns of Accumulibacter in the two reactors were correlated (correlation coefficient = 0.75, p<0.001) from 37 paired data points collected from R1 and R2 during over two years of operation before October 2007, despite seemingly random shifts in the Accumulibacter community composition. However, for about 3 months after December, 2007 R1 was dominated by clade IA, and R2 was dominated by clade IIA, although they achieved equally good P removal performance (data not shown). The relative abundances of clades IA and IIA measured by ppk1-qPCR were qualitatively verified by FISH with probes Acc-I-444 and Acc-II-444. No explanation as to why the two reactors exhibited very different Accumulibacter composition has been determined, nor do we have the ability to intentionally change the operational conditions to enrich for either clade. However, the contrasting clade distribution in the two reactors gave the opportunity to evaluate the ability of each clade to denitrify, and therefore test a previously published hypothesis that the two clades are physiologically different (He et al., 2007).
During the period with differential enrichment, batch tests were conducted to investigate whether the two sludges could couple nitrate reduction with P-uptake (Table 2). Briefly, 500 mL of sludge was collected from each bioreactor at the end of the anaerobic phase when soluble P was at its maximum, to establish four batch reactors, each consisting of 250 mL of sludge. Nitrogen gas was sparged into all four batch reactors to maintain an oxygen-free environment. Sodium nitrate was added to one batch reactor derived from R1 (BR1-N) and one batch reactor derived from R2 (BR2-N) to reach an initial nitrate concentration of 25 mg/L as nitrogen (NO3-N). The remaining two batch reactors (BR1-C and BR2-C) were maintained under anaerobic conditions as controls. During the 24-h incubation, pH was maintained at 7.3±0.1 and dissolved oxygen (DO) was measured at intervals to assure that DO was below detection in all batch experiments. Soluble inorganic phosphate (Pi) (by modified ascorbic acid method 4500-P E; (APHA et al., 1995)), acetate, NO3-N, and NO2-N [by a Shimadzu high performance liquid chromatography (Kyoto, Japan) equipped with an Alltech Previal Organic Acid Column (Deerfield, Illinois) and a photodiode array detector set at 210 and 214nm] were monitored at regular intervals. Total suspended solids (TSS) and volatile suspended solids (VSS) were measured to estimate biomass concentration (APHA et al., 1995) at the end of the batch tests.
Table 2.
Summary of Accumulibacter distribution and batch test results
| Sample | Batch | Total Acc1 (%) | Acc-IA2 (%) | Acc-IIA2 (%) | Initial P uptake rate3 (μmol/hr/g VSS) | Initial NO3-N uptake rate4 (μmol/hr/gVSS) | Effective P uptake rate5 (μmol/hr/gVSS) | Effective NO3-N uptake rate5 (μmol/hr/gVSS) | P uptake/N uptake6 (mol/mol) |
|---|---|---|---|---|---|---|---|---|---|
| BNA7 | BR1-N | 72±11 | 98±1 | 2±1 | 46 | 98 | 46 | 98 | 0.58 |
| BR1-C | −83 | NA | −83 | NA | NA | ||||
| BR2-N | 82±11 | 33±2 | 67±2 | −68 | 78 | −35 | 53 | −0.68 | |
| BR2-C | −98 | NA | −73 | NA | NA | ||||
| ANA7 | BR1-N | 72±11 | 93±1 | 7±1 | 95 | 154 | 67 | 103 | 0.63 |
| BR1-C | −62 | NA | −62 | NA | NA | ||||
| BR2-N | 82±11 | 39±1 | 61±1 | 114 | 160 | 30 | 78 | 0.29 | |
| BR2-C | −120 | NA | −55 | NA | NA | ||||
Relative abundance of total Accumulibacter estimated by FISH with PAOMIX probes as a proportion of total DAPI stained cells. The values reported are average and standard deviation.
Relative abundance of Accumulibacter clade IA and IIA determined by ppk1-qPCR. The values reported are average and standard deviation.
Net measured P-uptake (positive values) or release (negative values) rates calculated as the slope of the initial linear portion of the profile (Figure 2), normalized by VSS.
Rates were calculated as the slope of the initial linear portion of the NO3-N profile (Figure 2), normalized by VSS.
Effective P-uptake (or release) and NO3-N-uptake calculated using linear regressions fit to the profiles in Figure 2. Two separate regressions were used in some cases, to capture the biphasic structure of the profiles. The effective rate was calculated based on the time-weighted average of the two slopes.
Stoichiometric ratio of total P-uptake (or release) to NO3-N-uptake across the entire batch experiment.
Results for batch test samples collected one day before (BNA) and one day after the 24-hr nitrate acclimation (ANA). Batch test samples labeled –N had nitrate amendment and samples with –C were control batch tests operated anaerobically with no added electron acceptor.
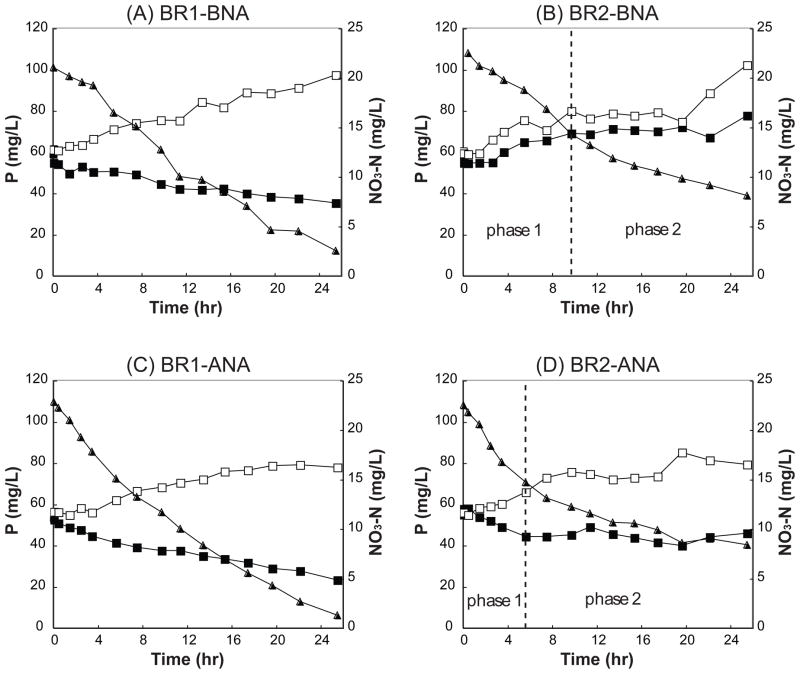
In the controls we observed secondary P-release instead of P-uptake, and the release was not coupled to acetate uptake since acetate was below detection at the start of the batch test. This phenomenon was documented previously (Stephens and Stensel, 1998). In the nitrate-amended batch reactors (BR1-N and BR2-N), both sludges consumed nitrate (Figure 2A and 2B), but a much higher effective nitrate consumption rate was observed in BR1-N across the entire experiment (Samples BNA in Table 2). No nitrite accumulation was observed during the batch tests, unlike as previously observed by Carvalho and colleagues (2007). Net P-uptake only occurred in BR1-N (Figure 2A), with both initial nitrate consumption and P-uptake rates comparable to those obtained by Carvalho and colleagues (2007). BR2-N did not exhibit a net P-uptake (Figure 2B and Table 2). However, the differences between BR2-N and BR2-C suggested that P-uptake and secondary P-release occurred simultaneously, probably mediated by the two respective populations of Accumulibacter. The lack of net P-uptake in BR2-N was not likely due to the limitation of carbon storage, since P-uptake occurred rapidly when oxygen was introduced after the 24-hour incubation (data not shown). Nearly identical results were obtained during another set of experiments conducted under similarly enriched conditions (no statistically significant difference in rates) (data not shown).
Figure 2.
Batch test results using sludge from R1 and R2 one day before (BNA) and one day after nitrate acclimation (ANA), showing P profiles in the nitrate-amended (■) and control (□) batches, and NO3-N profiles in the nitrate-amended batch (▲). NO3-N in the control and NO2-N in both nitrate-amended and control batches were below 0.3 mg/L, and therefore not plotted in this figure. Profiles from R2-derived batch tests are divided into two phases, to account for the differences in kinetics observed initially (first 10 hours in panel B and first 6 hours in panel D) and during the remainder of the experiment.
Previous studies suggested that nitrate reduction rates were higher following acclimatization with nitrate (Zeng et al., 2003; Carvalho et al., 2007). To evaluate this possibility, batch tests were also conducted on sludges that had been exposed to nitrate by addition of 4 mg/L NO3-N into both reactors at the beginning of each anaerobic phase for four consecutive cycles (24 hours) prior to the batch tests. This concentration was chosen because we had previously determined that 4 mg/L NO3-N did not affect reactor P removal performance (unpublished data). To minimize the effect of potential microbial community composition change, we compared the batch tests one day before (Samples BNA) and one day after the nitrate acclimatization (Samples ANA). Initial P-uptake rates in both BR1-N and BR2-N batch tests were found to increase significantly relative to the BNA experiments, after the acclimatization (p < 0.01 for both BR1-N and BR2-N, respectively) (Figure 2C and 2D and Table 2). Indeed, BR2-N exhibited a small net P-uptake by the end of the 24 hour testing period. Interestingly, the initial P-uptake rates for BR1-N and BR2-N after acclimation were not statistically different from one another. However, the uptake profile for BR2-N appeared to be strongly biphasic with a significantly shallower slope after 6 hours (Figure 2D).
The assumption that clade IIA is not able to reduce nitrate is further supported by the fact that nitrate consumption continued throughout the entire 24 hour cycle period for clade IA-dominant sludge (Figure 2A and 2C) while nitrate uptake rate decreased during phase 2 in clade IIA-dominant sludge (Figure 2B and 2D) indicating a limiting nitrate reduction capacity. We hypothesize that the period of more rapid P-uptake and nitrate reduction during phase 1 was controlled by the clade IA population that quickly depleted its PHA reserves, upon which P-uptake and nitrate reduction slowed substantially (phase 2).
Taking the genomic and experimental evidence together, we conclude that members of clade IA, but not clade IIA, can use nitrate as an electron acceptor for P-uptake. It is possible that clade IIA can use nitrite generated by clade IA as a denitrification intermediate for P-uptake, because genes enabling nitrite reduction to N2 were found in the genome of clade IIA (Garcia Martin et al., 2006), but how significant this contributed to the overall P-uptake cannot be clearly assessed by this study.
As noted in Table 2, roughly 20–30% of the microbial community in our bioreactors was not Accumulibacter. While it is possible that some of this flanking community can reduce nitrate, the operational conditions in the bioreactor likely excluded a significant contribution of the flanking community to nitrate reduction since all of the readily available carbon (acetate) was sequestered during the previous anaerobic phase. This ability to sequester carbon anaerobically provides Accumulibacter a competitive advantage activated sludge systems (Oehmen et al., 2007). We cannot rule out the presence of glycogen accumulating non-polyphosphate organisms capable of sequestering acetate as PHA and using it to fuel denitrification, but high calculated ratios of anaerobic P-release to acetate-uptake in our systems (0.61 ± 0.02 mmole-P/mmole-C) provide strong evidence that Accumulibacter sequestered nearly all of the acetate (Schuler and Jenkins, 2003).
The observed difference in anoxic P uptake supports the previously proposed hypothesis that different Accumulibacter clades have different metabolic capabilities (Carvalho et al., 2007; He et al., 2007). Therefore, we emphasize the importance of studying both genetic and physiological differences among different Accumulibacter populations. It also confirms the lack of nitrate reduction capability of clade IIA, which was inferred from the metagenomic analysis (Garcia Martin et al., 2006). The hypothesis that some Accumulibacter clades can denitrify, while others cannot, resolves conflicting observations about Accumulibacter’s denitrification capabilities previously reported in the literature (Kong et al., 2004; Freitas et al., 2005; Carvalho et al., 2007). A better understanding of Accumulibacter ecophysiology will help to better explain and predict the clade distribution in EBPR systems, and provides important information about whether or not simultaneous P and total nitrogen removal can be achieved under certain Accumulibacter community compositions.
Acknowledgments
The authors graciously acknowledge Gilda Carvalho and Phil Hugenholtz for insightful discussions; Norbert Tavares for help with batch reactor experiments; Nick Bartolerio and Forrest Bishop for help with bioreactor maintenance. This work was supported by funding from the National Science Foundation (award BES 0332136 to KDM) and was conducted in part under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, and by the National Institutes of Health Biotechnology Training Program Grant 5 T32 G08349.
References
- APHA, AWWA, and WEF. Standard methods for the examination of water and waste water. Washington, DC: American Public Health Association, American Water Works Association and Water Environment Federation; 1995. [Google Scholar]
- Carvalho G, Lemos PC, Oehmen A, Reis MAM. Denitrifying phosphorus removal: Linking the process performance with the microbial community structure. Water Research. 2007;41:4383–4396. doi: 10.1016/j.watres.2007.06.065. [DOI] [PubMed] [Google Scholar]
- Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D, Blackall LL. Identification of polyphosphate accumulating organisms and the design of 16s rrna-directed probes for their detection and quantitation. Applied and Environmental Microbiology. 2000;66:1175–1182. doi: 10.1128/aem.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabert P, Sialve B, Delgenès JP, Moletta R, Godon JJ. Characterisation of the microbial 16s rdna diversity of an aerobic phosphorus-removal ecosystem and monitoring of its transition to nitrate respiration. Applied Microbiology and Biotechnology. 2001;55:500–509. doi: 10.1007/s002530000529. [DOI] [PubMed] [Google Scholar]
- Freitas F, Temudo M, Reis MAM. Microbial population response to changes of the operating conditions in a dynamic nutrient-removal sequencing batch reactor. Bioprocess and Biosystems Engineering. 2005;28:199–209. doi: 10.1007/s00449-005-0029-9. [DOI] [PubMed] [Google Scholar]
- Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, et al. Metagenomic analysis of two enhanced biological phosphorus removal (ebpr) sludge communities. Nature Biotechnology. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- Guisasola A, Qurie M, Vargas MD, Casas C, Baeza JA. Failure of an enriched nitrite-dpao population to use nitrate as an electron acceptor. Process Biochemistry. 2009;44:689–695. [Google Scholar]
- He S, Gu AZ, McMahon KD. Fine-scale differences between accumulibacter-like bacteria in enhanced biological phosphorus removal activated sludge. Water Science and Technology. 2006;54:111–117. doi: 10.2166/wst.2006.378. [DOI] [PubMed] [Google Scholar]
- He S, Gall DL, McMahon KD. “Candidatus accumulibacter” Population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Applied and Environmental Microbiology. 2007;73:5865–5874. doi: 10.1128/AEM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gu AZ, McMahon KD. Progress toward understanding the distribution of accumulibacter among full-scale enhanced biological phosphorus removal systems. Microbial Ecology. 2008;55:229–236. doi: 10.1007/s00248-007-9270-x. [DOI] [PubMed] [Google Scholar]
- Hesselmann RPX, Werlen C, Hahn D, van der Meer JR, Zehnder AJB. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Systematic and Applied Microbiology. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- Hooper AB, Terry KR. Specific inhibitors of ammonia oxidation in nitrosomonas. Journal of Bacteriology. 1973;115:480–485. doi: 10.1128/jb.115.2.480-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Microautoradiographic study of rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol. 2004;70:5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehmen A, Lemos PC, Carvalho G, Yuan Z, Keller J, Blackall LL, Reis MAM. Advances in enhanced biological phosphorus removal: From micro to macro scale. Water Research. 2007;41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Peterson SB, Warnecke F, Madejska J, McMahon KD, Hugenholtz P. Environmental distribution and population biology of the genus accumulibacter, a primary agent of biological phosphorus removal in activated sludge. Environmental Microbiology. 2008;10:2692–2703. doi: 10.1111/j.1462-2920.2008.01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler AJ, Jenkins D. Enhanced biological phosphorus removal from wastewater by biomass with different phosphorus contents, part i: Experimental methods and results. Water Environment Research. 2003;75:485–498. doi: 10.2175/106143003x141286. [DOI] [PubMed] [Google Scholar]
- Stephens HL, Stensel HD. Effect of operating conditions on biological phosphorus removal. Water Environment Research. 1998;70:362. [Google Scholar]
- Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B, et al. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME Journal. 2008 doi: 10.1038/ismej.2008.38. [DOI] [PubMed] [Google Scholar]
- Yilmaz LS, Noguera DR. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Applied and Environmental Microbiology. 2004;70:7126–7139. doi: 10.1128/AEM.70.12.7126-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz LS, Noguera DR. Development of thermodynamic models for simulating probe dissociation profiles in fluorescence in situ hybridization. Biotechnology and Bioengineering. 2007;96:349–363. doi: 10.1002/bit.21114. [DOI] [PubMed] [Google Scholar]
- Yilmaz LS, Okten HE, Noguera DR. Making all parts of the 16s rrna of escherichia coli accessible in situ to single DNA oligonucleotides. Applied and Environmental Microbiology. 2006;72:733–744. doi: 10.1128/AEM.72.1.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng RJ, Saunders AM, Yuan Z, Blackall LL, Keller J. Identification and comparison of aerobic and denitrifying polyphosphate-accumulating organisms. Biotechnology and Bioengineering. 2003;83:140–148. doi: 10.1002/bit.10652. [DOI] [PubMed] [Google Scholar]
- Zilles JL, Peccia J, Kim MW, Hung CH, Noguera DR. Involvement of rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Applied and Environmental Microbiology. 2002;68:2763–2769. doi: 10.1128/AEM.68.6.2763-2769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]