Abstract
We develop fast algorithms for estimation of generalized linear models with convex penalties. The models include linear regression, two-class logistic regression, and multinomial regression problems while the penalties include ℓ1 (the lasso), ℓ2 (ridge regression) and mixtures of the two (the elastic net). The algorithms use cyclical coordinate descent, computed along a regularization path. The methods can handle large problems and can also deal efficiently with sparse features. In comparative timings we find that the new algorithms are considerably faster than competing methods.
1 Introduction
The lasso [Tibshirani, 1996] is a popular method for regression that uses an ℓ1 penalty to achieve a sparse solution. In the signal processing literature, the lasso is also known as basis pursuit [Chen et al., 1998]. This idea has been broadly applied, for example to generalized linear models [Tibshirani, 1996] and Cox’s proportional hazard models for survival data [Tibshirani, 1997]. In recent years, there has been an enormous amount of research activity devoted to related regularization methods:
The grouped lasso [Yuan and Lin, 2007, Meier et al., 2008], where variables are included or excluded in groups;
The Dantzig selector [Candes and Tao, 2007, and discussion], a slightly modified version of the lasso;
The elastic net [Zou and Hastie, 2005] for correlated variables, which uses a penalty that is part ℓ1, part ℓ2;
ℓ1 regularization paths for generalized linear models [Park and Hastie, 2007];
Regularization paths for the support-vector machine [Hastie et al., 2004].
The graphical lasso [Friedman et al., 2008] for sparse covariance estimation and undirected graphs
Efron et al. [2004] developed an efficient algorithm for computing the entire regularization path for the lasso. Their algorithm exploits the fact that the coefficient profiles are piecewise linear, which leads to an algorithm with the same computational cost as the full least-squares fit on the data (see also Osborne et al. [2000]).
In some of the extensions above [2,3,5], piecewise-linearity can be exploited as in Efron et al. [2004] to yield efficient algorithms. Rosset and Zhu [2007] characterize the class of problems where piecewise-linearity exists—both the loss function and the penalty have to be quadratic or piecewise linear.
Here we instead focus on cyclical coordinate descent methods. These methods have been proposed for the lasso a number of times, but only recently was their power fully appreciated. Early references include Fu [1998], Shevade and Keerthi [2003] and Daubechies et al. [2004]. Van der Kooij [2007] independently used coordinate descent for solving elastic-net penalized regression models. Recent rediscoveries include Friedman et al. [2007] and Wu and Lange [2008a]. The first paper recognized the value of solving the problem along an entire path of values for the regularization parameters, using the current estimates as warm starts. This strategy turns out to be remarkably efficient for this problem. Several other researchers have also re-discovered coordinate descent, many for solving the same problems we address in this paper—notably Shevade and Keerthi [2003], Krishnapuram and Hartemink [2005], Genkin et al. [2007] and Wu et al. [2009].
In this paper we extend the work of Friedman et al. [2007] and develop fast algorithms for fitting generalized linear models with elastic-net penalties. In particular, our models include regression, two-class logistic regression, and multinomial regression problems. Our algorithms can work on very large datasets, and can take advantage of sparsity in the feature set. We provide a publicly available R package glmnet. We do not revisit the well-established convergence properties of coordinate descent in convex problems [Tseng, 2001] in this article.
Lasso procedures are frequently used in domains with very large datasets, such as genomics and web analysis. Consequently a focus of our research has been algorithmic efficiency and speed. We demonstrate through simulations that our procedures outperform all competitors — even those based on coordinate descent.
In section 2 we present the algorithm for the elastic net, which includes the lasso and ridge regression as special cases. Section 3 and 4 discuss (two-class) logistic regression and multinomial logistic regression. Comparative timings are presented in Section 5.
2 Algorithms for the Lasso, Ridge Regression and the Elastic Net
We consider the usual setup for linear regression. We have a response variable Y ∈ ℝ and a predictor vector X ∈ ℝp, and we approximate the regression function by a linear model E(Y|X = x) = β0 + xT β. We have N observation pairs (xi, yi). For simplicity we assume the xij are standardized: Our algorithms generalize naturally to the unstandardized case. The elastic net solves the following problem
| (1) |
where
| (2) |
| (3) |
is the elastic-net penalty [Zou and Hastie, 2005]. Pα is a compromise between the ridge-regression penalty (α = 0) and the lasso penalty (α = 1). This penalty is particularly useful in the p ≫ N situation, or any situation where there are many correlated predictor variables.
Ridge regression is known to shrink the coefficients of correlated predictors towards each other, allowing them to borrow strength from each other. In the extreme case of k identical predictors, they each get identical coefficients with 1/kth the size that any single one would get if fit alone. From a Bayesian point of view, the ridge penalty is ideal if there are many predictors, and all have non-zero coefficients (drawn from a Gaussian distribution).
Lasso, on the other hand, is somewhat indifferent to very correlated predictors, and will tend to pick one and ignore the rest. In the extreme case above, the lasso problem breaks down. The Lasso penalty corresponds to a Laplace prior, which expects many coefficients to be close to zero, and a small subset to be larger and nonzero.
The elastic net with α = 1 − ε for some small ε > 0 performs much like the lasso, but removes any degeneracies and wild behavior caused by extreme correlations. More generally, the entire family Pα creates a useful compromise between ridge and lasso. As α increases from 0 to 1, for a given λ the sparsity of the solution to (1) (i.e. the number of coefficients equal to zero) increases monotonically from 0 to the sparsity of the lasso solution.
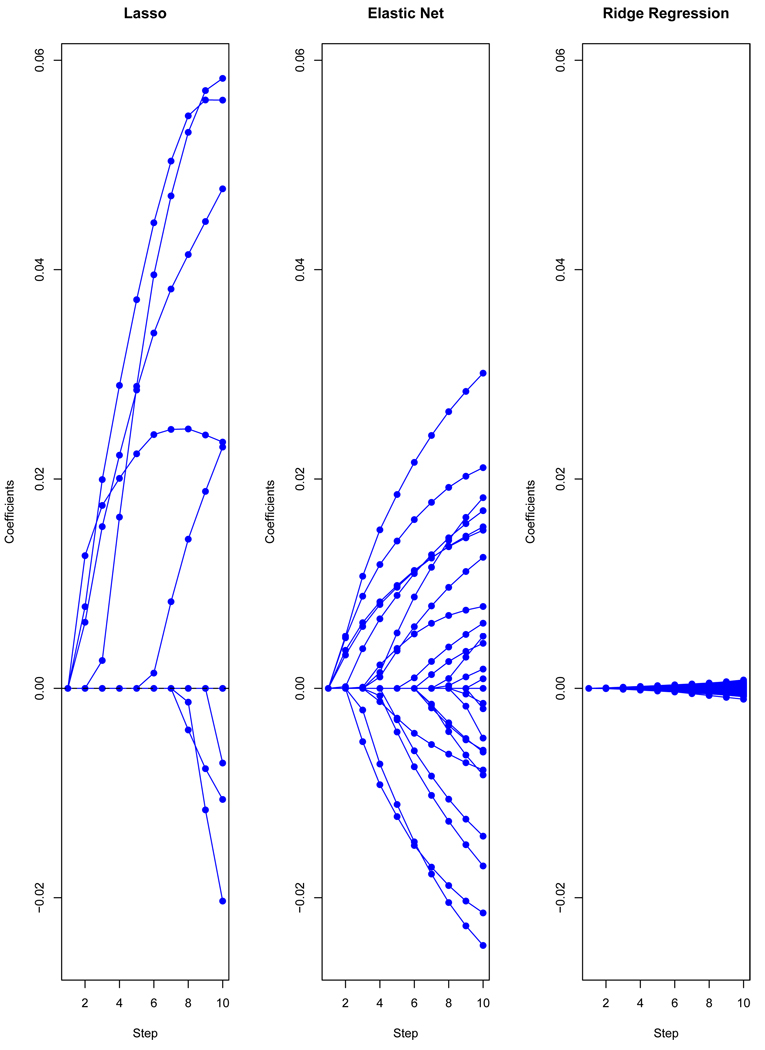
Figure 1 shows an example. The dataset is from [Golub et al., 1999b], consisting of 72 observations on 3571 genes measured with DNA microarrays. The observations fall in two classes: we treat this as a regression problem for illustration. The coefficient profiles from the first 10 steps (grid values for λ) for each of the three regularization methods are shown. The lasso admits at most N = 72 genes into the model, while ridge regression gives all 3571 genes non-zero coefficients. The elastic net provides a compromise between these two methods, and has the effect of averaging genes that are highly correlated and then entering the averaged gene into the model. Using the algorithm described below, computation of the entire path of solutions for each method, at 100 values of the regularization parameter evenly spaced on the log-scale, took under a second in total. Because of the large number of non-zero coefficients for ridge regression, they are individually much smaller than the coefficients for the other methods.
Figure 1.
Leukemia data: profiles of estimated coefficients for three methods, showing only first 10 steps (values for λ) in each case. For the elastic net, α = 0.2.
Consider a coordinate descent step for solving (1). That is, suppose we have estimates β̃0 and β̃ℓ for ℓ ≠ j, and we wish to partially optimize with respect to βj. Denote by R(β0, β) the objective function in (1). We would like to compute the gradient at βj = β̃j, which only exists if β̃j ≠ 0. If β̃j > 0, then
| (4) |
A similar expression exists if β̃j < 0, and β̃j = 0 is treated separately. Simple calculus shows [Donoho and Johnstone, 1994] that the coordinate-wise update has the form
| (5) |
where
is the fitted value excluding the contribution from xij, and hence the partial residual for fitting βj. Because of the standardization, is the simple least-squares coefficient when fitting this partial residual to xij.
- S(z, γ) is the soft-thresholding operator with value
(6)
The details of this derivation are spelled out in Friedman et al. [2007].
Thus we compute the simple least-squares coefficient on the partial residual, apply soft-thresholding to take care of the lasso contribution to the penalty, and then apply a proportional shrinkage for the ridge penalty. This algorithm was suggested by Van der Kooij [2007].
2.1 Naive Updates
Looking more closely at (5), we see that
| (7) |
where ŷi is the current fit of the model for observation i, and hence ri the current residual. Thus
| (8) |
because the xj are standardized. The first term on the right-hand side is the gradient of the loss with respect to βj. It is clear from (8) why coordinate descent is computationally efficient. Many coefficients are zero, remain zero after the thresholding, and so nothing needs to be changed. Such a step costs O(N) operations— the sum to compute the gradient. On the other hand, if a coefficient does change after the thresholding, ri is changed in O(N) and the step costs O(2N). Thus a complete cycle through all p variables costs O(pN) operations. We refer to this as the naive algorithm, since it is generally less efficient than the covariance updating algorithm to follow. Later we use these algorithms in the context of iteratively reweighted least squares (IRLS), where the observation weights change frequently; there the naive algorithm dominates.
2.2 Covariance Updates
Further efficiencies can be achieved in computing the updates in (8). We can write the first term on the right (up to a factor 1/N) as
| (9) |
where Hence we need to compute inner products of each feature with y initially, and then each time a new feature xk enters the model (for the first time), we need to compute and store its inner product with all the rest of the features (O(Np) operations). We also store the p gradient components (9). If one of the coefficients currently in the model changes, we can update each gradient in O(p) operations. Hence with m non-zero terms in the model, a complete cycle costs O(pm) operations if no new variables become non-zero, and costs O(Np) for each new variable entered. Importantly, O(N) calculations do not have to be made at every step. This is the case for all penalized procedures with squared error loss.
2.3 Sparse Updates
We are sometimes faced with problems where the N × p feature matrix X is extremely sparse. A leading example is from document classification, where the feature vector uses the so-called “bag-of-words” model. Each document is scored for the presence/absence of each of the words in the entire dictionary under consideration (sometimes counts are used, or some transformation of counts). Since most words are absent, the feature vector for each document is mostly zero, and so the entire matrix is mostly zero. We store such matrices efficiently in sparse column format, where we store only the non-zero entries and the coordinates where they occur.
Coordinate descent is ideally set up to exploit such sparsity, in an obvious way. The O(N) inner-product operations in either the naive or covariance updates can exploit the sparsity, by summing over only the non-zero entries. Note that in this case scaling of the variables will not alter the sparsity, but centering will. So scaling is performed up front, but the centering is incorporated in the algorithm in an efficient and obvious manner.
2.4 Weighted Updates
Often a weight wi (other than 1/N) is associated with each observation. This will arise naturally in later sections where observations receive weights in the IRLS algorithm. In this case the update step (5) becomes only slightly more complicated:
| (10) |
If the xj are not standardized, there is a similar sum-of-squares term in the denominator (even without weights). The presence of weights does not change the computational costs of either algorithm much, as long as the weights remain fixed.
2.5 Pathwise Coordinate Descent
We compute the solutions for a decreasing sequence of values for λ, starting at the smallest value λmax for which the entire vector β̂ = 0. Apart from giving us a path of solutions, this scheme exploits warm starts, and leads to a more stable algorithm. We have examples where it is faster to compute the path down to λ (for small λ) than the solution only at that value for λ.
When β̃ = 0, we see from (5) that β̃j will stay zero if Hence Nαλmax = maxℓ |〈xℓ, y〉|. Our strategy is to select a minimum value λmin = ελmax, and construct a sequence of K values of λ decreasing from λmax to λmin on the log scale. Typical values are ε = 0.001 and K = 100.
2.6 Other Details
Irrespective of whether the variables are standardized to have variance 1, we always center each predictor variable. Since the intercept is not regularized, this means that β̂0 = ȳ, the mean of the yi, for all values of α and λ.
It is easy to allow different penalties λj for each of the variables. We implement this via a penalty scaling parameter γj ≥ 0. If γj > 0, then the penalty applied to βj is λj = λγj. If γj = 0, that variable does not get penalized, and always enters the model unrestricted at the first step and remains in the model. Penalty rescaling would also allow, for example, our software to be used to implement the adaptive lasso [Zou, 2006].
Considerable speedup is obtained by organizing the iterations around the active set of features—those with nonzero coefficients. After a complete cycle through all the variables, we iterate on only the active set till convergence. If another complete cycle does not change the active set, we are done, otherwise the process is repeated. Active-set convergence is also mentioned in Meier et al. [2008] and Krishnapuram and Hartemink [2005].
3 Regularized Logistic Regression
When the response variable is binary, the linear logistic regression model is often used. Denote by G the response variable, taking values in 𝒢 = {1, 2} (the labeling of the elements is arbitrary). The logistic regression model represents the class-conditional probabilities through a linear function of the predictors
| (11) |
Alternatively, this implies that
| (12) |
Here we fit this model by regularized maximum (binomial) likelihood. Let p(xi) = Pr(G = 1|xi) be the probability (11) for observation i at a particular value for the parameters (β0, β), then we maximize the penalized log likelihood
| (13) |
Denoting yi = I(gi = 1), the log-likelihood part of (13) can be written in the more explicit form
| (14) |
a concave function of the parameters. The Newton algorithm for maximizing the (unpenalized) log-likelihood (14) amounts to iteratively reweighted least squares. Hence if the current estimates of the parameters are (β̃0, β̃), we form a quadratic approximation to the log-likelihood (Taylor expansion about current estimates), which is
| (15) |
where
| (16) |
| (17) |
and p̃(xi) is evaluated at the current parameters. The last term is constant. The Newton update is obtained by minimizing ℓQ.
Our approach is similar. For each value of λ, we create an outer loop which computes the quadratic approximation ℓQ about the current parameters (β̃0, β̃). Then we use coordinate descent to solve the penalized weighted least-squares problem
| (18) |
This amounts to a sequence of nested loops:
OUTER LOOP: Decrement λ.
MIDDLE LOOP: Update the quadratic approximation ℓQ using the current parameters (β̃0, β̃).
INNER LOOP: Run the coordinate descent algorithm on the penalized weighted-least-squares problem (18).
There are several important details in the implementation of this algorithm.
When p ≫ N, one cannot run λ all the way to zero, because the saturated logistic regression fit is undefined (parameters wander off to ±∞ in order to achieve probabilities of 0 or 1). Hence the default λ sequence runs down to λmin = ελmax > 0.
Care is taken to avoid coefficients diverging in order to achieve fitted probabilities of 0 or 1. When a probability is within ε = 10−5 of 1, we set it to 1, and set the weights to ε. 0 is treated similarly.
Our code has an option to approximate the Hessian terms by an exact upper-bound. This is obtained by setting the wi in (17) all equal to 0.25 [Krishnapuram and Hartemink, 2005].
We allow the response data to be supplied in the form of a two-column matrix of counts, sometimes referred to as grouped data. We discuss this in more detail in Section 4.2.
The Newton algorithm is not guaranteed to converge without step-size optimization [in Lee et al., 2006]. Our code does not implement any checks for divergence; this would slow it down, and when used as recommended we do not feel it is necessary.. We have a closed form expression for the starting solutions, and each subsequent solution is warm-started from the previous close-by solution, which generally makes the quadratic approximations very accurate. We have not encountered any divergence problems so far.
4 Regularized Multinomial Regression
When the categorical response variable G has K > 2 levels, the linear logistic regression model can be generalized to a multi-logit model. The traditional approach is to extend (12) to K − 1 logits
| (19) |
Here βℓ is a p-vector of coefficients. As in Zhu and Hastie [2004], here we choose a more symmetric approach. We model
| (20) |
This parametrization is not estimable without constraints, because for any values for the parameters give identical probabilities (20). Regularization deals with this ambiguity in a natural way; see Section 4.1 below.
We fit the model (20) by regularized maximum (multinomial) likelihood. Using a similar notation as before, let pℓ(xi) = Pr(G = ℓ|xi), and let gi ∈ {1, 2, …, K} be the ith response. We maximize the penalized log-likelihood
| (21) |
Denote by Y the N × K indicator response matrix, with elements yiℓ = I (gi = ℓ). Then we can write the log-likelihood part of (21) in the more explicit form
| (22) |
The Newton algorithm for multinomial regression can be tedious, because of the vector nature of the response observations. Instead of weights wi as in (17), we get weight matrices, for example. However, in the spirit of coordinate descent, we can avoid these complexities. We perform partial Newton steps by forming a partial quadratic approximation to the log-likelihood (22), allowing only (β0ℓ, βℓ) to vary for a single class at a time. It is not hard to show that this is
| (23) |
where as before
| (24) |
| (25) |
Our approach is similar to the two-class case, except now we have to cycle over the classes as well in the outer loop. For each value of λ, we create an outer loop which cycles over ℓ and computes the partial quadratic approximation ℓQℓ about the current parameters (β̃0, β̃). Then we use coordinate descent to solve the penalized weighted least-squares problem
| (26) |
This amounts to the sequence of nested loops:
OUTER LOOP: Decrement λ.
MIDDLE LOOP (OUTER): Cycle over ℓ ∈ {1, 2, …, K, 1, 2, …}.
MIDDLE LOOP (INNER): Update the quadratic approximation ℓQℓ using the current parameters
INNER LOOP: Run the co-ordinate descent algorithm on the penalized weighted-least-squares problem (26).
4.1 Regularization and Parameter Ambiguity
As was pointed out earlier, if characterizes a fitted model for (20), then gives an identical fit (c is a p-vector). Although this means that the log-likelihood part of ((21) is insensitive to (c0, c), the penalty is not. In particular, we can always improve an estimate (w.r.t. (21)) by solving
| (27) |
This can be done separately for each coordinate, hence
| (28) |
Theorem 1 Consider problem (28) for values α ∈ [0, 1]. Let β̄j be the mean of the βjℓ, and a median of the βjℓ (and for simplicity assume Then we have
| (29) |
with the left endpoint achieved if α = 0, and the right if α = 1.
The two endpoints are obvious. The proof of Theorem 1 is given in the appendix. A consequence of the theorem is that a very simple search algorithm can be used to solve (28). The objective is piecewise quadratic, with knots defined by the βjℓ. We need only evaluate solutions in the intervals including the mean and median, and those in between.
We recenter the parameters in each index set j after each INNER MIDDLE LOOP step, using the the solution cj for each j.
Not all the parameters in our model are regularized. The intercepts β0ℓ are not, and with our penalty modifiers γj (section 2.6) others need not be as well. For these parameters we use mean centering.
4.2 Grouped and Matrix Responses
As in the two class case, the data can be presented in the form of a N × K matrix miℓ of non-negative numbers. For example, if the data are grouped: at each xi we have a number of multinomial samples, with miℓ falling into category ℓ. In this case we divide each row by the row-sum mi = ∑ℓ miℓ, and produce our response matrix yiℓ = miℓ/mi. mi becomes an observation weight. Our penalized maximum likelihood algorithm changes in a trivial way. The working response (24) is defined exactly the same way (using yiℓ just defined). The weights in (25) get augmented with the observation weight mi:
| (30) |
Equivalently, the data can be presented directly as a matrix of class proportions, along with a weight vector. From the point of view of the algorithm, any matrix of positive numbers and any non-negative weight vector will be treated in the same way.
5 Timings
In this section we compare the run times of the coordinate-wise algorithm to some competing algorithms. These use the lasso penalty (α = 1) in both the regression and logistic regression settings. All timings were carried out on an Intel Xeon 2.80GH processor.
We do not perform comparisons on the elastic net versions of the penalties, since there is not much software available for elastic net. Comparisons of our glmnet code with the R package elasticnet will mimic the comparisons with lars for the lasso, since elasticnet is built on the lars package.
5.1 Regression with the Lasso
We generated Gaussian data with N observations and p predictors, with each pair of predictors Xj, Xj′ having the same population correlation ρ. We tried a number of combinations of N and p, with ρ varying from zero to 0.95. The outcome values were generated by
| (31) |
where βj = (−1)j exp(−2(j − 1)/20), Z ~ N (0, 1) and k is chosen so that the signal-to-noise ratio is 3.0. The coefficients are constructed to have alternating signs and to be exponentially decreasing.
Table 1 shows the average CPU timings for the coordinate-wise algorithm, and the LARS procedure [Efron et al., 2004]. All algorithms are implemented as R language functions. The coordinate-wise algorithm does all of its numerical work in Fortran, while LARS (written by Efron and Hastie) does much of its work in R, calling Fortran routines for some matrix operations. However comparisons in [Friedman et al., 2007] showed that LARS was actually faster than a version coded entirely in Fortran. Comparisons between different programs are always tricky: in particular the LARS procedure computes the entire path of solutions, while the coordinate-wise procedure solves the problem for a set of pre-defined points along the solution path. In the orthogonal case, LARS takes min(N, p) steps: hence to make things roughly comparable, we called the latter two algorithms to solve a total of min(N, p) problems along the path. Table 1 shows timings in seconds averaged over three runs. We see that glmnet is considerably faster than LARS; the covariance-updating version of the algorithm is a little faster than the naive version when N > p and a little slower when p > N. We had expected that high correlation between the features would increase the run time of glmnet, but this does not seem to be the case.
Table 1.
Timings (secs) for glmnet and lars algorithms for linear regression with lasso penalty. The first line is glmnet using naive updating while the second uses covariance updating. Total time for 100 λ values, averaged over 3 runs.
| Linear Regression — Dense Features |
||||||
|---|---|---|---|---|---|---|
| Correlation | ||||||
| 0 | 0.1 | 0.2 | 0.5 | 0.9 | 0.95 | |
| N = 1000, p = 100 | ||||||
| glmnet-naive | 0.05 | 0.06 | 0.06 | 0.09 | 0.08 | 0.07 |
| glmnet-cov | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 |
| lars | 0.11 | 0.11 | 0.11 | 0.11 | 0.11 | 0.11 |
| N = 5000, p = 100 | ||||||
| glmnet-naive | 0.24 | 0.25 | 0.26 | 0.34 | 0.32 | 0.31 |
| glmnet-cov | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 |
| lars | 0.29 | 0.29 | 0.29 | 0.30 | 0.29 | 0.29 |
| N = 100, p = 1000 | ||||||
| glmnet-naive | 0.04 | 0.05 | 0.04 | 0.05 | 0.04 | 0.03 |
| glmnet-cov | 0.07 | 0.08 | 0.07 | 0.08 | 0.04 | 0.03 |
| lars | 0.73 | 0.72 | 0.68 | 0.71 | 0.71 | 0.67 |
| N = 100, p = 5000 | ||||||
| glmnet-naive | 0.20 | 0.18 | 0.21 | 0.23 | 0.21 | 0.14 |
| glmnet-cov | 0.46 | 0.42 | 0.51 | 0.48 | 0.25 | 0.10 |
| lars | 3.73 | 3.53 | 3.59 | 3.47 | 3.90 | 3.52 |
| N = 100, p = 20000 | ||||||
| glmnet-naive | 1.00 | 0.99 | 1.06 | 1.29 | 1.17 | 0.97 |
| glmnet-cov | 1.86 | 2.26 | 2.34 | 2.59 | 1.24 | 0.79 |
| lars | 18.30 | 17.90 | 16.90 | 18.03 | 17.91 | 16.39 |
| N = 100, p = 50000 | ||||||
| glmnet-naive | 2.66 | 2.46 | 2.84 | 3.53 | 3.39 | 2.43 |
| glmnet-cov | 5.50 | 4.92 | 6.13 | 7.35 | 4.52 | 2.53 |
| lars | 58.68 | 64.00 | 64.79 | 58.20 | 66.39 | 79.79 |
5.2 Lasso-logistic regression
We used the same simulation setup as above, except that we took the continuous outcome y, defined p = 1/(1 + exp(−y)) and used this to generate a two-class outcome z with Prob(z = 1) = p, Prob(z = 0) = 1 − p. We compared the speed of glmnet to the interior point method l1lognet proposed by Koh et al. [2007], Bayesian Logistic Regression (BBR) due to Genkin et al. [2007] and the Lasso Penalized Logistic (LPL) program supplied by Ken Lange [Wu and Lange, 2008b]. The latter two methods also use a coordinate descent approach.
The BBR software automatically performs ten-fold cross-validation when given a set of λ values. Hence we report the total time for ten-fold cross-validation for all methods using the same 100 λ values for all. Table 2 shows the results; in some cases, we omitted a method when it was seen to be very slow at smaller values for N or p.
Table 2.
Timings (seconds) for logistic models with lasso penalty. Total time for tenfold cross-validation over a grid of 100 λ values.
| Logistic Regression — Dense Features |
||||||
|---|---|---|---|---|---|---|
| Correlation | ||||||
| 0 | 0.1 | 0.2 | 0.5 | 0.9 | 0.95 | |
| N = 1000, p = 100 | ||||||
| glmnet | 1.65 | 1.81 | 2.31 | 3.87 | 5.99 | 8.48 |
| l1lognet | 31.475 | 31.86 | 34.35 | 32.21 | 31.85 | 31.81 |
| BBR | 40.70 | 47.57 | 54.18 | 70.06 | 106.72 | 121.41 |
| LPL | 24.68 | 31.64 | 47.99 | 170.77 | 741.00 | 1448.25 |
| N = 5000, p = 100 | ||||||
| glmnet | 7.89 | 8.48 | 9.01 | 13.39 | 26.68 | 26.36 |
| l1lognet | 239.88 | 232.00 | 229.62 | 229.49 | 22.19 | 223.09 |
| N = 100, 000, p = 100 | ||||||
| glmnet | 78.56 | 178.45 | 205.94 | 274.33 | 552.48 | 638.50 |
| N = 100, p = 1000 | ||||||
| glmnet | 1.06 | 1.07 | 1.09 | 1.45 | 1.72 | 1.37 |
| l1lognet | 25.99 | 26.40 | 25.67 | 26.49 | 24.34 | 20.16 |
| BBR | 70.19 | 71.19 | 78.40 | 103.77 | 149.05 | 113.87 |
| LPL | 11.02 | 10.87 | 10.76 | 16.34 | 41.84 | 70.50 |
| N = 100, p = 5000 | ||||||
| glmnet | 5.24 | 4.43 | 5.12 | 7.05 | 7.87 | 6.05 |
| l1lognet | 165.02 | 161.90 | 163.25 | 166.50 | 151.91 | 135.28 |
| N = 100, p = 100, 000 | ||||||
| glmnet | 137.27 | 139.40 | 146.55 | 197.98 | 219.65 | 201.93 |
Again we see that glmnet is the clear winner: it slows down a little under high correlation. The computation seems to be roughly linear in N, but grows faster than linear in p.
Table 3 shows some results when the feature matrix is sparse: we randomly set 95% of the feature values to zero. Again, the glmnet procedure is significantly faster than l1lognet.
Table 3.
Timings (seconds) for logistic model with lasso penalty and sparse features (95% zero). Total time for ten-fold cross-validation over a grid of 100 λ values.
| Logistic Regression — Sparse Features |
||||||
|---|---|---|---|---|---|---|
| Correlation | ||||||
| 0 | 0.1 | 0.2 | 0.5 | 0.9 | 0.95 | |
| N = 1000, p = 100 | ||||||
| glmnet | 0.77 | 0.74 | 0.72 | 0.73 | 0.84 | 0.88 |
| l1lognet | 5.19 | 5.21 | 5.14 | 5.40 | 6.14 | 6.26 |
| BBR | 2.01 | 1.95 | 1.98 | 2.06 | 2.73 | 2.88 |
| N = 100, p = 1000 | ||||||
| glmnet | 1.81 | 1.73 | 1.55 | 1.70 | 1.63 | 1.55 |
| l1lognet | 7.67 | 7.72 | 7.64 | 9.04 | 9.81 | 9.40 |
| BBR | 4.66 | 4.58 | 4.68 | 5.15 | 5.78 | 5.53 |
| N = 10, 000, p = 100 | ||||||
| glmnet | 3.21 | 3.02 | 2.95 | 3.25 | 4.58 | 5.08 |
| l1lognet | 45.87 | 46.63 | 44.33 | 43.99 | 45.60 | 43.16 |
| BBR | 11.80 | 11.64 | 11.58 | 13.30 | 12.46 | 11.83 |
| N = 100, p = 10, 000 | ||||||
| glmnet | 10.18 | 10.35 | 9.93 | 10.04 | 9.02 | 8.91 |
| l1lognet | 130.27 | 124.88 | 124.18 | 129.84 | 137.21 | 159.54 |
| BBR | 45.72 | 47.50 | 47.46 | 48.49 | 56.29 | 60.21 |
5.3 Real data
Table 4 shows some timing results for four different datasets.
Cancer [Ramaswamy et al., 2001]: gene-expression data with 14 cancer classes.
Leukemia [Golub et al., 1999a]: gene-expression data with a binary response indicating type of leukemia—AML vs ALL. We used the preprocessed data of Dettling [2004].
Internet-Ad [Kushmerick, 1999]: document classification problem with mostly binary features. The response is binary, and indicates whether the document is an advertisement. Only 1.2% nonzero values in the predictor matrix.
Newsgroup [Lang, 1995]: document classification problem. We used the training set cultured from these data by Koh et al. [2007]. The response is binary, and indicates a subclass of topics; the predictors are binary, and indicate the presence of particular tri-gram sequences. The predictor matrix has 0.05% nonzero values.
Table 4.
Timings (seconds, unless stated otherwise) for some real datasets. For the Cancer, Leukemia and Internet-Ad datasets, times are for ten-fold cross-validation over 100 λ values; for Newsgroup we performed a single run with 100 values of λ, with λmin = 0.05 λmax.
| Name | Type | N | p | glmnet | l1logreg | BBR/BMR |
|---|---|---|---|---|---|---|
| Dense | ||||||
| Cancer | 14 class | 144 | 16,063 | 2.5 mins | 2.1 hrs | |
| Leukemia | 2 class | 72 | 3571 | 2.50 | 55.0 | 450 |
| Sparse | ||||||
| Internet-Ad | 2 class | 2359 | 1430 | 5.0 | 20.9 | 34.7 |
| Newsgroup | 2 class | 11,314 | 777,811 | 2 mins | 3.5 hrs | |
All four datasets are available as saved R data objects at http://www-stat.stanford.edu/~hastie/glmnet/ (the latter two in sparse format using the Matrix package).
For the Leukemia and Internet-Ad datasets, the BBR program used fewer than 100 λ values so we estimated the total time by scaling up the time for smaller number of values. The Internet-Ad and Newsgroup datasets are both sparse: 1% nonzero values for the former, 0.06% for the latter. Again glmnet is considerably faster than the competing methods.
6 Discussion
Cyclical coordinate descent methods are a natural approach for solving convex problems with ℓ1 or ℓ2 constraints, or mixtures of the two (elastic net). Each coordinate-descent step is fast, with an explicit formula for each coordinate-wise minimization. The method also exploits the sparsity of the model, spending much of its time evaluating only inner products for variables with non-zero coefficients. Its computational speed both for large N and p are quite remarkable.
A public domain R language package glmnet is available from the CRAN website. Matlab functions (written by Rahul Mazumder), as well as the real datasets used in Section 5.3, can be downloaded from the second author’s website at http://www-stat.stanford.edu/~hastie/glmnet/
Acknowledgments
We would like to thank Holger Hoefling for helpful discussions, and Rahul Mazumder for writing the Matlab interface to our Fortran routines. We thank the associate editor and two referees who gave useful comments on an earlier draft of this article.
Friedman was partially supported by grant DMS-97-64431 from the National Science Foundation. Hastie was partially supported by grant DMS-0505676 from the National Science Foundation, and grant 2R01 CA 72028-07 from the National Institutes of Health. Tibshirani was partially supported by National Science Foundation Grant DMS-9971405 and National Institutes of Health Contract N01-HV-28183.
Appendix
Proof of theorem 1. We have
| (32) |
Suppose α ∈ (0, 1). Differentiating w.r.t. t (using a sub-gradient representation), we have
| (33) |
where sjℓ = sign(βjℓ − t) if βjℓ ≠ t and sjℓ ∈ [−1, 1] otherwise. This gives
| (34) |
It follows that t cannot be larger than since then the second term above would be negative and this would imply that t is less than β̄j. Similarly t cannot be less than β̄j, since then the second term above would have to be negative, implying that t is larger than
References
- Candes E, Tao T. The Dantzig selector: statistical estimation when p is much larger than n. Annals of Statistics. 2007;35(6):2313–2351. [Google Scholar]
- Chen SS, Donoho D, Saunders M. Atomic decomposition by basis pursuit. SIAM Journal on Scientific Computing. 1998;20(1):33–61. [Google Scholar]
- Daubechies I, Defrise M, De Mol C. An iterative thresholding algorithm for linear inverse problems with a sparsity constraint. Communications on Pure and Applied Mathematics. 2004;57:1413–1457. [Google Scholar]
- Dettling M. Bagboosting for tumor classification with gene expression data. Bioinformatics. 2004:3583–3593. doi: 10.1093/bioinformatics/bth447. [DOI] [PubMed] [Google Scholar]
- Donoho DL, Johnstone IM. Ideal spatial adaptation by wavelet shrinkage. Biometrika. 1994;81:425–455. [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Annals of Statistics. 2004;32(2):407–499. (with discussion) [Google Scholar]
- Friedman J, Hastie T, Hoefling H, Tibshirani R. Pathwise coordinate optimization. Annals of Applied Statistics. 2007;2(1):302–332. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W. Penalized regressions: the bridge vs. the lasso. Journal of Computational and Graphical Statistics. 1998;7(3):397–416. [Google Scholar]
- Genkin A, Lewis D, Madigan D. Large-scale bayesian logistic regression for text categorization. Technometrics. 2007;49(3):291–304. [Google Scholar]
- Golub T, Slonim D, Tamayo P, Huard C, Gaasenbeek M, Mesirov J, Coller H, Loh M, Downing J, Caligiuri M, Bloomfield C, Lander E. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999a;286:531–536. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Golub T, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999b;286:531–536. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Hastie T, Rosset S, Tibshirani R, Zhu J. The entire regularization path for the support vector machine. Journal of Machine Learning Research. 2004;(5):1391–1415. [Google Scholar]
- in Lee S, Lee H, Abneel P, Ng A. Efficient l1 logistic regression; Proceedings of the Twenty-First National Conference on Artificial Intelligence (AAAI-06); 2006. [Google Scholar]
- Koh K, Kim S-J, Boyd S. An interior-point method for large-scale l1-regularized logistic regression. Journal of Machine Learning Research. 2007;8:1519–1555. [Google Scholar]
- Krishnapuram B, Hartemink AJ. Sparse multinomial logistic regression: Fast algorithms and generalization bounds; IEEE Trans. Pattern Anal. Mach. Intell; 2005. pp. 957–968. ISSN 0162-8828. doi: http://dx.doi.org/10.1109/TPAMI.2005.127. Fellow-Lawrence Carin and Senior Member-Mario A. T. Figueiredo. [DOI] [PubMed] [Google Scholar]
- Kushmerick N. Learning to remove internet advertisements; Proceedings of AGENTS-99; 1999. [Google Scholar]
- Lang K. Newsweeder: Learning to filter netnews; Proceedings of the Twenty-First International Conference on Machine Learning (ICML); 1995. pp. 331–339. [Google Scholar]
- Meier L, van de Geer S, Bühlman P. The group lasso for logistic regression. Journal of the Royal Statistical Society B. 2008 February;70(1):53–71. [Google Scholar]
- Osborne M, Presnell B, Turlach B. A new approach to variable selection in least squares problems. IMA Journal of Numerical Analysis. 2000;20:389–404. [Google Scholar]
- Park MY, Hastie T. l1-regularization path algorithm for generalized linear models. Journal of the Royal Statistical Society Series B. 2007;69:659–677. [Google Scholar]
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang C, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov J, Poggio T, Gerald W, Loda M, Lander E, Golub T. Multiclass cancer diagnosis using tumor gene expression signature. PNAS. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset S, Zhu J. Adaptable, efficient and robust methods for regression and classification via piecewise linear regularized coefficient paths. 2007 Unpublished. [Google Scholar]
- Shevade K, Keerthi S. A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics. 2003;19:2246–2253. doi: 10.1093/bioinformatics/btg308. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, Series B. 1996;58:267–288. [Google Scholar]
- Tibshirani R. The lasso method for variable selection in the cox model. Statistics in Medicine. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Tseng P. Convergence of block coordinate descent method for nondifferentiable maximization. J. Opt. Theory and Applications. 2001;109(3):474–494. [Google Scholar]
- Van der Kooij A. Prediction accuracy and stability of regrsssion with optimal sclaing transformations. Leiden University: Dept. of Data Theory; Technical report. 2007
- Wu T, Lange K. Coordinate descent algorithms for lasso penalized regression. Annals of Applied Statistics. 2008a;2(1):224–244. doi: 10.1214/10-AOAS388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Lange K. Coordinate descent procedures for lasso penalized regression. Annals of Applied Statistics. 2008b;2(1):224–244. [Google Scholar]
- Wu T, Chen Y, Hastie T, Sobel E, Lange K. Genome-wide association analysis by penalized logistic regression. Bioinformatics. 2009 March;25(6):714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society, Series B. 2007;68(1):49–67. [Google Scholar]
- Zhu J, Hastie T. Classification of gene microarrays by penalized logistic regression. Biostatistics. 2004 doi: 10.1093/biostatistics/5.3.427. (to appear) [DOI] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J. Royal. Stat. Soc. B. 2005;67(2):301–320. [Google Scholar]