Abstract
In myocardial disease, elevated expression and activity of Na+/H+ exchanger isoform 1 (NHE1) are detrimental. To better understand the involvement of NHE1, transgenic mice with elevated heart-specific NHE1 expression were studied. N-line mice expressed wild-type NHE1, and K-line mice expressed activated NHE1. Cardiac morphology, interstitial fibrosis, and cardiac function were examined by histological staining and echocardiography. Differences in gene expression between the N-line or K-line and nontransgenic littermates were probed with genechip analysis. We found that NHE1 K-line (but not N-line) hearts developed hypertrophy, including elevated heart weight-to-body weight ratio and increased cross-sectional area of the cardiomyocytes, interstitial fibrosis, as well as depressed cardiac function. N-line hearts had modest changes in gene expression (50 upregulations and 99 downregulations, P < 0.05), whereas K-line hearts had a very strong transcriptional response (640 upregulations and 677 downregulations, P < 0.05). In addition, the magnitude of expression alterations was much higher in K-line than N-line mice. The most significant changes in gene expression were involved in cardiac hypertrophy, cardiac necrosis/cell death, and cardiac infarction. Secreted phosphoprotein 1 and its signaling pathways were upregulated while peroxisome proliferator-activated receptor γ signaling was downregulated in K-line mice. Our study shows that expression of activated NHE1 elicits specific pathways of gene activation in the myocardium that lead to cardiac hypertrophy, cell death, and infarction.
Keywords: transgenic mice, secreted phosphoprotein 1, peroxisome proliferator-activated receptor γ
the mammalian na+/h+ exchangers (NHEs) are a group of integral membrane transport proteins that mediate an electroneutral 1:1 exchange of intracellular H+ for extracellular Na+ and, in doing so, regulate intracellular pH (pHi) and cell volume. Ten isoforms of NHE have been described so far; however, NHE isoform 1 (NHE1) is the predominant plasma membrane isoform expressed in the myocardium (21). Besides being a regulator of pHi and ionic homeostasis, NHE1 also acts as an anchor for actin filaments and a scaffold for an ensemble of signaling complexes. Thus NHE1 regulates a diverse range of cell behaviors, including migration, proliferation, growth, differentiation, cytoskeleton dynamics, and survival (43).
NHE1 is of particular importance in the myocardium because it has been associated with myocardial ischemia-reperfusion (I/R) injury (33) and maladaptive heart hypertrophy (31). Enhanced activity of the NHE1 was first reported in the hypertrophic myocardium of spontaneously hypertensive rats by Perez et al. (47). Thereafter, NHE1 protein expression and activity have been demonstrated to be elevated in a variety of cardiovascular diseases, including in hypertensive, hypertrophied, and diabetic myocardium (7, 18, 30, 32), in hearts subjected to ischemia (24), in isolated cardiomyocytes subjected to chronic acidosis (16), and in patients with end-stage heart failure (55). Inhibition of NHE1 activity has demonstrated cardioprotective roles in many studies (1, 8, 19, 21, 33).
Because NHE1 expression and activity are elevated in cardiac disorders, we examined transgenic mice with cardiac-specific NHE1 elevation to elucidate NHE1-mediated signals that lead to myocardial injury. Two transgenic mouse lines were studied, the N-line carrying an elevated level of wild-type NHE1 and the K-line carrying an elevated level of activated NHE1. We evaluated the phenotypic effects of elevated NHE1 expression and activity on the heart, including cardiac morphology, histology, and function and also changes in gene expression pattern in NHE1 transgenic mice. The data revealed that expression of activated NHE1 induced pathological cardiac hypertrophy and dysfunction. Upregulation of secreted phosphoprotein 1 (SPP1) and downregulation of peroxisome proliferator-activated receptor (PPAR)γ likely form the effective signaling pathways that contribute to cardiac pathology when activated NHE1 is elevated.
MATERIALS AND METHODS
NHE1 transgenic mice.
Transgenic mice that specifically express NHE1 in the mouse myocardium under control of the cardiac-specific α-myosin heavy chain promoter have been described previously (2, 9, 29). The N-line expresses wild-type NHE1 protein, and a second K-line expresses NHE1 with a mutation in the cytoplasmic regulatory calmodulin binding domain of the protein (Lys641, Arg643, Arg645, and Arg647 were mutated to Glu residues). The mutation causes activation of NHE1 via an alkaline shift in NHE1 pH dependence (3, 9, 29). Male mice were used for all experiments, and age-matched nontransgenic littermates were used as controls. For phenotypic characterization mice were 10–12 wk of age, and for gene expression profiling mice were 2 wk old. All animal protocols were approved by the Animal Care Committee of the University of California San Diego, by the University of Alberta Animal Policy and Welfare Committee, and by the Canadian Council on Animal Care.
Heart-to-body weight ratio.
Mice were euthanized with halothane, and the hearts were isolated. All extracardiac structures were excised, and hearts were washed in PBS, blotted, and weighed (10). Cardiac hypertrophy was evaluated by measuring heart-to-body weight ratio (HW/BW), in milligrams per gram.
Histology.
Hearts were fixed in 10% buffered formalin overnight, embedded in paraffin, and serially sectioned into 5-μm slices (6, 40). Samples were stained with hematoxylin and eosin (H & E) to evaluate gross morphology and cross-sectional area (CSA) or with picrosirius red to assess fibrosis (41). CSA was measured from digitized images with Image ProPlus version 4.5 (Media Cybermetics, Silver Spring, MD). Digitized images were gathered with a Leica DMIRB microscope (Leica Microsystems, Bannockburn, IL) equipped with a Photometrics CoolSNAPfx camera (Photometrics, Surrey, BC, Canada). Dimensions of at least 100 cells from 3 or 4 sections per heart were measured. Visual fields were accepted for quantification if nuclei were visible and cell membranes were intact (40, 41). Interstitial fibrosis (IF) was measured from digitized images with Openlab 3.5 (Improvision, Waltham, MA). Digitized images were gathered with a QImaging camera (QImaging, Surrey, BC, Canada) equipped with a Leica DMLA microscope (Leica Microsystems). Ten fields were randomly selected from three or four sections per heart. The maximum fibrosis observed for any section was calculated as the area occupied by red-stained connective tissue divided by the areas occupied by connective tissue plus cardiac myocytes × 100. Intramural vessels, perivascular collagen, endocardium, and trabeculae were excluded (26).
Echocardiography.
Echocardiography was performed on mice anesthetized with isoflurane with the Vevo High-Resolution Imaging System (VisualSonics, Toronto, ON, Canada) equipped with a 35-MHz transducer. Short-axis M-mode analysis, at the level of the papillary muscle, was used for measurement of diastolic and systolic diameters of the left ventricle as well as for the measurement of wall thickness. Pulsed wave Doppler, performed at the levels of the left ventricular outflow tract, the right ventricular outflow tract, and the mitral valve tips, was used to monitor flow pattern and velocity. Myocardial performance index or Tei index was calculated with the following equation: (isovolumic contraction time + isovolumic relaxation time)/ejection time (13).
Gene expression profiling.
Two-week-old mice were chosen to determine early gene expression changes that lead to later phenotypic alterations. At this early age, mice had not developed major pathology. Total RNA was extracted from frozen hearts of wild-type control, N-line, and K-line mice (n = 3/group) with an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA quality was examined with the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA (300 ng) was used to synthesize biotin-labeled complementary RNA (cRNA) probes with an Illumina RNA amplification kit (Ambion, Austin, TX) as previously described (57). Illumina Sentrix mouse-6 expression genechips (Illumina, San Diego, CA) were used to determine differences in gene expression (34). Biotin-labeled cRNA (1.5 μg) was added to the chip and incubated for 16–20 h at 55°C. The bound biotin-labeled cRNA was then stained with streptavidin-Cy3. After hybridization, the genechips were washed, dried, and scanned by the Illumina BeadArray Reader (Illumina); the absolute intensity of each probe on the image was generated with BeadStudio software (Illumina). The difference in expression level was calculated by comparing N-line or K-line with control samples from each chip after cubic spline normalization. The significant threshold was fold change greater than 1.50 and differential score either greater than 13 for upregulation or less than −13 for downregulation (i.e., adjusted P values < 0.05). The data sets of chip analyses can be traced under Gene Expression Omnibus (GEO) series access number GSE19819 in the National Center for Biotechnology Information GEO database (http://www.ncbi.nlm.nih.gov/geo). Functional categorization of gene alterations was created with the Ingenuity Pathways Analysis (IPA) program (Ingenuity Systems, Redwood City, CA) (57). The P value for each network or function was calculated with a right-tailed Fisher's exact test. The score for each network or function was shown as −log10[P value], which indicates the likelihood of finding a set of focus genes in the network or function by random chance. The significance threshold was set to a score of 1.3 (i.e., P ≤ 0.05).
Quantitative real-time RT-PCR.
Seven differentially expressed genes were randomly selected from genechip experiments and validated by quantitative real-time RT-PCR, as described previously (58). The primer sets for specific genes were designed with the Primer3 program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi/) and synthesized by Invitrogen (Carlsbad, CA) (Supplemental Table S1).1 The fold change in expression level for each specific gene was calculated by the 2−ΔΔCt method (where Ct is threshold cycle) (39), after normalization to β-actin (loading controls). The final result represents the mean fold change of three individual transgenic samples over controls.
Statistical analysis.
For HW/BW, histology, and echocardiography, all values are expressed as means ± SE or as % of control ± %SE. Groups were compared by both Student's t-test and Wilcoxon signed-rank test. For quantitative real-time RT-PCR, groups were compared by the one-way ANOVA with Dunnett posttest. Statistical significance was set to P < 0.05.
RESULTS
Cardiac hypertrophy, fibrosis, and dysfunction caused by elevation of activated NHE1.
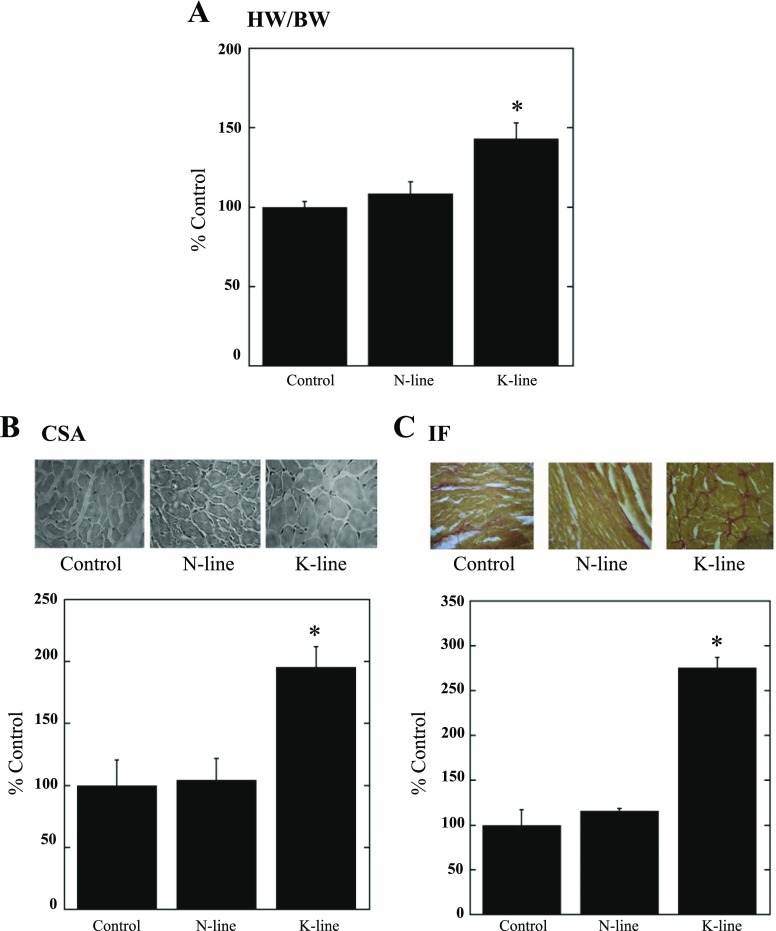
N-line and K-line mice have previously been shown to possess approximately two- and threefold increases in NHE1 activity, respectively (9, 29). Since our previous data also showed that NHE1 transgenic hearts had an enlargement in overall size (29), in this study we further characterized the cardiac phenotype by examining HW/BW and CSA of the ventricular cardiomyocytes. Figure 1A demonstrates that HW/BW was significantly increased in K-line mice (143 ± 10.0%, P < 0.05) compared with both control and N-line mice (108.4 ± 7.5%). No significant difference was found in HW/BW between N-line and control mice. The variations in HW/BW between groups were due to changes in heart weight, as there were no significant changes in body weight (data not shown). Figure 1B examines the CSA of control, N-line, and K-line mice. Figure 1B, top, is representative of myocardial cross sections stained with H & E, and Fig. 1B, bottom, is a quantitative summary of the results. Whereas the N-line mice were not significantly different from the control mice, the K-line mice had significantly increased CSA (195.6 ± 16.4%, P <0.05).
Fig. 1.
Elevation of activated Na+/H+ exchanger isoform 1 (NHE1) induces larger cardiomyocytes, interstitial fibrosis (IF), and cardiac dysfunction. A: heart-to-body weight ratio (HW/BW). *P < 0.05 for control vs. K-line or N-line vs. K-line; n = 7–13. B: cross-sectional area (CSA) of cardiomyocytes: histological analysis of control, N-line, and K-line heart cross sections (×40) stained with hematoxylin and eosin (H & E). Top: examples of cross sections. Bottom: summary of CSA per group, expressed as % of control. *P < 0.05 for control vs. K-line. C: picrosirius red-stained cross sections assessed for left ventricular IF. Top: typical cross sections. Bottom: quantitative analysis. *P < 0.05 for control vs. K-line (3 or 4 sections/heart, n = 4).
To determine whether increased NHE1 expression and activity promoted myocardial fibrosis, a manifestation of hearts subjected to intrinsic and external stress, cross sections were stained with picrosirius red (41). Figure 1C, top, is representative of picrosirius red-stained cross sections of control, N-line, and K-line hearts, and Fig. 1C, bottom, is a quantitative summary of the results. Elevated expression of activated NHE1 (K-line mice) resulted in significantly increased IF (275.4 ± 11.6%, P <0.05), although a significant increase in IF was not observed in N-line mice.
In vivo echocardiographic assessment of cardiac morphology confirmed our previous indication of NHE1-dependent cardiac hypertrophy. Left ventricular posterior wall thickness, intraventricular septal wall thickness, and left ventricular mass, all indicators of hypertrophy, were significantly greater in K-line mice versus both control and N-line mice (Table 1). Moreover, K-line mice showed a decrease in systolic function demonstrated by a decrease in left ventricular fractional shortening and a decrease in diastolic dysfunction demonstrated by an increase in ratio of peak E wave to peak A wave mitral valve velocity. The global deterioration in myocardial performance was further confirmed by the significant increase in the Tei index in K-line compared with control and N-line mice (Table 1).
Table 1.
Echocardiographic analysis of heart parameters of N- and K-line mice
| Parameter | Control | N-Line | K-Line |
|---|---|---|---|
| IVSTd, mm | 0.79 ± 0.02 | 0.75 ± 0.01 | 0.87 ± 0.03*‡ |
| LVPWd, mm | 0.74 ± 0.02 | 0.74 ± 0.09 | 0.86 ± 0.03*‡ |
| LVM, mg | 101.6 ± 3.8 | 95.6 ± 2.4 | 142.8 ± 11.0*§ |
| LVIDd, mm | 4.37 ± 0.1 | 4.28 ± 0.06 | 4.81 ± 0.2 |
| LVIDs, mm | 2.78 ± 0.1 | 2.85 ± 0.1 | 3.75 ± 0.4*‡ |
| FS, % | 36.5 ± 1.1 | 33.5 ± 1.5 | 23.1 ± 3.8*‡ |
| EF, % | 66.4 ± 1.5 | 62.3 ± 2.1 | 45.2 ± 6.9*‡ |
| MV E/A | 1.98 ± 0.27 | 1.94 ± 0.15 | 3.56 ± 0.60*‡ |
| Tei index | 0.49 ± 0.02 | 0.51 ± 0.02 | 0.78 ± 0.03†§ |
Values are expressed as means ± SE (n = 7-8/group). IVSTd, diastolic interventricular septal wall thickness; LVPWd, diastolic left ventricular posterior wall thickness; LVM, left ventricular mass; LVIDd and LVIDs, left ventricular internal diameter during diastole and systole; FS, left ventricular % fractional shortening; EF, left ventricular % ejection fraction; MV E/A, ratio of peak E wave mitral valve velocity to peak A wave mitral valve velocity.
P < 0.05 vs. Control;
P < 0.001 vs. Control;
P < 0.05 for N-line vs. K-line;
P < 0.001 for N-line vs. K-line.
Gene expression profile in hearts with elevated NHE1 expression and activity.
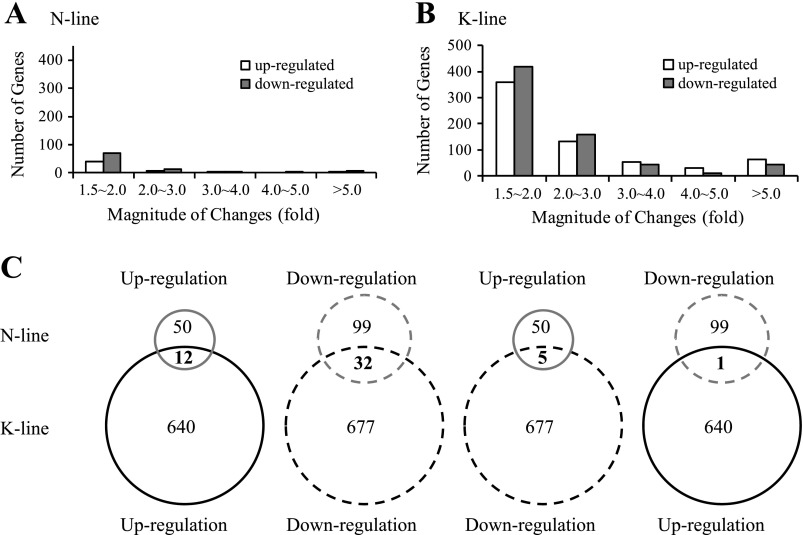
Since the onset of progressive hypertrophy in NHE1 transgenic hearts was detected at the age of 20 days (44), NHE1 transgene-induced early gene expression changes were examined at the early age of 2 wk. The fold change of each gene expression was calculated by comparing the ratio of K-line and N-line gene expression to that of age-matched controls (Supplemental Tables S2 and S3). We observed a marked difference in the transcriptional response between N-line and K-line mice. N-line mice demonstrated modest changes in gene expression in the myocardium (50 upregulations and 99 downregulations, P < 0.05) (Supplemental Table S2), whereas K-line mice showed a strong transcriptional response (640 upregulations and 677 downregulations, P < 0.05) (Supplemental Table S3), approximately nine times the number of genes induced in the N-line mice. In addition, the magnitude of expression alterations was much higher in K-line mice: 538 genes had >2-fold changes in K-line mice, whereas the number was reduced to 39 genes in N-line mice (Fig. 2, A and B), indicating that both expression and activation of NHE1 transgene are critical for the induction of transcriptional alterations.
Fig. 2.
Cluster analysis of gene expression profiles in mouse hearts with elevated NHE1. A and B: number and magnitude of gene changes in N-line (A) and K-line (B) mouse hearts. C: comparison of gene changes between N-line and K-line mouse hearts. Gray circle, N-line; dark circle, K-line; solid circle, upregulation; dashed circle, downregulation.
Of the 149 significantly altered genes in the N-line mice, the most upregulated genes were SPP1 (secreted phosphoprotein 1) followed by ITGB6 (integrin β6) and CCL12 (C-C motif chemokine ligand 12), while the most downregulated genes were ZBTB16 (zinc finger protein 145), GDF15 (growth differentiation factor 15), and CNKSR1 (connector enhancer of kinase suppressor of Ras 1) (Supplemental Table S2). Of the 1,317 significantly altered genes in the K-line mice, the most upregulated genes were SPP1 followed by PIRA6 (paired-Ig-like receptor A6) and XIST (inactive X specific transcripts), while the most downregulated genes were HAMP (hepcidin antimicrobial peptide), MYBPH1 (myosin binding protein H-like), and PRSS35 (serine protease 35) (Supplemental Table S3). Unlike N-line mice, K-line mice showed >100-fold changes in some extreme cases of up- or downregulated genes.
Interestingly, there were 12 genes commonly upregulated and 32 genes commonly downregulated in N-line and K-line mice (Fig. 2C and Supplemental Table S4). The most pronounced upregulated genes were SPP1, ITGB6, and CCL12, and the most pronounced downregulated genes were ZBTB16, PPARGC1A (peroxisome proliferator-activated receptor γ, coactivator 1α), and MAP3K6 (mitogen-activated protein kinase kinase kinase 6). Only six genes were altered in an opposite direction in the two NHE1 transgenic groups (Fig. 2C), and the alterations were small, less than twofold.
Functional categorization of differentially expressed genes.
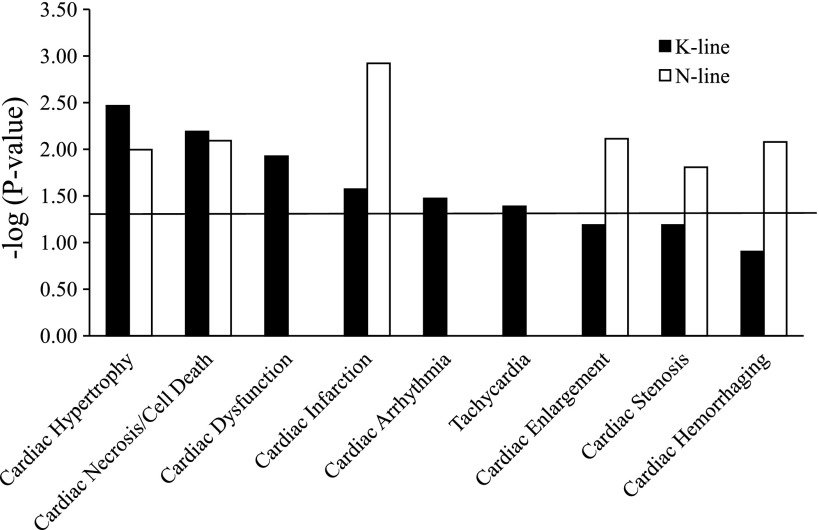
The data were further analyzed with IPA to better understand the biological significance of these gene changes induced by NHE1. Since NHE1 transgenic mice exhibited cardiac hypertrophy, fibrosis, and dysfunction, our focus centered on the gene changes involved in cardiac pathological processes. Accordingly, 80 genes that have been correlated previously with cardiac pathological function were altered by the NHE1 transgene (Supplemental Table S5). The cardiac pathological processes significantly altered in both N-line and K-line mouse hearts were cardiac hypertrophy, cardiac necrosis/cell death, and cardiac infarction (Fig. 3). Moreover, N-line mice had gene alterations involved in cardiac enlargement, cardiac stenosis, and cardiac hemorrhaging processes (Fig. 3), while K-line mice had additional gene changes in cardiac dysfunction, cardiac arrhythmia, and tachycardia (Fig. 3). These data suggest that the type of cardiac pathology varies based on whether NHE1 is expressed as wild type (N-line) or activated form (K-line). Since NHE1 transgenic mice showed interstitial fibrosis at the age of 2.5 mo, cardiac fibrosis-related gene changes were also included in Supplemental Table S5, although these changes do not reach statistical significance when they are considered as a functional group at the examined age of 2 wk. Additionally, 6-wk-old NHE1 transgenic mice showed more gene alterations involved in cardiac injury and failure than 2-wk-old mice (data not shown), demonstrating a disease progression from early compensatory growth/proliferation to later tissue damage and functional failure.
Fig. 3.
Functional categorization by cardiac pathology. Genes significantly altered by NHE1 transgene were classified into associated cardiac pathological functions (as depicted in x-axis) with Ingenuity Pathway Analysis (IPA) software. Functions are listed from most significant to least. y-Axis depicts −log10[P value]. The significance threshold was set to 1.3 (P ≤ 0.05), as delineated by the horizontal line.
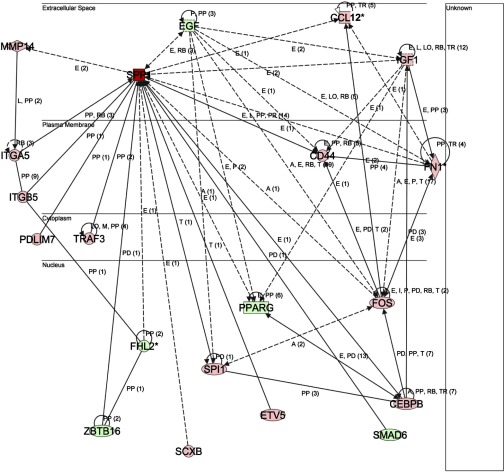
Of the 80 genes related to cardiac pathology, 5 showed the same trend in expression changes when hearts of N-line were compared with K-line mice. These were SPP1, VCAM1 (vascular cell adhesion molecule 1), EDN1 (endothelin 1), HIF3A (hypoxia-inducible factor 3, α-subunit) and PPARGC1A (Supplemental Table S5), with SPP1 being the most dramatically altered. Intriguingly, the expression of several genes associated with or regulating SPP1 were also altered in K-line mice (Table 2), including 1) genes regulated by SPP1, e.g., MMP14 (matrix metalloproteinase 14), CD44, and EGF (epidermal growth factor) (25, 48, 54); 2) SPP1-interacting receptors or proteins, e.g., ITGA5, ITGB5 (integrin receptor α5, β5), CD44, and TRAF3 (TNF receptor-associated factor 3) (5, 27, 37); 3) signaling molecules of SPP1, such as PRKCB, PRKCD (protein kinase Cβ, δ), PIK3CG (phosphoinositide 3-kinase, catalytic, γ polypeptide), AKT2 (thymoma viral protooncogene 2), and MAP3K6 (45, 49–51); 4) the transcriptional regulators of SPP1, e.g., ETV5 (ets variant 5), SCXB (scleraxis homolog B), and CEBPB [CCAAT/enhancer binding protein (C/EBP), β] (17, 38, 42); and 5) the molecules previously reported to increase expression of SPP1 mRNA (28, 53), e.g., IGF1 (insulin-like growth factor 1) and FN1 (fibronectin 1). Figure 4 illustrates the relationships between these genes and SPP1. Most of these associated gene changes were not detected in N-line hearts.
Table 2.
Differentially expressed genes related to SPP1 in K-line mouse transgenic hearts
| Symbol | Entrez Gene Name | GenBank Accession No. | Fold Change | Family |
|---|---|---|---|---|
| SPP1 | Secreted phosphoprotein 1 | NM_009263 | 1,548.82 | Cytokine |
| CCL12 | Chemokine (C-C motif) ligand 12 | NM_011331 | 6.84 | Cytokine |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | NM_010234 | 3.46 | Transcription regulator |
| PRKCB | Protein kinase C, β | NM_008855 | 2.85 | Kinase |
| ETV5 | ets variant 5 | NM_023794 | 2.32 | Transcription regulator |
| CD44 | CD44 molecule (Indian blood group) | NM_009851 | 1.99 | Other |
| FN1 | Fibronectin 1 | XM_129845 | 1.91 | Enzyme |
| SCXB | Scleraxis homolog B (mouse) | NM_198885 | 1.89 | Other |
| PIK3CG | Phosphoinositide 3-kinase, catalytic, γ polypeptide (Pik3cg) | NM_020272 | 1.89 | Kinase |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), β | NM_009883 | 1.87 | Transcription regulator |
| PRKCD | Protein kinase C, δ (Prkcd) | NM_011103 | 1.84 | Kinase |
| MMP14 | Matrix metallopeptidase 14 (membrane-inserted) | NM_008608 | 1.73 | Peptidase |
| SPI1 | Spleen focus forming virus (SFFV) proviral integration oncogene spi1 | NM_011355 | 1.70 | transcription regulator |
| TRAF3 | TNF receptor-associated factor 3 | NM_011632 | 1.69 | Other |
| ITGB5 | Integrin, β5 | NM_010580 | 1.62 | Other |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | NM_010512 | 1.62 | Growth factor |
| ITGA5 | Integrin, α5 (fibronectin receptor, α polypeptide) | NM_010577 | 1.62 | Other |
| PDLIM7 | PDZ and LIM domain 7 (enigma) | NM_026131 | 1.61 | Other |
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 (Map3k5) | NM_008580 | 1.53 | Kinase |
| AKT2 | Thymoma viral proto-oncogene 2 (Akt2) | NM_007434 | −1.64 | Kinase |
| ZBTB16 | Zinc finger and BTB domain containing 16 | XM_134826 | −1.92 | Transcription regulator |
| PPARG | Peroxisome proliferator-activated receptor γ | NM_011146 | −1.94 | Ligand-dependent nuclear receptor |
| MAP4K2 | Mitogen-activated protein kinase kinase kinase kinase 2 (Map4k2) | NM_009006 | −2.03 | Kinase |
| MAP3K6 | Mitogen-activated protein kinase kinase kinase 6 (Map3k6) | NM_016693 | −2.15 | Kinase |
| EGF | Epidermal growth factor (β-urogastrone) | NM_010113 | −2.21 | Growth factor |
| FHL2 | Four and a half LIM domains 2 | AK052936 | −2.46 | Other |
| SMAD6 | SMAD family member 6 | NM_008542 | −2.48 | Transcription regulator |
The differentially expressed genes in K-line transgenic hearts were probed with Illumina mouse-6 expression genechips and determined by comparing K-line with controls. The significant threshold was fold change >1.50 and differential score either >13 for upregulation or < −13 for downregulation (i.e., adjusted P < 0.05). Positive fold change represents upregulation, while negative fold change represents downregulation.
Fig. 4.
The network between secreted phosphoprotein 1 (SPP1) and its related differentially expressed genes generated by IPA. The network is displayed graphically as nodes (genes/proteins) and edges (biological relationships between nodes). The intensity of the node color indicates the degree of upregulation (red) or downregulation (green). Edges with various labels indicate the nature of the relationship between the nodes as follows: A, activation; E, expression; I, inhibition; L, proteolysis; LO, localization; M, biochemical modification; P, phosphorylation/dephosphorylation; PD, protein-DNA binding; PP, protein-protein binding; PR, protein-RNA binding; RB, regulation of binding; T, transcription; TR, translocation. Straight lines denote direct interactions, and dashed lines denote indirect interactions.
PPARs are ligand-activated nuclear receptors that play an important role in myocardial metabolism, inflammation, and cardiomyopathic remodeling by transcriptionally regulating gene expression (15). Expression of PPARG (peroxisome proliferator-activated receptor γ), its heterodimeric partner RXRG (retinoid X receptor γ), and coactivator PPARGC1A were all decreased in K-line hearts (Supplemental Table S3). Furthermore, the target genes of PPARG (11, 36, 46, 56), e.g., SPP1, NPPB (natriuretic peptide precursor B), VCAM1, and TPM2 (tropomyosin 2β), were also significantly altered (Supplemental Table S3) in K-line mice. These data indicate that PPARG pathway may contribute to cardiac pathology in K-line mice. For N-line hearts, only PPARGC1A, SPP1, and VCAM1 showed significant expression changes among the genes related to the PPARG pathway.
NHE1 is known to be a key downstream mediator of cardiac hypertrophy produced by endothelin-1 via endothelin receptor type A (ETA) (22). In K-line mice, expression of both EDN1 and ETA were reduced, possibly reflecting a negative-feedback regulatory loop (Supplemental Table S3).
To gain insight into the molecular mechanisms of NHE1 action, we also classified the differentially expressed genes on the basis of molecular and cellular functions. The top-ranked functions were cell death, cellular movement, cell-to-cell signaling and interaction, cellular growth and proliferation, and cellular function and maintenance, etc. in both N-line and K-line mice (data not shown). These results indicate that elevated NHE1 expression and activity induces gene changes leading to cell death.
Gene expression of NHE1 functionally related molecules.
Gene expression of NHE1 functionally related molecules was examined to determine whether elevated expression of NHE1 in the myocardium affected the expression of these genes. No significant difference was detected among N- and K-lines and controls regarding gene expression of proteins involved in pHi regulation and Na+ and Ca2+ fluxes, including Na+-HCO3− cotransporter (NBCn1), Cl−/HCO3− exchangers (AEs), Na+/Ca2+ exchanger (NCX), sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA), phospholamban, ryanodine receptors, calsequestrin, Na+-K+-ATPase (data not shown). However, the genes encoding a component of H+ transporting, vacuolar ATPase (V-ATPase) A and C subunits were significantly increased in K-line mice (Supplemental Table S3). NHE1 anchors actin filaments by direct binding to the ezrin-radixin-moesin (ERM) family. The expression of moesin gene was elevated only in K-line mice (Supplemental Table S3), while there were no evident changes in expression of ezrin and radixin genes. Among the kinases that modulate NHE1 activity, expression of Rho-activated kinase 1 (ROCK1), PIK3CG, PRKCB, PRKCD, and mitogen-activated protein kinase kinase kinase 5 (MAP3K5) were upregulated only in K-line mice (Supplemental Table S3). In contrast, expression of MAP3K6 and AKT2 were downregulated (Supplemental Table S3). No apparent differences were found for the genes encoding p38 mitogen-activated protein kinase, p90 ribosomal s6 kinase (P90RSK), and protein kinase A (PKA) (data not shown).
Validation of genechip results with quantitative real-time RT-PCR.
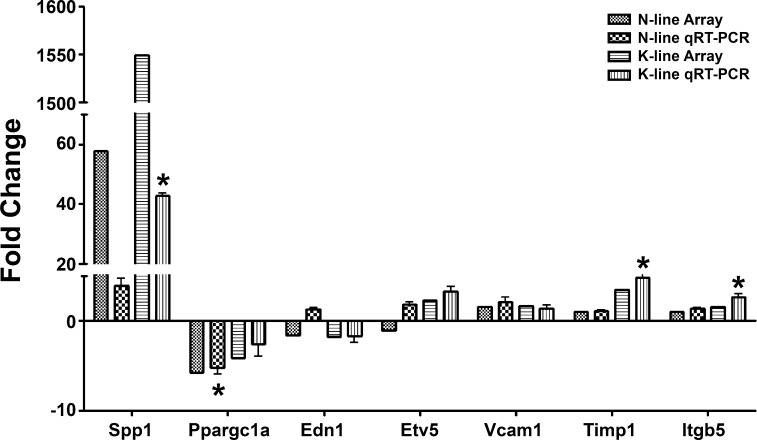
The genechip results were validated by quantitative real-time RT-PCR. As presented in Fig. 5, the data from real-time RT-PCR demonstrated the same trend of changes as in the genechip studies.
Fig. 5.
Verification of genechip results with quantitative real-time RT-PCR (qRT-PCR). Seven significantly altered genes from the genechip study were randomly selected and tested by real-time RT-PCR. β-Actin was used to normalized the gene expression levels. Values are means ± SE, n = 3. *Statistical significance compared with controls (P < 0.05, 1-way ANOVA with Dunnett posttest).
DISCUSSION
NHE1 has been demonstrated to play pathological roles in cardiovascular disorders, including in I/R injury, hypertrophy, and heart failure (1, 21, 33). Our studies (present data) and the studies of Nakamura's group (44) have shown that increased expression of constitutively active NHE1 is important in mediating cardiac hypertrophy. A variety of stimuli can lead to cardiac hypertrophy, including elevated arterial blood pressure, myocardial infarction, valvular heart disease, and cardiomyopathy (23). NHE1 inhibition has been proven to prevent or induce regression of hypertrophy in several models of cardiac hypertrophy (6, 7, 18, 19, 32, 35, 40). However, it is still largely unknown by what mechanism(s) elevated NHE1 propagates hypertrophic injury, although a number of pathways have been proposed (20, 44). Therefore, to address this question, transgenic mice with elevated myocardial NHE1 were generated and we assessed the early causal pathways involved in the detrimental effects of NHE1 expression.
The initial characterization of the transgenic mice demonstrated that elevated expression of NHE1 generated a prohypertrophic effect but only in mice that expressed an activated form of NHE1. K-line mice showed increased cardiomyocyte size (CSA), increased HW/BW, and IF. In addition, these K-line mice developed a number of functional abnormalities. These results are in general agreement with a recent study (44) that expressed activated NHE1 in transgenic mice by using a large deletion of the 637–656 bp region. However, in our study we used specific point mutations rather than a large deletion. In addition, we made a transgenic mouse that expressed a wild-type NHE1 protein with normal activity, and these mice remained principally equivalent to control mice. This indicates that an activated form of NHE1 was required to induce the majority of the detrimental effects.
The examination of the gene expression profile in NHE1 transgenic mice at the age of 2 wk revealed that elevated NHE1 activity elicited large-scale gene expression changes before the development of hypertrophy and fibrosis. Elevated expression of the NHE1 protein alone without activation induced fewer gene alterations. At least three general novel observations were made in the present study. First, SPP1 signaling was significantly upregulated by increases in activated NHE1. SPP1, also known as osteopontin, is a matricellular protein that can be synthesized by several cell types, including cardiac myocytes, macrophages, microvascular endothelial cells, smooth muscle cells, and fibroblasts (45, 49–51). Expression of SPP1 is high during embryogenesis but is almost absent in postnatal healthy myocardium (49). Reinduction of SPP1 expression is observed in various cardiac pathologies including in heritable cardiomyopathy of Syrian hamsters, in spontaneous hypertensive and aortic banded rats, in pressure-overload hypertrophy, in myocardial infarction, in patients with advanced heart failure, and in atherosclerosis (45, 49–51). SPP1 is important in myocardial remodeling, cardiac hypertrophy, and fibrosis (45, 49–51). It recruits macrophages and T cells to sites of injury and inflammation and also regulates the production of inflammatory cytokines and nitric oxide in macrophages. SPP1 directly interacts with fibronectin and certain types of collagen, promotes collagen synthesis and deposition, and also regulates expression and activity of matrix metalloproteinases (MMPs), thus mediating extracellular matrix reorganization (remodeling) after tissue injury. SPP1 modulates myocardial hypertrophy, probably via integrin-associated signaling, and is required for myofibroblast differentiation and activation induced by transforming growth factor-β (TGF-β) (45, 49–51). Interestingly, the gene expression of all these SPP1-related molecules, including MMP, fibronectin, procollagens, integrin receptors, CD44, TGF-β receptor, and signaling kinases, were elevated with expression of activated NHE1 (Table 2). The results strongly suggest that SPP1 and its associated signaling are key players in NHE1-induced cardiac pathology. Figure 6 outlines potential mechanisms by which increased SPP1 may play a role in myocardial remodeling, fibrosis, and hypertrophy.
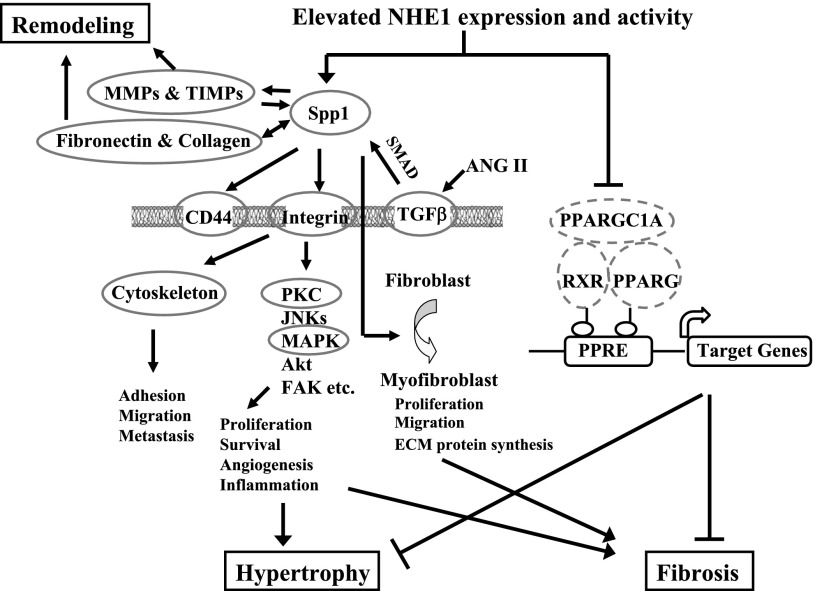
Fig. 6.
Schematic illustration of potential pathways mediated by SPP1 or peroxisome proliferator-activated receptor γ (PPARG), which underlie prohypertrophic and profibrotic effects of elevated activated NHE1. Increased SPP1 by elevation of activated NHE1 promotes collagen synthesis and deposition and also regulates expression/activity of matrix metalloproteinases (MMPs), thus mediating extracellular matrix remodeling after tissue injury. Through interaction with integrin receptors or hyaluronic receptor CD44, SPP1 induces cytoskeletal rearrangement and consequent deadhesion, thus facilitating cell migration, infiltration, and spreading. SPP1 also activates downstream signaling and modulates cell growth, proliferation, and survival, which contribute to cardiac hypertrophy and fibrosis. Furthermore, SPP1 mediates transforming growth factor-β (TGF-β)-induced myofibroblast differentiation/activation and plays a pivotal role in fibrosis. Upon ligand activation, PPARG heterodimerizes with its obligate partner retinoid X receptor (RXR) and recruits transcriptional coactivator peroxisome proliferator-activated receptor γ, coactivator 1α (PPARGC1A), then binds to PPAR response elements (PPRE) in the promoter regions of target genes, thus controlling gene expression that protects myocardium from inflammation, hypertrophy, fibrosis, and ischemia-reperfusion injury. Present data reveal that elevated activated NHE1 reduced PPARG/RXR/PPARGC1A expression, thereby promoting the development of cardiac hypertrophy and fibrosis. Gene expression of circled molecules (solid circle, upregulation; dashed circle, downregulation) were simultaneously altered in NHE1 K-line hearts.
Second, PPARG, its heterodimeric partner RXR, and coactivator PPARGC1A were all significantly downregulated in K-line hearts. When engaged by its ligand, PPARG heterodimerizes with its obligate partner RXR and recruits transcriptional coactivator PPARGC1A (Fig. 6). This complex then binds to specific PPAR response elements (PPREs) in the promoter regions of target genes and controls gene expression that protects the myocardium from inflammation, hypertrophy, fibrosis, and I/R injury (15, 52). Cardiomyocyte-specific PPAR-γ knockout mice (4, 12, 14) develop progressive cardiac hypertrophy accompanied by increased expression of cardiac embryonic genes and elevated nuclear factor-κB activity (14) or mitochondrial oxidative damage (12). Most of these knockout mice die from dilated cardiomyopathy or heart failure (12). Therefore, in the present study, reduced expression of PPARG, RXR, and PPARGC1A induced by elevated expression of activated NHE1 could contribute to the observed cardiac hypertrophy in K-line mice. Intriguingly, it has been reported that SPP1 expression in cardiomyocytes and/or macrophages is affected by PPARG (4). Although it is unknown whether SPP1 gene expression is regulated by PPARG in NHE1 K-line mice, both increased SPP1 and decreased PPARG can lead to cardiac hypertrophy and fibrosis (Fig. 6).
A third novel observation of this study is that elevated expression and activity of NHE1 did not significantly change mRNA levels of other regulatory proteins involved in pHi regulation and Na+, Ca2+ flux, including Na+-HCO3− cotransporter (NBCn1), Cl−-HCO3− exchangers (AEs), Na+/Ca2+ exchanger (NCX), SERCA, phospholamban, ryanodine receptors, calsequestrin, and Na+-K+-ATPase. These data correlate with their protein expression in NHE1 transgenic mice (29, 44). Nakamura et al. (44) have demonstrated that enhanced NHE1 activity elevated intracellular Na+ concentration ([Na+]i), elevated systolic and diastolic intracellular Ca2+ concentration ([Ca2+]i), and altered sarcoplasmic reticulum Ca2+ handling in their transgenic myocytes. Thus they propose that [Na+]i and [Ca2+]i trigger hypertrophic changes in the myocardium. They also showed that calcineurin and CaMKII were highly activated in transgenic hearts, accompanied by nuclear translocation of NFAT (nuclear factor of activated T cells) and nuclear export of HDAC4 (histone deacetylase 4), which can alter hypertrophy-associated gene expression (44). In our transgenic mice, changes in gene expression of calcineurin, CaMKII, NFAT, and HDAC4 were not evident. The data suggest that modifications of calcineurin-NFAT and CaMKII-HDAC4 pathways by elevated NHE1 occurred at a posttranscriptional level.
Our present study has demonstrated that increased expression of activated NHE1 alone is sufficient to induce myocardial injury, including cardiac hypertrophy, IF, cardiac dysfunction, and, in the long-term, development of heart failure and increased mortality with aging. Analysis of gene expression pattern in NHE1 transgenic hearts enabled us to identify potentially disease-causing genes, such as SPP1, PPARG, and their associated genes. In addition, we propose pathways mediated by these molecules that may underlie myocardial injury (Fig. 6). The question remains as to how NHE1 orchestrates these gene expression changes. Are these changes adaptive or maladaptive? Do SPP1 and PPARG act additively or synergistically? We still do not have clear answers for these questions at present.
GRANTS
This work was supported by National Institutes of Health Grants PO1-HD-32573 and RO1-NS-037756 to G. G. Haddad, by a Parker B. Francis Fellowship grant to J. Xue, by American Heart Association Grant 0835188N to D. Zhou, by Canadian Institutes of Health Research (CIHR) Grant MOP-97816 and an Alberta Heritage Foundation for Medical Research (AHFMR) Senior Scientist award to L. Fliegel, and by CIHR and AHFMR to F. Mraiche. M. Karmazyn is a Canada Research Chair in Experimental Cardiology.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Orit Gavrialov for technical assistance.
Footnotes
Supplemental Material for this article is available online at the Journal website.
REFERENCES
- 1. Avkiran M, Cook AR, Cuello F. Targeting Na+/H+ exchanger regulation for cardiac protection: a RSKy approach? Curr Opin Pharmacol 8: 133– 140, 2008. [DOI] [PubMed] [Google Scholar]
- 2. Baczko I, Mraiche F, Light PE, Fliegel L. Diastolic calcium is elevated in metabolic recovery of cardiomyocytes expressing elevated levels of the Na+/H+ exchanger. Can J Physiol Pharmacol 86: 850– 859, 2008. [DOI] [PubMed] [Google Scholar]
- 3. Bertrand B, Wakabayashi S, Ikeda T, Pouyssegur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J Biol Chem 269: 13703– 13709, 1994. [PubMed] [Google Scholar]
- 4. Caglayan E, Stauber B, Collins AR, Lyon CJ, Yin F, Liu J, Rosenkranz S, Erdmann E, Peterson LE, Ross RS, Tangirala RK, Hsueh WA. Differential roles of cardiomyocyte and macrophage peroxisome proliferator-activated receptor gamma in cardiac fibrosis. Diabetes 57: 2470– 2479, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen K, Huang J, Gong W, Iribarren P, Dunlop NM, Wang JM. Toll-like receptors in inflammation, infection and cancer. Int Immunopharmacol 7: 1271– 1285, 2007. [DOI] [PubMed] [Google Scholar]
- 6. Chen L, Chen CX, Gan XT, Beier N, Scholz W, Karmazyn M. Inhibition and reversal of myocardial infarction-induced hypertrophy and heart failure by NHE-1 inhibition. Am J Physiol Heart Circ Physiol 286: H381– H387, 2004. [DOI] [PubMed] [Google Scholar]
- 7. Chen L, Gan XT, Haist JV, Feng Q, Lu X, Chakrabarti S, Karmazyn M. Attenuation of compensatory right ventricular hypertrophy and heart failure following monocrotaline-induced pulmonary vascular injury by the Na+-H+ exchange inhibitor cariporide. J Pharmacol Exp Ther 298: 469– 476, 2001. [PubMed] [Google Scholar]
- 8. Cingolani HE, Rebolledo OR, Portiansky EL, Perez NG, Camilion de Hurtado MC. Regression of hypertensive myocardial fibrosis by Na+/H+ exchange inhibition. Hypertension 41: 373– 377, 2003. [DOI] [PubMed] [Google Scholar]
- 9. Coccaro E, Mraiche F, Malo M, Vandertol-Vanier H, Bullis B, Robertson M, Fliegel L. Expression and characterization of the Na+/H+ exchanger in the mammalian myocardium. Mol Cell Biochem 302: 145– 155, 2007. [DOI] [PubMed] [Google Scholar]
- 10. Dawn B, Xuan YT, Marian M, Flaherty MP, Murphree SS, Smith TL, Bolli R, Jones WK. Cardiac-specific abrogation of NF-kappaB activation in mice by transdominant expression of a mutant IkappaBalpha. J Mol Cell Cardiol 33: 161– 173, 2001. [DOI] [PubMed] [Google Scholar]
- 11. Diep QN, El Mabrouk M, Cohn JS, Endemann D, Amiri F, Virdis A, Neves MF, Schiffrin EL. Structure, endothelial function, cell growth, and inflammation in blood vessels of angiotensin II-infused rats: role of peroxisome proliferator-activated receptor-gamma. Circulation 105: 2296– 2302, 2002. [DOI] [PubMed] [Google Scholar]
- 12. Ding G, Fu M, Qin Q, Lewis W, Kim HW, Fukai T, Bacanamwo M, Chen YE, Schneider MD, Mangelsdorf DJ, Evans RM, Yang Q. Cardiac peroxisome proliferator-activated receptor gamma is essential in protecting cardiomyocytes from oxidative damage. Cardiovasc Res 76: 269– 279, 2007. [DOI] [PubMed] [Google Scholar]
- 13. Dolinsky VW, Chan AY, Robillard Frayne I, Light PE, Des Rosiers C, Dyck JR. Resveratrol prevents the prohypertrophic effects of oxidative stress on LKB1. Circulation 119: 1643– 1652, 2009. [DOI] [PubMed] [Google Scholar]
- 14. Duan SZ, Ivashchenko CY, Russell MW, Milstone DS, Mortensen RM. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circ Res 97: 372– 379, 2005. [DOI] [PubMed] [Google Scholar]
- 15. Duan SZ, Ivashchenko CY, Usher MG, Mortensen RM. PPAR-gamma in the cardiovascular system. PPAR Res 2008: 745804, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dyck JR, Maddaford TG, Pierce GN, Fliegel L. Induction of expression of the sodium-hydrogen exchanger in rat myocardium. Cardiovasc Res 29: 203– 208, 1995. [PubMed] [Google Scholar]
- 17. El-Tanani M, Platt-Higgins A, Rudland PS, Campbell FC. Ets gene PEA3 cooperates with beta-catenin-Lef-1 and c-Jun in regulation of osteopontin transcription. J Biol Chem 279: 20794– 20806, 2004. [DOI] [PubMed] [Google Scholar]
- 18. Engelhardt S, Hein L, Keller U, Klambt K, Lohse MJ. Inhibition of Na+-H+ exchange prevents hypertrophy, fibrosis, and heart failure in beta1-adrenergic receptor transgenic mice. Circ Res 90: 814– 819, 2002. [DOI] [PubMed] [Google Scholar]
- 19. Ennis IL, Escudero EM, Console GM, Camihort G, Dumm CG, Seidler RW, Camilion de Hurtado MC, Cingolani HE. Regression of isoproterenol-induced cardiac hypertrophy by Na+/H+ exchanger inhibition. Hypertension 41: 1324– 1329, 2003. [DOI] [PubMed] [Google Scholar]
- 20. Ennis IL, Garciarena CD, Escudero EM, Perez NG, Dulce RA, Camilion de Hurtado MC, Cingolani HE. Normalization of the calcineurin pathway underlies the regression of hypertensive hypertrophy induced by Na+/H+ exchanger-1 (NHE-1) inhibition. Can J Physiol Pharmacol 85: 301– 310, 2007. [DOI] [PubMed] [Google Scholar]
- 21. Fliegel L. Regulation of the Na+/H+ exchanger in the healthy and diseased myocardium. Expert Opin Ther Targets 13: 55– 68, 2009. [DOI] [PubMed] [Google Scholar]
- 22. Fliegel L, Karmazyn M. The cardiac Na-H exchanger: a key downstream mediator for the cellular hypertrophic effects of paracrine, autocrine and hormonal factors. Biochem Cell Biol 82: 626– 635, 2004. [DOI] [PubMed] [Google Scholar]
- 23. Frey N, Katus HA, Olson EN, Hill JA. Hypertrophy of the heart: a new therapeutic target? Circulation 109: 1580– 1589, 2004. [DOI] [PubMed] [Google Scholar]
- 24. Gan XT, Chakrabarti S, Karmazyn M. Modulation of Na+/H+ exchange isoform 1 mRNA expression in isolated rat hearts. Am J Physiol Heart Circ Physiol 277: H993– H998, 1999. [DOI] [PubMed] [Google Scholar]
- 25. Gao C, Guo H, Downey L, Marroquin C, Wei J, Kuo PC. Osteopontin-dependent CD44v6 expression and cell adhesion in HepG2 cells. Carcinogenesis 24: 1871– 1878, 2003. [DOI] [PubMed] [Google Scholar]
- 26. Henderson NC, Mackinnon AC, Farnworth SL, Poirier F, Russo FP, Iredale JP, Haslett C, Simpson KJ, Sethi T. Galectin-3 regulates myofibroblast activation and hepatic fibrosis. Proc Natl Acad Sci USA 103: 5060– 5065, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu DD, Lin EC, Kovach NL, Hoyer JR, Smith JW. A biochemical characterization of the binding of osteopontin to integrins alphavbeta1 and alphavbeta5. J Biol Chem 270: 26232– 26238, 1995. [DOI] [PubMed] [Google Scholar]
- 28. Hu WY, Fukuda N, Satoh C, Jian T, Kubo A, Nakayama M, Kishioka H, Kanmatsuse K. Phenotypic modulation by fibronectin enhances the angiotensin II-generating system in cultured vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 20: 1500– 1505, 2000. [DOI] [PubMed] [Google Scholar]
- 29. Imahashi K, Mraiche F, Steenbergen C, Murphy E, Fliegel L. Overexpression of the Na+/H+ exchanger and ischemia-reperfusion injury in the myocardium. Am J Physiol Heart Circ Physiol 292: H2237– H2247, 2007. [DOI] [PubMed] [Google Scholar]
- 30. Jandeleit-Dahm K, Hannan KM, Farrelly CA, Allen TJ, Rumble JR, Gilbert RE, Cooper ME, Little PJ. Diabetes-induced vascular hypertrophy is accompanied by activation of Na+-H+ exchange and prevented by Na+-H+ exchange inhibition. Circ Res 87: 1133– 1140, 2000. [DOI] [PubMed] [Google Scholar]
- 31. Karmazyn M, Kilic A, Javadov S. The role of NHE-1 in myocardial hypertrophy and remodelling. J Mol Cell Cardiol 44: 647– 653, 2008. [DOI] [PubMed] [Google Scholar]
- 32. Karmazyn M, Liu Q, Gan XT, Brix BJ, Fliegel L. Aldosterone increases NHE-1 expression and induces NHE-1-dependent hypertrophy in neonatal rat ventricular myocytes. Hypertension 42: 1171– 1176, 2003. [DOI] [PubMed] [Google Scholar]
- 33. Karmazyn M, Sawyer M, Fliegel L. The Na+/H+ exchanger: a target for cardiac therapeutic intervention. Curr Drug Targets Cardiovasc Haematol Disord 5: 323– 335, 2005. [DOI] [PubMed] [Google Scholar]
- 34. Kuhn K, Baker SC, Chudin E, Lieu MH, Oeser S, Bennett H, Rigault P, Barker D, McDaniel TK, Chee MS. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res 14: 2347– 2356, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kusumoto K, Haist JV, Karmazyn M. Na+/H+ exchange inhibition reduces hypertrophy and heart failure after myocardial infarction in rats. Am J Physiol Heart Circ Physiol 280: H738– H745, 2001. [DOI] [PubMed] [Google Scholar]
- 36. Liang F, Wang F, Zhang S, Gardner DG. Peroxisome proliferator activated receptor (PPAR)alpha agonists inhibit hypertrophy of neonatal rat cardiac myocytes. Endocrinology 144: 4187– 4194, 2003. [DOI] [PubMed] [Google Scholar]
- 37. Lin YH, Yang-Yen HF. The osteopontin-CD44 survival signal involves activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. J Biol Chem 276: 46024– 46030, 2001. [DOI] [PubMed] [Google Scholar]
- 38. Liu Y, Watanabe H, Nifuji A, Yamada Y, Olson EN, Noda M. Overexpression of a single helix-loop-helix-type transcription factor, scleraxis, enhances aggrecan gene expression in osteoblastic osteosarcoma ROS17/2.8 cells. J Biol Chem 272: 29880– 29885, 1997. [DOI] [PubMed] [Google Scholar]
- 39. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−DeltaDeltaCT method. Methods 25: 402– 408, 2001. [DOI] [PubMed] [Google Scholar]
- 40. Marano G, Vergari A, Catalano L, Gaudi S, Palazzesi S, Musumeci M, Stati T, Ferrari AU. Na+/H+ exchange inhibition attenuates left ventricular remodeling and preserves systolic function in pressure-overloaded hearts. Br J Pharmacol 141: 526– 532, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsui Y, Jia N, Okamoto H, Kon S, Onozuka H, Akino M, Liu L, Morimoto J, Rittling SR, Denhardt D, Kitabatake A, Uede T. Role of osteopontin in cardiac fibrosis and remodeling in angiotensin II-induced cardiac hypertrophy. Hypertension 43: 1195– 1201, 2004. [DOI] [PubMed] [Google Scholar]
- 42. Matsumoto M, Sakao Y, Akira S. Inducible expression of nuclear factor IL-6 increases endogenous gene expression of macrophage inflammatory protein-1alpha, osteopontin and CD14 in a monocytic leukemia cell line. Int Immunol 10: 1825– 1835, 1998. [DOI] [PubMed] [Google Scholar]
- 43. Meima ME, Mackley JR, Barber DL. Beyond ion translocation: structural functions of the sodium-hydrogen exchanger isoform-1. Curr Opin Nephrol Hypertens 16: 365– 372, 2007. [DOI] [PubMed] [Google Scholar]
- 44. Nakamura TY, Iwata Y, Arai Y, Komamura K, Wakabayashi S. Activation of Na+/H+ exchanger 1 is sufficient to generate Ca2+ signals that induce cardiac hypertrophy and heart failure. Circ Res 103: 891– 899, 2008. [DOI] [PubMed] [Google Scholar]
- 45. Okamoto H. Osteopontin and cardiovascular system. Mol Cell Biochem 300: 1– 7, 2007. [DOI] [PubMed] [Google Scholar]
- 46. Oyama Y, Akuzawa N, Nagai R, Kurabayashi M. PPARgamma ligand inhibits osteopontin gene expression through interference with binding of nuclear factors to A/T-rich sequence in THP-1 cells. Circ Res 90: 348– 355, 2002. [DOI] [PubMed] [Google Scholar]
- 47. Perez NG, Alvarez BV, Camilion de Hurtado MC, Cingolani HE. pHi regulation in myocardium of the spontaneously hypertensive rat. Compensated enhanced activity of the Na+-H+ exchanger. Circ Res 77: 1192– 1200, 1995. [DOI] [PubMed] [Google Scholar]
- 48. Philip S, Bulbule A, Kundu GC. Osteopontin stimulates tumor growth and activation of promatrix metalloproteinase-2 through nuclear factor-kappaB-mediated induction of membrane type 1 matrix metalloproteinase in murine melanoma cells. J Biol Chem 276: 44926– 44935, 2001. [DOI] [PubMed] [Google Scholar]
- 49. Schellings MW, Pinto YM, Heymans S. Matricellular proteins in the heart: possible role during stress and remodeling. Cardiovasc Res 64: 24– 31, 2004. [DOI] [PubMed] [Google Scholar]
- 50. Singh M, Ananthula S, Milhorn DM, Krishnaswamy G, Singh K. Osteopontin: a novel inflammatory mediator of cardiovascular disease. Front Biosci 12: 214– 221, 2007. [DOI] [PubMed] [Google Scholar]
- 51. Singh M, Foster CR, Dalal S, Singh K. Osteopontin: role in extracellular matrix deposition and myocardial remodeling post-MI. J Mol Cell Cardiol 48: 538– 543, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takano H, Komuro I. Peroxisome proliferator-activated receptor gamma and cardiovascular diseases. Circ J 73: 214– 220, 2009. [DOI] [PubMed] [Google Scholar]
- 53. Tanaka H, Wakisaka A, Ogasa H, Kawai S, Liang CT. Effect of IGF-I and PDGF administered in vivo on the expression of osteoblast-related genes in old rats. J Endocrinol 174: 63– 70, 2002. [DOI] [PubMed] [Google Scholar]
- 54. Tuck AB, Hota C, Wilson SM, Chambers AF. Osteopontin-induced migration of human mammary epithelial cells involves activation of EGF receptor and multiple signal transduction pathways. Oncogene 22: 1198– 1205, 2003. [DOI] [PubMed] [Google Scholar]
- 55. Yokoyama H, Gunasegaram S, Harding SE, Avkiran M. Sarcolemmal Na+/H+ exchanger activity and expression in human ventricular myocardium. J Am Coll Cardiol 36: 534– 540, 2000. [DOI] [PubMed] [Google Scholar]
- 56. Yu S, Matsusue K, Kashireddy P, Cao WQ, Yeldandi V, Yeldandi AV, Rao MS, Gonzalez FJ, Reddy JK. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J Biol Chem 278: 498– 505, 2003. [DOI] [PubMed] [Google Scholar]
- 57. Zhou D, Wang J, Zapala MA, Xue J, Schork NJ, Haddad GG. Gene expression in mouse brain following chronic hypoxia: role of sarcospan in glial cell death. Physiol Genomics 32: 370– 379, 2008. [DOI] [PubMed] [Google Scholar]
- 58. Zhou D, Xue J, Gavrialov O, Haddad GG. Na+/H+ exchanger 1 deficiency alters gene expression in mouse brain. Physiol Genomics 18: 331– 339, 2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.