Abstract
Naturally occurring cystic fibrosis (CF)-causing mutations in the CFTR gene have not been identified in any nonhuman animal species. Since domestic dogs are known to develop medical conditions associated with atypical CF in humans (e.g., bronchiectasis and pancreatitis), we hypothesized that dogs with these disorders likely have a higher expression rate of CFTR mutations than the at-large population. Temporal temperature-gradient gel electrophoresis (TTGE) was used to screen canine CFTR in 400 animals: 203 dogs diagnosed with pancreatitis, 23 dogs diagnosed with bronchiectasis, and 174 dogs admitted to clinics for any illness (at-large dogs). Twenty-eight dogs were identified with one of four CFTR missense mutations. P1281T and P1464H mutations occur in relatively unconserved residues. R1456W is analogous to the human R1453W mutation, which has ∼20% of normal CFTR function and is associated with pancreatitis and panbronchiolitis. R812W disrupts a highly conserved protein kinase A recognition site within the regulatory domain. We conclude that naturally occurring CFTR mutations are relatively common in domestic dogs and can be detected with TTGE. No substantive differences in mutation frequency were observed between the at-large, pancreatitis, and bronchiectasis dogs.
Keywords: cystic fibrosis, missense mutation, temporal temperature-gradient gel electrophoresis, pancreatitis, bronchiectasis
cystic fibrosis (CF) is caused by mutations in the gene that codes for the cystic fibrosis transmembrane conductance regulator (CFTR) (20), which functions as a cAMP-activated anion channel (2). The clinical features of CF in its most severe form include pancreatic insufficiency, lung disease, male infertility, intestinal obstruction, and elevated salt concentration in sweat. Although multiple organ systems are affected in CF, lung disease is the leading cause of mortality (16). CF lung disease is typified by production of abnormally thick airway mucus, chronic airway infections, reductions in airway clearance, and development of bronchiectasis. Despite intense research efforts over the last several decades, the etiology of CF lung disease remains incompletely understood.
CF is inherited as a Mendelian recessive trait. Full manifestation of CF disease requires essentially complete loss of CFTR anion channel function. Some CF patients, however, are pancreatitic sufficient (PS) and express somewhat less severe lung disease (25). These PS CF patients have mutation combinations that apparently allow slightly more CFTR channel function than patients with severe CF disease. Clinical confirmation of CF in patients relies heavily on physiological tests, such as measurements of nasal potential difference and sweat chloride concentrations (16). Unfortunately, these tests often do not detect mild to moderate CFTR-related disease presumably due to the presence of less severe mutations or disease modifiers (22). Indeed, it has become apparent that persons expressing CFTR mutations whose net channel function is above the detectable threshold for these physiological tests comprise significant fractions of individuals diagnosed with diseases that had not been previously considered to be related to CF, such as idiopathic bronchiectasis (17), idiopathic pancreatitis (15), and congenital bilateral absence of the vas deferens (CBAVD) in men (5). Many of these patients have two identifiable CFTR mutations, often consisting of a severe and a mild mutation.
To date, CF-causing CFTR mutations have never been identified in a nonhuman species. Both sheep and monkeys have been the subjects of large-scale screening efforts to identify severe CFTR mutation carriers for the purpose of breeding a nonhuman animal model of CF (11, 23, 26). Unfortunately, neither of these groups reported finding a disease-causing CFTR mutation. We recently considered that domestic dogs might carry CF-causing CFTR mutations because these animals are known to have diseases that in humans harbor relatively high frequencies of CFTR mutations: bronchiectasis (9), pancreatic insufficiency (14), pancreatitis (24), and atresia of the vas deferens (6). Because dogs are popular pets and commonly receive medical treatment, we reasoned that animals afflicted with these various disorders could be identified through veterinary hospitals and screened for CFTR mutations. This information could be valuable in the treatment of these canine diseases as well as identifying animals that could be bred to produce a large-animal model of CF disease.
MATERIALS AND METHODS
Blood sample collection.
Whole blood samples from dogs (∼0.5–3.0 ml) were collected from multiple sources. Most blood samples were obtained as discarded clinical specimens from veterinary clinics and laboratories that were initially collected for diagnostic purposes. Some blood samples were obtained by venipuncture specifically for our study from dogs at North Carolina State University Veterinary Teaching Hospital (NCSU-VTH). This procedure was approved by the NCSU Institutional Animal Care and Use Committee. Blood from dogs that carry the pancreatic acinar atrophy (PAA) trait, which is associated with exocrine pancreatic insufficiency (EPI), were provided by the animals' primary care veterinarian in accordance with a protocol approved by the Clinical Research Review Committee of the College of Veterinary Medicine of Texas A&M University.
Four groups of dogs were recognized based on their diagnosis: pancreatitis, PAA, bronchiectasis, and at-large animals. The diagnosis of pancreatitis was based on one or more of the following criteria: appropriate historic and physical findings, elevations in one or more serum markers of pancreatitis (pancreatic lipase immunoreactivity or lipase or amylase activities), and characteristic radiographic or ultrasonographic abnormalities. PAA is a degenerative recessive genetic disease of the exocrine pancreas that causes EPI in the homozygous state (14). Clinical EPI diagnosis was made in an animal with a family history of PAA and serum canine trypsinlike immunoreactivity (cTLI) levels ≤2.5 μg/l. An asymptomatic dog that was heterozygous for the PAA trait was also included in this group. Bronchiectasis diagnosis, which included cylindrical or saccular forms of the disease, was based on radiographic evidence of airways that do not taper normally. “At-large” animals were dogs that were admitted to clinics without regard to diagnosis. We were unable to locate dogs diagnosed with atresia of the vas deferens.
For most of the DNA samples obtained, breed information was unavailable. Therefore, it was not possible to determine breed predisposition for the mutations.
DNA isolation and mutation detection.
Genomic DNA was purified from peripheral blood lymphocytes by standard isolation procedures using the Puregene Blood Kit (Gentra Systems, Minneapolis, MN). Typical DNA yields per sample were between 45 and 70 μg. Genomic DNA served as a template for the amplification of the entire coding region of the CFTR gene, which was amplified as 27 separate PCR products that included exons and splice junctions. Primers were designed from the published genomic sequence (Canis familiaris target 1 genomic scaffold, accession no. DP000236) and purchased from Integrated DNA Technologies (Coralville, IA). Sequences of PCR primers that were used to amplify CFTR exons are shown in Table 1. Each PCR product spanned the exon and its splice junctions, except for exon 14. Exon 14 (727 bp), as the largest exon, was amplified as two smaller overlapping fragments (520 and 597 bp) to increase the sensitivity of mutation detection. The PCR products, which ranged in length from 219 to 1,136 bp, were amplified with Promega GoTaq PCR MasterMix. The PCR conditions for most amplicons included an initial 96°C denaturation for 2 min, followed by 10 cycles (95°C for 30 s, 50°C for 30 s, and 72°C for 40 s) and then by another 25 cycles of amplification (96°C for 30 s, 50°C for 30 s, and 72°C for 40 s plus 5 s/cycle). These conditions were modified for some exons to improve separation and resolution of the bands. For exon 1, the denaturation step was lengthened to 1 min during the first 10 cycles, and the annealing temperature was increased to 53°C. For exons 8 and 13, the annealing temperature was increased to 58°C. The largest amplicon, 1,136 bp spanning exons 16 and 17, required an annealing temperature of 52°C and an initial extension time of 75 s. After amplification, all PCR reactions were denatured at 95°C, slowly cooled to 40°C at a rate of 1°C/min, and then held at 4°C.
Table 1.
PCR primer sequences for canine CFTR mutation analysis
| Exon | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| 1 | GAAGGAGCGAAAGCAAAG | AAAACCCAGCAACTACACAC |
| 2 | TCTAGTATTTATGCACCCACC | TAAGCCATTAATAAGCTAGTTCTCTATCCT |
| 3 | CACTTGGGTTAATCTCCTTG | TGTATTTGGAGTTGAATTAGTCC |
| 4 | AAGCCCATCTTAAATCCTTC | TGAACTGTATTGATTACCAAGG |
| 5 | GCTGTGGTACTATGTTACTTATTCC | TAACTGACCCAGGAAAACC |
| 6 | ATTTTATGACCTTGCTGTGC | GACACCCAGTACGTAGATCC |
| 7 | TGAAGTGTCCTACACTGGAC | AAATATTAAACAGGGACACCC |
| 8 | GTTTTACATTCCAAGATGCC | CAAGACCAACACTTCATCTCC |
| 9 | GATAAATCCTAGTGCTTGGC | GAAAACCAGATTCAACAAGG |
| 10 | TGTACTCTAGAACCCACATGC | ATATCCAAGCGTTCTCCTAC |
| 11 | GGATGATAGTTGGAGGCG | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGCTGACTGTTGAATAGGAGGCTA |
| 12 | ACGGTGTGACCTACGATTAC | TTAAAGGTGATTCTCAACCC |
| 13 | CTAAGGCAGGCGCTAAAC | GGTAAAATGCAGCCTATGAG |
| 14a | TGCAATATTTCAAGAAGACACC | TTTCACCAAACTCTCCAGTCTGT |
| 14b | TATCATGCTACGTATTGCCTTTCA | TATGGACTCTGGACCTTCCG |
| 15 | CCCACATTCAACATAAAACC | TCTGCTTCTCCCTCTGCCTTTCTC |
| 16,17 | CTGCAATCCTAGCTCTTCTGTG | TCATTACGAATCTTCAACGGG |
| 18 | ACACGAGCGTTTTCTAAGTC | CAACTCCCAAAATGAAAAATAAGC |
| 19 | TCTTTGGAGCTACACCTCTG | TTGGGGGGAATGGGAAGAGTAG |
| 20 | TTCAGGTCCCTGGTAGTAAG | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGTTGCAATTGAATGGTCAGTCAC |
| 21 | CTTGAATAGTTCATACGCCC | GAGAGACACAGTAGCCTTCG |
| 22 | TTTTGTAAGGAAACATCACAC | CTGTAATAATAATAATGCTACCAATGGAC |
| 23 | AACGCCAGATTACATTTGTG | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGAGTTCAATCACTGGATGGAG |
| 24 | ATACGAAGGTGTTTAGGTTATTC | ATCTCGGTTATGACAACCAC |
| 25 | CTTCTCGAGTGTCTTTCACC | TTATCCCAGGAAATAAGCAC |
| 26 | CATGGAAACCTAGAATCTGC | ACTGTTGTGTTGTGATTTGC |
| 27 | TAGAGCCCCTTTCACACAC | CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGACGTCCTCCTCCACCTTGATG |
CFTR, cystic fibrosis transmembrane conductance regulator.
Bold type indicates GC clamps that have been added to the primers to facilitate temporal temperature-gradient gel electrophoreses (TTGE) analysis.
To detect mutations, temporal temperature-gradient gel electrophoresis (TTGE) analysis was performed with the DCode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA). TTGE has been shown to be a sensitive screening tool for detecting novel mutations in the human CFTR gene (27). This technique can detect single base substitutions, insertions, and deletions. With heterozygous mutations, four bands are typically resolved from reannealed PCR products, which form homoduplex and heteroduplex pairs. Homozygous mutations can also be detected by TTGE if the mutation alters the melting properties of the PCR product.
The PCR products were separated on 6–8.5% polyacrylamide gels with a ratio of 37.5:1 acrylamide to bisacrylamide that contained at least 6 M urea. In addition to urea, gels for exons 5, 25, 26, and 27 contained 5% formamide to enhance the resolution of homoduplex and heteroduplex bands. The gels were run at 140 V in 1.25× TAE buffer [in mM: 50 Tris base, 25 acetic acid, 1.25 EDTA (pH 8.0)] for up to 9 h with temperature increasing between 0.9 and 1.5°C/h. The TTGE conditions for each PCR product were optimized empirically. For some reverse primers, a GC clamp of ∼40 bases was added to the 5′ ends to raise the melting temperature of the PCR product and increase the resolution of the homoduplex and heteroduplex bands. TTGE conditions for each PCR product are shown in Table 2. After electrophoresis, the gels were stained with ethidium bromide (2 μg/ml) for 5 min and destained in distilled water for at least 30 min before the gels were imaged. The gels were imaged with a Fluor-S MultiImager (Bio-Rad Laboratories) and Quantity One software (Bio-Rad Laboratories). When a gel banding pattern differed from the reference, the PCR product was purified with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI) and sequenced (Eurofins MWG Operon, Huntsville, AL). To aid in the identification of mutations, the sequences of the PCR products were aligned with ClustalX 2.0.5 to the corresponding regions of the canine CFTR gene. To confirm the presence of heterozygous mutations, the individual sequence chromatograms were scanned with ChromasPro version 1.42.
Table 2.
TTGE conditions for CFTR mutation detection in CFTR exons
| Exon | Exon Length, bp | Amplicon, bp | Temp Range, °C | Ramp Rate, °C/h | Run Time, h | Gel, % | Urea, M |
|---|---|---|---|---|---|---|---|
| 1 | 53 | 395 | 55–65 | 1.5 | 7 | 7 | 8 |
| 2 | 111 | 355 | 45–54 | 1.3 | 8 | 7 | 6 |
| 3 | 109 | 367 | 44–50 | 1.0 | 8 | 7 | 7 |
| 4 | 216 | 466 | 45–56 | 1.4 | 8 | 6 | 7 |
| 5 | 90 | 243 | 44–52 | 1.2 | 7 | 8 | 7* |
| 6 | 164 | 369 | 48–58 | 1.3 | 8 | 7 | 7 |
| 7 | 126 | 601 | 45–52 | 0.9 | 8 | 6 | 7 |
| 8 | 247 | 415 | 47–56 | 1.0 | 8 | 7 | 7 |
| 9 | 93 | 357 | 43–52 | 1.0 | 7 | 6 | 7 |
| 10 | 183 | 459 | 42–53 | 1.4 | 8 | 7 | 7 |
| 11 | 192 | 399 | 48–56 | 1.0 | 8 | 7 | 7 |
| 12 | 95 | 340 | 48–55 | 1.0 | 7 | 7 | 7 |
| 13 | 87 | 256 | 48–53 | 1.0 | 6 | 8 | 6 |
| 14 | 727 | 520 | 45–53 | 1.0 | 8 | 6 | 6 |
| 14 | 727 | 597 | 48–57 | 1.2 | 8 | 6.5 | 6 |
| 15 | 129 | 339 | 48–54 | 0.9 | 7 | 8 | 6 |
| 16 & | 38 | 1136 | 43–56 | 1.5 | 9 | 6 | 6 |
| 17 | 251 | ||||||
| 18 | 80 | 346 | 43–50 | 0.9 | 8 | 7.5 | 6 |
| 19 | 151 | 338 | 48–56 | 1.0 | 8 | 8 | 8 |
| 20 | 228 | 338 | 49–55 | 0.9 | 7 | 8 | 6 |
| 21 | 101 | 341 | 48–57 | 1.3 | 7 | 7 | 7 |
| 22 | 255 | 538 | 45–54 | 1.3 | 7 | 7 | 6 |
| 23 | 156 | 489 | 44–51 | 1.0 | 8 | 6 | 7 |
| 24 | 90 | 219 | 45–51 | 1.0 | 6 | 8.5 | 7 |
| 25 | 173 | 443 | 47–54 | 1.0 | 7 | 7 | 6* |
| 26 | 106 | 399 | 47–54 | 1.0 | 8 | 7 | 7* |
| 27 | 201 | 320 | 54–62 | 1.0 | 8 | 8 | 8* |
Gels contained 5% formamide.
RESULTS
In the intronic regions, point mutations were common and a few small deletions and insertions were found as well. Far fewer mutations were identified in the CFTR exons. Sequence variants, which did not cause a change in amino acid sequence, were also detected throughout the gene. Of the total of 400 screened dogs, 28 dogs carried one of four missense mutations. None of the animals was found to possess two missense mutations.
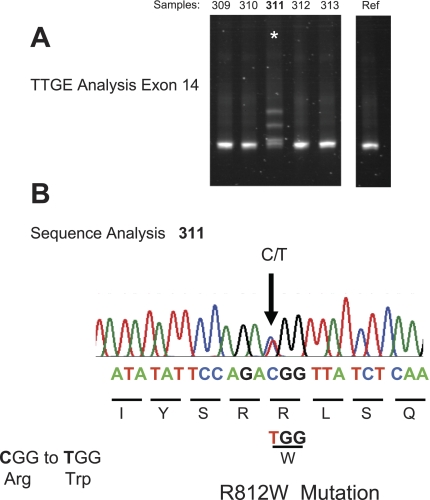
Figure 1A demonstrates TTGE detection of a mutation in exon 14. The four band pattern (2 heteroduplex and 2 homoduplex bands) that typified heterozygotes is apparent. Samples from four other dogs including the reference animal do not show this pattern. When the lane 311 PCR product was sequenced, a single base substitution was identified (Fig. 1B) indicating that two bases, C and T, occur at the same position. The presence of a T (thymidine) at this position changes the amino acid at this codon from arginine to tryptophan, thus identifying this mutation as an R812W missense mutation. Three other missense mutations were detected in the screened population. P1281T was present in exon 23, and two mutations, R1453W and P1464H, were present in exon 27. No frameshift, insertion, or deletion mutations were detected in any of the animals.
Fig. 1.
Temporal temperature-gradient gel electrophoresis (TTGE) detection of R812W missense mutation. A: image of TTGE gel showing 4-band pattern characteristic of heterozygous mutation in lane 311. B: sequence analysis identifying the mutation as a missense R812W mutation.
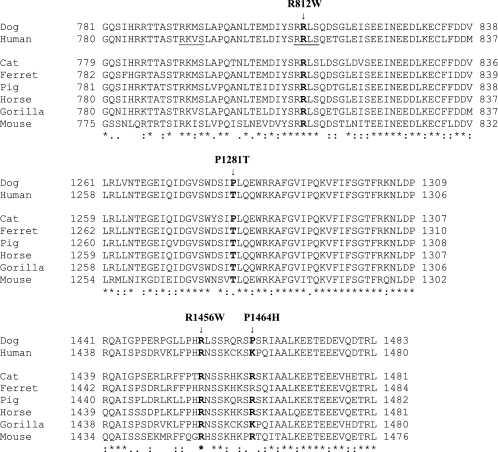
To determine whether the missense CFTR mutations occur at conserved positions of the protein, the amino acid sequences from several species were obtained from the National Center for Biotechnology Information and aligned with ClustalX 2.0.5 (Fig. 2). Both R812W and R1456W mutations occur at fully conserved sites. P1281T occurs at a weakly conserved site, whereas P1464H occurs at an unconserved site.
Fig. 2.
Locations of canine missense mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) amino acid sequence: comparisons with sequences in other species. Positions of the 4 canine CFTR mutations in the amino acid sequence are shown with arrows above the amino acid alignment. The positions of the mutated canine residues and the corresponding positions in the other species are indicated in bold. The underlined human sequences are protein kinase A phosphorylation recognition sites. Fully conserved residues are designated by asterisks. Conservation of similar residues based on Gonnet PAM250 matrix similarity scores are indicated by colons (strong similarity) and dots (weak similarity). The multiple sequence alignment was generated with Clustal X version 2.0.5. The aligned protein sequences, identified by species and accession number, include cat (Felis catus, NP_001041474), dog (Canis familiaris, AAV40962), ferret (Mustela putorius furo, ABI93660), pig (Sus scrofa, Q6PQZ2), gorilla (Gorilla gorilla gorilla, Q2IBF6), horse (Equus caballus, Q2QLA3), human (Homo sapiens, NP_000483), and mouse (Mus musculus, AAA37417).
Table 3 shows the distribution of the mutations by group. All four mutations were found in the pancreatitis animals, which constituted the largest group (n = 203). The at-large animals were the next largest group (n = 174), which expressed all but the P1281T mutation. Only two P1464H mutations were found in the bronchiectasis animals, which comprised the smallest group (n = 23). The rank order of the groups for heterozygote mutation frequency was bronchiectasis > pancreatitis > at-large animals (Table 4). The differences between these three groups of dogs are small and do not support the hypothesis that dogs with pancreatitis and bronchiectasis express higher than normal frequencies of CFTR mutations. No mutation homozygotes or compound heterozygotes were identified in this study. However, from the observed carrier frequencies for the four missense mutations, we estimate the probabilities of a dog carrying any two of these CFTR mutations (i.e., mutation homozygotes or compound heterozygotes) as being 1:1,001 for at-large animals, 1:733 for pancreatitis animals, and 1:529 for bronchiectasis animals. These calculations emphasize that small differences in mutation carrier frequencies, as seen in the present study, can have large effects on mutation homozygote or compound heterozygote frequencies.
Table 3.
Mutation detection by category
| Category |
|||
|---|---|---|---|
| Mutation | At large (n = 174) | Pancreatitis (n = 203) | Bronchiectasis (n = 23) |
| R812W (exon 14) | 1 | 1 | |
| c.2434C>T | |||
| Arg → Trp | |||
| P1281T (exon 23) | 1 | ||
| c.3841C>A | |||
| Pro → Thr | |||
| R1456W(exon 27) | 7 | 7 | |
| c.4366C>T | |||
| Arg → Trp | |||
| P1464H (exon 27) | 3 | 6 | 2 |
| c.4391C>A | |||
| Pro → His |
Table 4.
CFTR mutation frequencies in canine populations
| At-Large | Pancreatitis | Bronchiectasis | |
|---|---|---|---|
| Mutation carrier:total animals | 1:15.8 | 1:13.5 | 1:12 |
No CFTR mutations were detected in the DNA screened from dogs heterozygous and homozygous for the PAA trait. We conclude that this disorder is not caused by inheritance of CFTR mutations.
DISCUSSION
These results confirm that CFTR mutations are present in the canine genome and that it is feasible to screen large numbers of dogs for these mutations. With the use of TTGE as a screening tool, 28 dogs were identified with one of four missense CFTR mutations out of a total of 400 screened animals. Pancreatitis animals and bronchiectasis animals expressed carrier mutation frequencies that were similar to those in the at-large dogs. Thus we found no substantive support for the hypothesis that dogs with these disorders express higher CFTR mutation frequencies than at-large dogs.
It is very difficult to accurately predict the severity of a single amino acid change in the channel function of the CFTR. However, some insights can be gained about the possible impact of these canine mutations from the human CFTR mutation literature. One of the canine mutations that was discovered, R1456W, has a human analog, R1453W, that is associated with disease. The different reference number arises from canine CFTR being three amino acids longer than human CFTR. The R1453 residue is highly conserved among mammalian species. One human patient that was homozygous for the R1453W mutation was diagnosed with pancreatitis (8). Another patient, who was compound heterozygous for the R1453W and 5T splice variant of the polyT region of CFTR, was diagnosed with panbronchiolitis (7). Panbronchiolitis is a progressive lung disease that typically develops in middle age and expresses characteristics similar to mild CF, such as mucus occlusion of bronchioles and colonization with Pseudomonas aeruginosa (18). The human R1453W CFTR mutant has a normal unitary conductance, but the open probability (Po) of the channel is only ∼20% of normal (12).
The R812W mutation has no human analog, but it is located in the regulatory domain of CFTR within a protein kinase A recognition site that is highly conserved among mammalian species (4). In the human protein, S813 is one of four functionally relevant phosphorylation sites (700, 737, 768, and 813) among the nine dibasic protein kinase A sites in the R domain (21). The loss of a single phosphorylation site in the regulatory domain would not be expected to fully disable the channel (10, 21). Nonetheless, a human patient who expressed the S813P mutation with the D110E mutation was reported to have mild CF and the R810G mutation and severe loss of function mutation F508del were present in a patient with CBAVD (7).
Phenotypes for the other two missense mutations, P1281T and P1464H, are less certain since human mutations have not been documented at or near these positions. The P1281T mutation occurs at a weakly conserved residue, between the Walker A box and the Q loop, in the second nucleotide binding domain (NBD2). It is not obvious from its position whether this mutation will have any effect on ATP binding or channel gating. The P1464H mutation is found at a weakly conserved position within the carboxy terminal region. In addition to the PDZ binding motif, other sequences have been mapped to the carboxy terminal region that contribute to apical localization of CFTR (13); however, neither R1456W nor P1464H falls within these regions. These mutations do not disrupt three endocytic motifs identified in the carboxy terminal region (1).
We also screened two German shepherd dogs that were heterozygous and homozygous for the PAA trait, a heritable form of EPI. In humans the major causes of EPI are CF and chronic pancreatitis (19). Therefore, despite some differences in the pattern of pancreatic pathology (e.g., CF produces a ductal pattern of atropy), we considered that this condition could represent canine CF, as previously speculated by Bishop et al. (3). However, no deleterious CFTR mutations were detected in either dog. We conclude that CFTR mutations do not play any role in the development of this genetic disorder.
We believe that our initial results demonstrate the utility of this approach for identifying CFTR mutations in dogs. To date, we have not identified unambiguously severe mutations, i.e., frameshifts, insertions, or deletions that would generate premature stop codons and protein truncation. If CFTR mutations exist in the canine genome in approximately the same frequency as the Japanese population, where CF occurs in only ∼1 in 350,000 individuals (28), we expect that a CF-causing mutation would be present in ∼1 in 300 dogs. Our screening total to date is 400 dogs, which is slightly higher than this threshold. We expect that CF-causing mutations are present in the canine population and speculate that a continued screening effort will successfully identify the animals that carry these mutations.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL-063302 and Cystic Fibrosis Foundation Grant BALLAR07G0.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
The authors acknowledge the excellent technical assistance of Julie M. Bradley. The authors also acknowledge the assistance of Dr. Wesley Denny, who provided helpful advice regarding the use of the TTGE assay for mutation detection.
Present address for K. E. Murphy and L. A. Clark: Dept. of Genetics and Biochemistry, College of Agriculture, Forestry, and Life Sciences, Clemson University, Clemson, SC 29634-0318.
REFERENCES
- 1.Ameen N, Silvis M, Bradbury NA. Endocytic trafficking of CFTR in health and disease. J Cyst Fibros 6: 1–14, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MP, Gregory RJ, Thompson S, Sousa DW, Paul S, Mulligan RC, Smith AE, Welsh MJ. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253: 202–205, 1991 [DOI] [PubMed] [Google Scholar]
- 3.Bishop MA, Steiner JM, Moore LE, Williams DA. Evaluation of the cationic trypsinogen gene for potential mutations in miniature schnauzers with pancreatitis. Can J Vet Res 68: 315–318, 2004 [PMC free article] [PubMed] [Google Scholar]
- 4.Chen TJ, Boles RG, Wong LJC. Detection of mitochondrial DNA mutations by temporal temperature gradient gel electrophoresis. Clin Chem 45: 1162–1167, 1999 [PubMed] [Google Scholar]
- 5.Chillon M, Casals T, Mercier B, Bassas L, Lissens W, Silber S, Romey MC, Ruiz-Romero J, Verlingue C, Claustres M, Nunes V, Ferec C, Estivill X. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N Engl J Med 332: 1475–1480, 1995 [DOI] [PubMed] [Google Scholar]
- 6.Copland MD, MacLachlan NJ. Aplasia of the epididymis and vas deferens in the dog. J Small Anim Pract 17: 443–449, 1976 [DOI] [PubMed] [Google Scholar]
- 7.Cystic Fibrosis Genetic Analysis Consortium Cystic Fibrosis Mutation Database (CFMDB). Updated March 2, 2007 ( http://www.genet.sickkids.on.ca/cftr/) [Accessed Jan 22, 2010].
- 8.Fujiki K, Ishiguro H, Ko SBH, Mizuno N, Suzuki Y, Takemura T, Yamamoto A, Yoshikawa T, Kitagawa M, Hayakawa T, Sakai Y, Takayama T, Saito M, Kondo T, Naruse S. Genetic evidence for CFTR dysfunction in Japanese: background for chronic pancreatitis. J Med Genet 41: e55, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins EC, Basseches J, Berry CR, Stebbins ME, Ferris KK. Demographic, clinical, and radiographic features of bronchiectasis in dogs. J Am Vet Med Assoc 223: 1628–1635, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Hegedus T, Aleksandrov A, Mengos A, Cui L, Jensen TJ, Riordan JR. Role of individual R domain phosphorylation sites in CFTR regulation by protein kinase A. Biochim Biophys Acta 1788: 1341–1349, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Kuo E, Hurlock G, Potocnik U, Ravnik-Glavac M, Robinson C, Dean M, Glavac D, Wine JJ. Genomic sequences of CFTR in three species of nonhuman primates (abstract). Pediatr Pulmonol Suppl 19: 210, 1999 [Google Scholar]
- 12.Lee JH, Choi JH, Namkung W, Hanrahan JW, Chang J, Song SY, Park SW, Kim DS, Yoon JH, Suh Y, Jang IN, Nam JH, Kim SJ, Cho MO, Lee JE, Kim KH, Lee MG. A haplotype-based molecular analysis of CFTR mutations associated with respiratory and pancreatic diseases. Hum Mol Genet 12: 2321–2332, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Milewski MI, Mickle JE, Forrest JK, Stafford DM, Moyer BD, Cheng J, Guggino WB, Stanton BA, Cutting GR. A PDZ-binding motif is essential but not sufficient to localize the C terminus of CFTR to the apical membrane. J Cell Sci 114: 719–726, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Moeller EM, Steiner JM, Clark LA, Murphy KE, Famula TR, Williams DA, Stankovics ME, Vose AS. Inheritance of pancreatic acinar atrophy in German shepherd dogs. Am J Vet Res 63: 1429–1434, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Noone PG, Zhou Z, Silverman LM, Jowell PS, Knowles MR, Cohn JA. Cystic fibrosis gene mutations and pancreatitis risk: relation to epithelial ion transport and trypsin inhibitor gene mutations. Gastroenterology 121: 1310–1319, 2001 [DOI] [PubMed] [Google Scholar]
- 16.O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet 373: 1891–1904, 2009 [DOI] [PubMed] [Google Scholar]
- 17.Pignatti PF, Bombieri C, Marigo C, Benetazzo M, Luisetti M. Increased incidence of cystic fibrosis gene mutations in adults with disseminated bronchiectasis. Hum Mol Genet 4: 635–639, 1995 [DOI] [PubMed] [Google Scholar]
- 18.Poletti V, Casoni G, Chilosi M, Zompatori M. Diffuse panbronchiolitis. Eur Respir J 28: 862–871, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Pongprasobchai S, DiMagno EP. Treatment of pancreatic insufficiency. In: Pancreatitis and Its Complications, edited by Forsmark CE, Totowa, NJ: Humana, 2005, chapt. 16, p. 295–312 [Google Scholar]
- 20.Riordan JR, Rommens JM, Kerem BS, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui LC. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245: 1066–1073, 1989 [DOI] [PubMed] [Google Scholar]
- 21.Seibert FS, Chang XB, Aleksandrov AA, Clarke DM, Hanrahan JW, Riordan JR. Influence of phosphorylation by protein kinase A on CFTR at the cell surface and endoplasmic reticulum. Biochim Biophys Acta 1461: 275–283, 1999 [DOI] [PubMed] [Google Scholar]
- 22.Taylor CJ, Hardcastle J, Southern KW. Physiological measurements confirming the diagnosis of cystic fibrosis: the sweat test and measurements of transepithelial potential difference. Paediatr Respir Rev 10: 220–226, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Tebbutt SJ, Lakeman MB, Wilson-Wheeler JC, Hill DF. Genetic variation within the ovine cystic fibrosis transmembrane conductance regulator gene. Mutat Res 382: 93–98, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Watson PJ, Roulois AJA, Scase T, Johnston PEJ, Thompson H, Herrtage ME. Prevalence and breed distribution of chronic pancreatitis at post-mortem examination in first-opinion dogs. J Small Anim Pract 48: 609–618, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Wilschanski M, Durie PR. Patterns of GI disease in adulthood associated with mutations in the CFTR gene. Gut 56: 1153–1163, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wine JJ, Dean M, Glavac D. Natural animal models of human genetic diseases. Methods Mol Med 70: 31–46, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Wong LJC, Alper OM. Detection of CFTR mutations using temporal temperature gradient gel electrophoresis. Electrophoresis 25: 2593–601, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Yamashiro Y, Shimizu T, Oguchi S, Shioya T, Nagata S, Ohtsuka Y. The estimated incidence of cystic fibrosis in Japan. J Pediatr Gastroenterol Nutr 24: 544–547, 1997 [DOI] [PubMed] [Google Scholar]