Abstract
The majority of BRCA1-associated breast cancers are basal cell–like, which is associated with a poor outcome. Using a spontaneous mouse mammary tumor model, we show that platinum compounds, which generate DNA breaks during the repair process, are more effective than doxorubicin in Brca1/p53–mutated tumors. At 0.5 mg/kg of daily cisplatin treatment, 80% primary tumors (n = 8) show complete pathologic response. At greater dosages, 100% show complete response (n = 19). However, after 2 to 3 months of complete remission following platinum treatment, tumors relapse and become refractory to successive rounds of treatment. Approximately 3.8% to 8.0% (mean, 5.9%) of tumor cells express the normal mammary stem cell markers, CD29hi24med, and these cells are tumorigenic, whereas CD29med24–/lo and CD29med24hi cells have diminished tumorigenicity or are nontumorigenic, respectively. In partially platinum-responsive primary transplants, 6.6% to 11.0% (mean, 8.8%) tumor cells are CD29hi24med; these populations significantly increase to 16.5% to 29.2% (mean, 22.8%; P < 0.05) in platinum-refractory secondary tumor transplants. Further, refractory tumor cells have greater colony-forming ability than the primary transplant–derived cells in the presence of cisplatin. Expression of a normal stem cell marker, Nanog, is decreased in the CD29hi24med populations in the secondary transplants. Top2A expression is also down-regulated in secondary drug-resistant tumor populations and, in one case, was accompanied by genomic deletion of Top2A. These studies identify distinct cancer cell populations for therapeutic targeting in breast cancer and implicate clonal evolution and expansion of cancer stem-like cells as a potential cause of chemoresistance.
Introduction
Mutations in breast cancer susceptibility genes BRCA1 and BRCA2 and several other genes encoding BRCA1/2-interacting proteins are associated with hereditary breast and ovarian cancer syndrome (reviewed in refs. 1, 2). Hereditary breast cancer accounts for 5% to 10% of all breast cancers, and of these, BRCA1 mutations account for approximately half of the cases. In sporadic breast cancer, BRCA1 mutations are rare; however, significant percentages of sporadic cancers show reduced or absent expression of BRCA1 due to promoter hypermethylation (3). BRCA1 encodes a 220-kDa nuclear phosphoprotein that contains multiple functional domains that interact with proteins involved in different cellular processes, including ubiquitously expressed tumor suppressors, oncoproteins, DNA damage repair proteins, cell cycle regulators, and transcriptional activators and repressors (reviewed in refs. 4, 5).
In addition to interacting with ubiquitously expressed proteins, BRCA1 associates and regulates ubiquitination of steroid hormone receptors, estrogen receptor a and progesterone receptor (reviewed in ref. 6). Paradoxically, BRCA1-associated breast cancers are frequently high-grade, estrogen receptor α/progesterone receptor/Her2 (triple)–negative basal cell–like tumors (7), with frequent mutations of TP53 and PTEN (8). The triple-negative tumors are initially responsive to chemotherapy with a high percentage entering pathologic complete response; however, tumors that recur or do not enter complete remission progress rapidly, resulting in a poor outcome.
Roles of BRCA1 in both homologous recombination and nonhomologous end joining DNA repair have been shown. Therapeutic strategies that explore the DNA repair defect in BRCA mutants have been proposed and are showing promise, in particular with poly(ADP-ribose) polymerase 1 inhibitors (9). BRCA1-mutated cell lines are more sensitive to cisplatin and less responsive to doxorubicin (10, 11). However, the in vivo response and long-term effects of platinum-based therapy in BRCA1-associated tumors have yet to be established.
Mice carrying somatic mutations of Brca1 and/or p53 alleles in mammary epithelial cells using the Cre/loxP system develop mammary tumors with high penetrance (12–15). Tumor latency in Brca1fp/fpp53fp/fpWAPCrec mice, which express exon 11–deleted Brca1 and exon 5– and exon 6–deleted p53, is slightly shorter than that of Brca1fp/fpp53fp/fp CK14Crec mice, which carry null alleles of Brca1 and p53 (13, 14). On the other hand, MMTVCrea target a small number of cells in the mammary gland and has much longer tumor latency (12). High tumor penetrance and consistent tumor latency of Brca1fp/fpp53fp/fpWAPCrec and p53fp/fpWAPCrec mice allow studies of therapeutic response to single chemotherapeutic agents such as cisplatin [cis-dichlorodiamineplatinum (CDDP)], carboplatin, and doxorubicin, in vivo. CDDP is a member of platinum-based compounds that form various intrastrand and interstrand adducts with DNA (reviewed in ref. 16). Repair of the platinum adduct is mediated by nucleotide excision repair and single-strand and double-strand DNA break repair pathways. The clinical use of this drug is limited due to the emergence of intrinsic and acquired resistance and severe peripheral neurotoxicity. Other platinum derivatives, such as carboplatin and oxaliplatin, have different toxicities and are more commonly used in the clinical setting. Doxorubicin, also known as Adriamycin, is the most widely used anthracycline antibiotic in the treatment of breast cancer. It forms a complex with DNA by intercalation between base pairs, leading to the formation of free radicals and subsequent inhibition of DNA topoisomerase II catalytic activity (17). In clinically aggressive breast cancer, doxorubicin is most lethal to cells that contain high levels of topoisomerase II and are undergoing high rates of DNA replication. Chemotherapy has improved survival rates among cancer patients, but chemoresistance, which results in failure in cancer treatment, remains a major challenge.
There is now much evidence that cancer stem cells, a minority tumor cell population with stem cell properties, are capable of maintaining continuous tumor growth (reviewed in ref. 18). Cancer stem cells were first identified in acute myelogenous leukemia and have recently been shown to be present in many solid tumors, including tumors of the breast, central nervous system, and lung adenocarcinoma (reviewed in ref. 19). In breast cancers, CD44+CD24–/low putative cancer stem cells were identified (20). In glioblastoma, a population of CD133+ cancer stem cells showed significant resistance to chemotherapeutic agents including temozolomide, carboplatin, paclitaxel, and etopo-side (21). In small-cell lung carcinoma, a small population of stem-like cells showing high clonogenic activity and coexpression of CD44 and multidrug resistance gene, MDR1, showed multidrug resistance (22).
Despite the clinical importance of chemoresistance, to date there have been no reports of chemoresistant stem cell populations in breast cancer. Identification and characterization of such a subpopulation of cells will help develop strategies to target these cells. We show here that spontaneous tumors that developed in the conditional Brca1/p53 knockout mice respond favorably to platinum treatment, but chemoresistance does occur and often emerges over time. Expansion of a subpopulation of cancer stem cells correlates with drug resistance.
Materials and Methods
Generation of Brca1fp/fpp53fp/fpCre mutant mice and spontaneous mammary tumor formation
Generation of Brca1fp/fpp53fp/fpCre and p53fp/fpCre mice has previously been described (12, 13). Briefly, Brca1 exon 11–floxed (Brca1fp/fp) mice were obtained from Dr. Chu-Xia Deng (NIH, Bethesda, MD; ref. 15). Brca1fp/fp mice were bred to p53 exon 5– and exon 6–floxed (p53fp/fp) mice to obtain Brca1fp/+p53fp/+ mice. Brca1fp/fpp53fp/fpMMTVCrea and Brca1fp/fpp53fp/fpWAPCrec mice were generated by crossing the heterozygous floxed mice with MMTVCrea or WAPrtTACrec transgenic mice followed by crosses of heterozygous mice. The WAPrtTACrec transgene expression was detected either before puberty as reported by Lin and colleagues (12) or in doxycycline-treated pregnant mice. Only the former group of mice were analyzed in the current study. PCR reaction was done to confirm exon 11 deletion of Brca1 gene and exon 5 and 6 deletions of p53 gene as previously reported (12, 13). The mice were in a C57BL/6 and 129/Sv mixed background. Mice were monitored for palpable tumors weekly. All animal experiments were in accordance with the guidelines of federal law and Institutional Animal Care and Use Committee at the University of California, Irvine.
Doxorubicin, cisplatin, and carboplatin treatment
Animals were treated with doxorubicin, cisplatin, or carboplatin when tumor diameter reached ~0.5 cm. Doxorubicin (Fluka) was prepared at a stock concentration of 58 mg/mL in DMSO and stored at 4°C. Doxorubicin was diluted in 40% polyethylene glycol 400 (PEG-400; Sigma) in saline at the time of treatment. Cisplatin (CDDP; Sigma) was prepared fresh daily in 40% PEG-400 and saline. Stock solution of carboplatin (Sigma) at 10 mg/mL in water was stored at 4°C; dilution in 40% PEG-400 and saline was made at the time of treatment. CDDP was administered i.p. every other day at 0.5, 1.5, 3, or 6 mg/kg daily for 7 d. Carboplatin was administered i.p. once every 3 d for 15 d at 32.5 or 60 mg/kg; doxorubicin was administered i.p. weekly at 1.25 or 5 mg/kg for 21 d. Tumor growth was monitored by daily caliper measurements in two perpendicular dimensions. Tumor volume (mm3) was calculated according to the formula (d2 × D) / 2, where d and D represent the shortest and longest diameters, respectively.
Histology and immunohistochemistry
Mice were sacrificed and tissues collected when tumors reached 0.75 to 1 cm3. The tissue was fixed in 4% paraformaldehyde (Sigma-Aldrich) at 4°C overnight followed by paraffin embedding. Paraffin sections were stained with H&E and examined by light microscopy. Immunostaining was done following the protocol described in the Vectastain Elite ABC kit (Vector Laboratories). To retrieve the antigen, slides were heated for 20 min in 10 mmol/L citrate buffer (pH 6.0) in a microwave oven. Caspase-3 (Cell Signaling Technology), phospho-H3 (Upstate), and Rad51 antibodies were used at 1:200, 1:2,000, and 1:50 dilutions, respectively.
Tissue disaggregation and cell preparation
Tumor tissues were processed according to Stingl and colleagues (23) with modifications. Briefly, tumors were minced into ~2-mm3 fragments with sterile scalpels and enzymatically disaggregated for 8 h at 37°C in EpiCult-B medium (StemCell Technologies, Inc.) with 5% fetal bovine serum (FBS), 300 units/mL collagenase, and 100 units/mL hyaluronidase. After centrifugation at 450 × g for 5 min, RBC were removed by pipetting in 0.64% NH4Cl. Single-cell suspensions were obtained by sequential pipetting for 1 to 2 min in 0.25% trypsin, followed by 2 min in 5 mg/mL dispase II plus 0.1 mg/mL DNase I (Sigma). The resulting suspension was then filtered through a 40-μm mesh and subjected to removal of lineage positive cells using anti-CD45, anti-TER119, and anti-CD31 antibodies to eliminate hematopoietic and endothelial cells. Magnetic beads were used to enrich mammary epithelial cells using the Mouse Mammary Stem Cell Enrichment Kit according to the manufacturer's protocol (StemCell Technologies). Following the enrichment step, the resulting cell suspension was kept on ice until use.
Cell labeling, flow cytometry, and sorting
Antibody staining was done in PBS supplemented with 1% bovine serum albumin (BSA). Cells were first incubated with 1% BSA for 20 min at a cell concentration of 1 × 106/mL. Cells were subsequently washed and stained with different conjugated antibodies for 30 min, at dilutions predetermined by titration experiments. Antibodies used in this study were antimouse CD24-phycoerythrin (clone M1/69; BD Biosciences) and antimouse CD29-FITC (clone Ha2/5; BD Biosciences). Following removal of excess unbound antibodies by washing twice in PBS, the cells were resuspended in sorting buffer containing 1% BSA and 1 mmol/L EDTA in PBS. The stained cells were subjected to filtration through a 40-μm mesh and kept on ice until sorted. Flow cytometry analysis and sorting was done with the MoFlo flow cytometer (DakoCytomation). Forward scatter area versus forward scatter width profiles were used to eliminate dead cells and cell doublets. Sorting was done to achieve a final average purity of >95%.
Mammary gland transplantation
The no. 4 inguinal mammary gland outgrowths, between nipple and lymph node, of Rag1–/– (The Jackson Laboratory) recipient mice were removed at 3 wk of age. Sorted cells were spun down by low-speed centrifugation (850 × g for 5 min) and resuspended in DMEM supplemented with 10% FBS and 2 mmol/L l-glutamine. In all experiments, cell number and viability were confirmed with the trypan blue dye exclusion test. Dilutions were done to achieve the required number of viable cells for appropriate injection doses. Cells were then mixed with BD Matrigel (BD Biosciences) at a 1:1 ratio and injected into cleared mammary fat pads of 6- to 8-wk-old Rag1–/– recipient mice. To minimize experimental variability due to potential differences in recipient mice, control cell populations sorted by fluorescence-activated cell sorting (FACS) were injected into the opposite flank of each animal.
In vitro epithelial progenitor assays
The colony-forming ability of mammary epithelial progenitors was determined as previously described (23–25). Sorted cells were cultured onto preseeded feeder layers, prepared from irradiated (5 × 103 cGy) NIH 3T3 cells, at a clonal density of ~800/cm2, in EpiCult-B medium (StemCell Technologies) supplemented with 5% FBS. After 24 h, the medium was replaced with serum-free EpiCult-B medium. In cases where CDDP was used, a predetermined GI50 value was added to the culture for 48 h. Five to seven days later, cell colonies were fixed with acetone/methanol (1:1), stained with Giemsa, and counted. Colonies were counted from three replicate experiments and tabulated as the average number of colonies per 100 primary cells.
Embryonic stem cell culture
E14 embryonic stem cell line was cultured as described (26). Total RNA was prepared for reverse transcription-PCR (RT-PCR) and used as a positive control for Oct4 expression.
PCR and RT-PCR
Semiquantitative PCR reactions were done with 100 ng of genomic DNA as template in a mixture containing 20 pmol of each gene-specific primer. The PCR cycling conditions were done with an initial denaturation step at 95°C for 1 min, followed by 35 cycles of 40 s at 94°Cfor denaturation, 40 s at 60°C for annealing, and 1-min extension at 72°C, followed by a final extension at 72°C for 7 min. Semiquantitative RT-PCR analysis of total RNAs was done with the MasterAmp RT-PCR Kit (Epicentre Biotechnologies). Fifty nanograms of total RNA were used as templates for a first strand synthesis at 60°C for 20 min. This single-tube reaction was then subjected to 40 cycles of denaturation at 95°C for 40 s, annealing at 60°C for 40 s, and extension at 71°C for 1 min with a final extension of 7 min at 72°C. Alternatively, 200 ng of total RNA were reverse transcribed into cDNA using Oligo dT primer and the ImProm-II Reverse Transcription System (Promega) according to the manufacturer's protocol. The resulting single-strand cDNAs (1 μL) were either added to 25 μL of PCR reaction or stored at –20°C. Primer sequences are listed in Supplementary Table S1.
Statistical analysis
Results are presented as the mean ± SD for at least three repeated individual experiments for each group. Analyses were done with the SPSS software. The percentages tumor-free mice were generated using Kaplan-Meier survival curves. Analyses were done with GraphPad Prism software, which combines the log-rank (Mantel-Cox) test and the Gehan-Breslow-Wilcoxon method.
Results and Discussion
To investigate whether Brca1 mutation alters tumor chemotherapy response in vivo, we established p53- and Brca1/p53–mutant mammary tumor models, respectively, using the Cre/loxP system (12, 13). Nulliparous Brca1fp/fpp53fp/fpMMTVCrea females developed mammary tumors with complete penetrance (11 of 11) between 11.8 and 16.7 months of age, with a median tumor latency of 14.1 months, as compared with nulliparous p53fp/fpMMTVCrea females (n = 18), which developed tumors with a median tumor latency of 16.9 months (P < 0.0001; Fig. 1A). Expression of MMTVCrea is restricted to a small number of cells in the mammary gland (12). As previously shown, mammary tumor latency varied depending partly on the numbers of targeted cells. In nulliparous Brca1fp/fpp53fp/fpWAPCrec mice, mammary tumors developed between 3.7 and 9.5 months of age, with a median tumor latency of 6.3 months and complete penetrance (39 of 39 mice), as compared with nulliparous p53fp/fpWAPCrec mice (n = 18), which have a median tumor latency of 10.6 months (P < 0.0001; Fig. 1A). The median tumor latency in Brca1fp/fpp53fp/fpK14Crec mice, described by Liu and colleagues (14), is ~7.0 months. Thus, double knockout mouse strains harboring WAPCrec have a slightly shorter median tumor latency compared with K14Cre mice. Whether this is due to the usage of different promoters or the expression of mutant p53 and BRCA1 proteins in our models is unclear. In contrast to MMTVCrea, the WAPCrec is expressed in >90% of cells in the adult mammary gland (12). The greater acceleration of tumorigenesis by Brca1 mutation in WAPCrec mice when compared with MMTVCrea is likely due to paracrine actions on cell proliferation by the larger number of Brca1/p53–mutated cells.
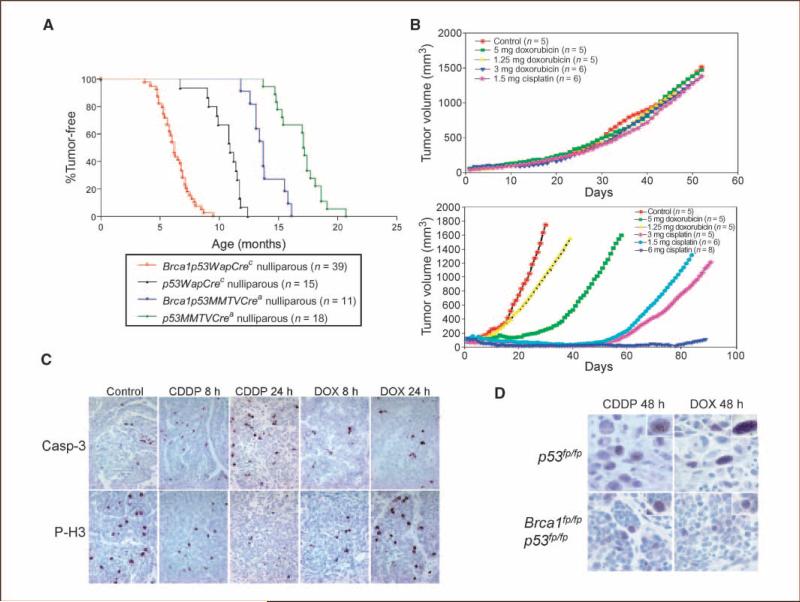
Figure 1.
Brca1/p53–mediated mammary tumors are sensitive to cisplatin treatment. A, proportion of tumor-free nulliparous Brca1fp/fpp53fp/fp and p53fp/fp;Cre mice. X axis, age of each mouse when a palpable tumor was first detected. The curves were plotted using the Kaplan-Meier method. n values indicate the number of mice analyzed of each genotype. B, response of p53-mediated (top) and Brca1/p53–mediated (bottom) mammary tumors to doxorubicin or CDDP in vivo. Data were obtained using both WAPCrec and MMTVCrea mice. Tumor growth was monitored by taking daily caliper measurements in two perpendicular dimensions as described in Materials and Methods. C, immunohistochemical staining of activated caspase-3 (Casp-3) and phospho-histone 3 (P-H3) in CDDP- or doxorubicin (DOX)-treated samples at the indicated time points. D, Rad51 foci formation in CDDP- or doxorubicin-treated tumors. Insets, higher magnifications of stained nuclei. A total of 8 Brca1/p53–mediated and 10 p53-mediated tumors were analyzed, and all showed similar results.
Most mammary tumors that develop in mice with Brca1 and p53 mutations are either adenocarcinomas or spindle cell carcinomas (Supplementary Data), with the vast majority of tumors being poorly differentiated adenocarcinomas, similar to that of human BRCA1-associated tumors. This observation correlates with the previously reported Brca1fp/fpp53fp/fpK14Crec mice (14). In p53fp/fpWAPCrec mice, soft tissue sarcomas, skin tumors, and osteosarcomas, in addition to mammary tumors, were detected in a small number of mice (data not shown). In contrast, all Brca1fp/fpp53fp/fpWAPCrec mice developed mammary tumors and only one case of osteosarcoma was found in a cohort of 39 mice. Thus, mutations of the Brca1 gene greatly shorten tumor latency as well as increase selectivity of mammary tumor development.
Tumor-bearing mice were treated with doxorubicin, CDDP, or carboplatin. p53fp/fp mouse mammary tumors were refractory to doxorubicin or CDDP treatment at all doses tested (Fig. 1B, top). Conversely, all Brca1fp/fpp53fp/fp mammary tumors (n = 19) responded to an initial cycle of four injections of 1.5, 3, or 6 mg/kg CDDP every other day for 7 days (Fig. 1B, bottom). Sensitivity to CDDP treatment was observed in spontaneous tumors developed in WAPCrec; MMTVCrea mice, or in tumor transplants in Rag1–/– mice, indicating that Brca1 mutations render tumor cells sensitive to platinum compounds regardless of which Cre transgenic mice are assessed. Brca1fp/fpp53fp/fp mammary tumors were also sensitive to treatment with 32.5 mg/kg (n = 3) or 60 mg/kg (n = 1) carboplatin (data not shown). Taken together, a differential sensitivity of spontaneous Brca1fp/fpp53fp/fp mammary tumors to platinum over doxorubicin was found in vivo, consistent with the reported observation in a BRCA1-deficient human breast cancer cell line (11).
Sensitivity to platinum treatment (24–48 hours posttreatment) correlated with increased immunostaining of activated caspase-3, which showed cytoplasmic and perinuclear localization (Fig. 1C). Phospho-histone 3 expression, marker of the mitotic phase of the cell cycle, was decreased in CDDP-treated cells (Fig. 1C). Rad51 foci that formed at DNA double-strand breaks were detected 48 hours after CDDP treatment in p53fp/fp tumors (Fig. 1D). In contrast, no Rad51 foci were detected in Brca1fp/fpp53fp/fp tumors. Thus, lack of homologous recombinational repair correlates with increased cell death and reduced mitotic cells in the favorable response to cisplatin. Recent studies showed that BRCA-deficient cancer cells are profoundly sensitized to inhibitors of poly(ADP-ribose) polymerase 1 in vitro (9). Poly(ADP-ribose) polymerase 1 binds to ssDNA breaks and may lead to persistent DNA lesions, preventing repair by homologous recombination pathways. Indeed, silencing of BRCA1 or BRCA2 network genes in p53-deficient, but not p53-proficient, cells enhances sensitivity to cisplatin (27). Taken together, BRCA1 and p53 deficiency leads to selective sensitivity to specific types of chemotherapeutic drug.
Because with doxorubicin treatment Brca1fp/fpp53fp/fp mammary tumors had only a transient slowing down of tumor growth, subsequent studies focused on CDDP-treated mice. Using a lower CDDP dose (0.5 mg/kg; n = 8), 80% of tumors had complete pathologic response to the initial treatment cycle (Fig. 2A) and 20% had only partial regression for 11 to 12 days followed by rapid growth (data not shown), indicating that there is heterogeneity in the initial cisplatin response at the lower CDDP dosage. Following the initial tumor shrinkage and complete pathologic response, tumors relapsed at the same site ~2 to 3 months posttreatment. Relapse also occurred in mice treated with higher dosages of CDDP (data not shown). To address whether recurrent tumors remain sensitive to platinum compounds, a second round of cisplatin treatment was done in three mice (Fig. 2A and data not shown). Tumor shrinkage was observed initially but tumors recurred with a more rapid growth rate than the original relapsed tumor following 1 week of complete regression (Fig. 2A). Rapid recurrence and growth suggest the existence of a subpopulation of cisplatin-resistant cells and selection of the resistant cells during successive platinum treatment.
Figure 2.
Chemoresistance to CDDP in vivo and the presence of cancer stem cells. A, sensitivity of primary and recurrent mammary tumors to CDDP treatment. CDDP was administered i.p., when tumors reached ~0.5 cm3, at 0.5 mg/kg daily for 7 d. Tumor sizes were monitored following detection of palpable tumors before, during, and after CDDP treatment (n = 8). Tumor that recurred in three mice underwent additional cycle of treatments. B, FACS sorting analyses done on cells dissociated from primary tumor. Cells sorted according to the indicated gatings were used for in vivo tumorigenicity assays. C, transplantation using 1,000 cells of the CD29hiCD24med subpopulation resulted in tumor formation (arrows) whereas CD29med24–/lo (arrowhead) or CD29med24hi (asterisk) did not. D, a representative FACS profile of the resulting tumors following CD29hiCD24med cell transplantations.
Previous studies have shown that cancer stem cells contribute to cancer recurrence and chemoresistance in several tumor types. To investigate whether cancer stem cells contribute to platinum resistance, we first tested whether markers for normal mammary stem cells could be used to enrich mammary cancer stem cells. CD24, a heat-stable antigen, is expressed in human breast tumors and neural stem cells. Sleeman and colleagues (28) divided the CD24-expressing normal mammary cells into three subgroups (i.e., CD24low, CD24med, and CD24hi) and showed that cells with the highest repopulation capacity resided in the CD24med population. CD29 (β1-integrin), a stem cell marker in skin, was found to be expressed at a very high level in normal mouse mammary stem cells (25). Thus, we characterized the CD29hiCD24med cells that represented 3.8% to 8.0% (mean, 5.9%) of tumor cell population in primary tumors that arose following CDDP treatment (Fig. 2B). Cells analyzed from untreated primary tumors had a mean of 1.5% CD29hiCD24med cells (data not shown). Injections of 1,000 (5 of 5) and 500 (4 of 6) CD29hiCD24med cells from the resistant primary tumors into fat pads of immunodeficient mice gave rise to tumors, whereas injections of 1,000 CD29med24hi cells (0 of 4) did not (Fig. 2C). One of five injections of CD29med24–/lo resulted in small tumors. This is perhaps due to the presence of small numbers of contaminating cells from the CD29hiCD24med population. Alternatively, the CD29med24–/lo population may contain a small number of cells with limited proliferative ability. Thus, cancer cells expressing high level of CD29 and intermediate level of CD24 are more likely to represent the cancer stem cell subpopulation because they showed a greater repopulation capacity than CD29med24–/lo or CD29med24hi. In the normal mammary fat pad, a single CD29hiCD24+ cell is capable of reconstituting a functional mammary gland (25). Importantly, the CD29hiCD24med cells give rise to tumors that express a wide range of CD29 and CD24 (Fig. 2D).
To systematically investigate the role of CD29hiCD24med cells in cisplatin resistance, we investigated whether tumors that developed in Rag1–/– mice, following transplantation of FACS-sorted tumor cells, respond to platinum treatment. Treatment of first-round primary transplants (n = 10) resulted only in partial regression (Fig. 3A, dotted line). Importantly, secondary tumor transplants (n = 7) generated from CD29hiCD24med cells were completely refractory to CDDP treatment (Fig. 3A, solid line), suggesting an increase in CDDP-resistant cells compared with the primary transplant tumor. Rottenberg and colleagues (29) reported that they were unable to identify resistance to CDDP when they performed transplantations using tumor pieces. They, however, also failed to eliminate the tumors, suggesting the potential existence of CDDP-resistant, tumor-initiating cells. In our study, we showed that these cells are enriched in the CD29hi24med subpopulation. To test whether there were expansions of these cancer stem cells, we compared profiles of CD29 and CD24 in tumor cells from the primary and secondary tumor transplants. The frequency of CD29hiCD24med cells ranged from 6.6% to 11.0% (mean, 8.8%; n = 10) in the primary transplants and from 16.5% to 29.2% (mean, 22.8%, n = 7) in the secondary tumor transplants (Fig. 3B and data not shown). Thus, there is an ~3-fold increase in the frequency of CD29hiCD24med cells in the secondary tumor transplants compared with the primary transplants, suggesting that the expansion of the CD29hiCD24med population may contribute to platinum resistance. Because frequencies of CD29hiCD24med cell populations in primary transplants are comparable to those in recurrent and primary tumors, it is unlikely that expansion of these populations in the secondary tumor transplants is due to transplantation per se. To confirm this, we carried out FACS analysis on primary and secondary transplant tumors that were not treated with CDDP. Results showed that the percentage of CD29hiCD24med in these tumors remained low, within the range of 3.5% to 9.8% (mean, 6.6%; n = 5). Importantly, in the CDDP-treated tumors, the CD29hiCD24med cells gave rise to heterogeneous cell populations, expressing wide ranges of CD24 and CD29 (Fig. 3B), indicating that the regenerated tumors contained phenotypically diverse populations of cells, as one would expect from cancer stem cells.
Figure 3.
Chemoresponse, FACS profiling, and colony-forming abilities of the primary and secondary Brca1fp/fpp53fp/fp tumor transplants. A, response to CDDP treatment of the primary and secondary tumor transplants, developed after transplantation with 1,000 CD29hiCD24med cells. The treatment was administered i.p., when tumors reached ~0.5 cm3,at 0.5 mg/kg daily for 7 d. Tumor volumes before, during, and after CDDP treatment were shown. Arrows are not drawn to scale. B, FACS analyses of cells dissociated from primary tumor transplant, which showed partial resistance to CDDP (left), and from secondary tumor transplant, which were refractory to CDDP (right). C, colony formation assay of sorted CD29hiCD24med, CD29med24–/lo, and CD29med24hi cells from the primary tumor transplant. D, colony formation of unsorted cells from primary and secondary tumor transplants in the presence or absence of CDDP. C and D, columns, mean number of colonies per 100 primary cells; bars, SD. *, P < 0.05, versus the control group.
In addition to assaying the potential of cancer stem cells to regenerate tumors in vivo, we also carried out in vitro colony-forming assays on CD29hiCD24med cells. Colony formation assay has previously been used for estimation of progenitor numbers in a cell population (23–25). CD29hiCD24med cells have ~3- and 6-fold higher colony-forming efficiencies on irradiated NIH 3T3 feeder layers than CD29med24–/lo and CD29med24hi cells, respectively (Fig. 3C). In a separate experiment, a low colony-forming ability was seen with unsorted cells (Fig. 3D, shaded column). These independent experiments provided further evidence that the CD29hiCD24med population contained higher numbers of mammary progenitor cells. In the unsorted population, cells harvested from the second transplant were 3-fold enriched for colony formation compared with cells from the first transplant (Fig. 3D). This observation corroborates the findings from the FACS analyses where a 3-fold increase in the CD29hiCD24med subpopulation was observed in the secondary transplant. These colonies were also larger (Fig. 3D, bottom), suggesting increased proliferation of cancer stem cells following secondary transplantation. To explore the possibility that this population of cells were also CDDP resistant, we added CDDP at a predetermined GI50 concentration to the culture in the colony assay. Interestingly, ~22% of the colonies from the second transplant were resistant to CDDP treatment, compared with <1% from the first transplant (Fig. 3D), strongly suggesting an expansion of CDDP-resistant progenitor cells in the second transplant population.
To investigate the involvement of key regulators in self-renewal and pluripotency in these CD29hiCD24med populations, we carried out semiquantitative RT-PCR for Nanog, Oct4, and Sox2, critical players for the pluripotent embryonic stem cells (reviewed in ref. 30). Sox2 and Oct4 expression was low in all the samples from primary as well as secondary tumor transplants, whereas Oct4 expression was readily detectable in the embryonic stem cells (Fig. 4A and B). Interestingly, Nanog was readily detectable in these cells, particularly in the primary tumor transplant, whereas Nanog expression was reduced in CD29hiCD24med populations of the secondary tumor transplants. Lin and colleagues (31) showed that Nanog expression is suppressed by p53, leading to mouse embryonic stem cell differentiation. In our mouse model, conditional deletion of p53 may contribute to Nanog up-regulation. On the other hand, in contrast to normal stem cells, Nanog may no longer be subject to Sox2-Oct4 transcriptional regulation in cancer stem cells.
Figure 4.
Selective gene expression profiles in recurrent primary and secondary tumor transplants. A, semiquantitative RT-PCR analyses of CD29hiCD24med, CD29med24–/lo, and CD29med24hi cells from the primary and secondary tumor transplants. B, a positive control for Oct4, using mouse embryonic stem (ES) cells. C, semiquantitative PCR analyses of genomic DNA of CD29hiCD24med cells from the primary and secondary tumor transplants; WT, wild-type. D, model illustrating changes in cancer stem cells during the course of chemotherapy.
We also examined MDR1 and Top2A expression in primary and secondary tumor transplants by semiquantitative RT-PCR. MDR1 expression was low or below detection in all cases (Fig. 4A). However, whereas high levels of Top2A expression were seen in primary transplants, expression was greatly reduced in all secondary transplants examined (Fig. 4A and data not shown). Genomic deletion of Top2A was seen in one of the secondary tumors (Fig. 4C). Whereas differential gene expression was detected in primary and secondary tumor transplants, mechanisms leading to chemoresistance remain to be addressed. It should be noted that down-regulation of Top2A has been observed in postchemotherapy samples compared with the primary tumors (32), suggesting its possible contribution to chemoresistance. However, it should also be noted that expression of Top2A has been associated with both cisplatin resistance and sensitivity in ovarian cancer in different studies (33, 34). Therefore, the predictive power of Top2A in BRCA1-associated tumors has yet to be elucidated.
Taken together, exposure to CDDP may lead to the emergence of a subpopulation of mouse tumor cells (i.e., CD29hiCD24med) that become resistant to CDDP (Fig. 4D). Alternatively, a small population of preexisting cells with inherent CDDP resistance survives CDDP treatment. Following subsequent exposure to CDDP, these resistant cells self-renew as well as differentiate into tumor masses containing cells of various phenotypes. Transplantation of this subpopulation of cells into immunodeficient mice led to its further expansion. Up-regulation of c-Myc was detected in the CD29hiCD24med population in the primary, but not secondary, tumor transplant. Whether c-myc and Nanog are involved in the expansion, but not maintenance, of the CD29hiCD24med cancer stem cell populations remains to be addressed.
Studies by Rottenberg and colleagues (29) used a model with null alleles of Brca1 and p53, whereas the model described here expressed exon 11–less isoforms of Brca1. The CDDP treatment regimen also differs in the two studies. A 20-day recovery preceded each single injection of CDDP reported by Rottenberg and colleagues (29). Tumors were not eliminated under the regimen, indicating the existence of chemoresistant cells. The spontaneous chemoresistant model described here allows a longitudinal follow-up of putative cancer stem cells that will allow delineation of chemoresistance. Clearly, the platinum-resistant CD29hiCD24med cancer stem cells not only expanded but presumably also increased their proliferation rates relative to the total population of tumor cells during the treatment course.
Supplementary Material
Acknowledgments
Grant support: Avon Foundation-American Association for Cancer Research International Scholar Award in Breast Cancer Research (N. Shafee and E.J. Stanbridge), Department of Defense Breast Cancer Center of Excellence grant DAMD17-02-1-0694, NIH grant CA04964 (E.Y. Lee), and Department of Defense predoctoral fellowship (C.R. Smith). N. Shafee is a Visiting Scholar from Universiti Putra Malaysia. C.R. Smith is inspired by his son Keagen.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25:5898–905. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 2.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–5. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 4.Ting NS, Lee WH. The DNA double-strand break response pathway: becoming more BRCAish than ever. DNA Repair (Amst) 2004;3:935–44. doi: 10.1016/j.dnarep.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Turner N, Tutt A, Ashworth A. Hallmarks of “BRCA-ness” in sporadic cancers. Nat Rev Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 6.Heine GF, Parvin JD. BRCA1 control of steroid receptor ubiquitination. Sci STKE. 2007;391:pe34. doi: 10.1126/stke.3912007pe34. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruvberger-Saal SK, Persson C, Lövgren K, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–7. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 11.Tassone P, Tagliaferri P, Perricelli A, et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br J Cancer. 2003;88:1285–91. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SC, Lee KF, Nikitin AY, et al. Somatic mutation of p53 leads to estrogen receptor α-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer Res. 2004;64:3525–32. doi: 10.1158/0008-5472.CAN-03-3524. [DOI] [PubMed] [Google Scholar]
- 13.Poole AJ, Li Y, Kim Y, Lin SC, Lee WH, Lee EY. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science. 2006;314:1467–70. doi: 10.1126/science.1130471. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Holstege H, van der Gulden H, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–6. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Wagner KU, Larson D, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 16.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 17.Tack DK, Palmieri FM, Perez EA. Anthracycline vs nonanthracycline adjuvant therapy for breast cancer. Oncology. 2004;18:1367–76. [PubMed] [Google Scholar]
- 18.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 19.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–5. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67–78. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutova M, Najbauer J, Gevorgyan A, et al. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS ONE. 2007;2:e243. doi: 10.1371/journal.pone.0000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stingl J, Emerman JT, Eaves CJ. Enzymatic dissociation and culture of normal human mammary tissue to detect progeny activity. In: Helgason CD, Miller CL, editors. Methods in molecular biology: basic cell culture protocols. Humana; New Jersey: 2005. pp. 249–63. [DOI] [PubMed] [Google Scholar]
- 24.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 25.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 26.Skarnes WC. Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Methods Enzymol. 2000;328:592–615. doi: 10.1016/s0076-6879(00)28420-6. [DOI] [PubMed] [Google Scholar]
- 27.Bartz SR, Zhang Z, Burchard J, et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–86. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottenberg S, Nygren AO, Pajic M, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci U S A. 2007;104:12117–22. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–84. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 31.Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–71. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 32.Jazaeri AA, Awtrey CS, Chandramouli GV, et al. Gene expression profiles associated with response to chemotherapy in epithelial ovarian cancers. Clin Cancer Res. 2005;11:6300–10. doi: 10.1158/1078-0432.CCR-04-2682. [DOI] [PubMed] [Google Scholar]
- 33.Helleman J, Jansen MPHM, Span PN, et al. Molecular profiling of platinum resistant ovarian cancer. Int J Cancer. 2005;118:1963–71. doi: 10.1002/ijc.21599. [DOI] [PubMed] [Google Scholar]
- 34.Cornarotti M, Capranico G, Bohm S, et al. Gene expression of topoisomerase I, IIα and IIβ and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Int J Cancer. 1996;67:479–84. doi: 10.1002/(SICI)1097-0215(19960807)67:4<479::AID-IJC3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.