Abstract
Objectives
The intravenous administration of a bolus dose of endotoxin to healthy human subjects triggers acute systemic inflammatory responses that include cytokine production and dynamic changes in gene expression in peripheral blood leukocytes (PBL). This study sought to determine the state of clock gene expression in human PBL, and leukocytes subpopulations, challenged with in vivo endotoxin at two circadian/diurnal phases of the clock.
Design
Clinical and laboratory investigation.
Setting
University-based research laboratory and clinical research center
Subjects
Human volunteers.
Interventions
Human subjects were administered a standard dose of endotoxin (2ng/kg) or saline at either 09:00 or 21:00 h. Blood samples were collected at selected time points pre- and post-infusion.
Measurements and Mains results
Clock gene expression was determined in human PBL, neutrophils, and monocytes, by quantitative real-time polymerase chain reaction. The fold change for each gene was determined using the 2(-ΔΔCt) method. We show that endotoxin causes profound suppression of circadian clock gene expression, clearly manifested in human PBL, neutrophils, and monocytes. Clock, Cry1-2, Per3, CSNK1ε, Rora and Rev-erb gene expression were all reduced by 80-90% with the nadir between 3 to 6 hours post-infusion. Per1 and Per2 reached an expression nadir between 13 and 17 hours post-infusion. The levels of plasma interleukin-6 and tumor necrosis factor peaked and then returned to baseline within 6 hours. In contrast, clock gene expression remained suppressed for up to 17 hours, irrespective of the phase of the clock at the time of the endotoxin challenge. Endotoxin did not perturb the melatonin secretory rhythm.
Conclusions
Circadian clock gene expression in PBL is dramatically altered, and possibly uncoupled from the activity of the central clock, during periods of acute systemic inflammation. The realignment of the central and peripheral clocks may constitute a previously unappreciated key factor affecting recovery from disease in humans.
Keywords: endotoxin, circadian clock, peripheral blood leukocytes, melatonin
Introduction
Numerous physiologic activities in higher organisms are synchronized with the 24 h rotation period of the Earth. The central “master” clock controlling behavioral circadian rhythms is located in the suprachiasmatic nucleus (SCN) within the brain hypothalamus [1, 2]. The molecular components of the circadian clock consist of transcription factors and proteins whose activity and/or availability cycle with a periodicity of approximately 24 h [3-5]. Circadian oscillations of clock-associated genes have been identified in many tissues (e.g., [6-9]), highlighting the probable existence of multiple peripheral clocks. The mechanisms by which the master clock and the peripheral clocks interact are not completely understood.
Circadian rhytmicity and host immunity are closely interrelated. The number of circulating red blood cells, platelets, as well as all human peripheral blood mononucleated cells subsets exhibit significant diurnal variation [10, 11]. Lange et al [12] reported that human monocytes obtained during the normal sleep hours produce interleukin-12 (IL-12) in response to ex-vivo endotoxin challenge, whereas monocytes obtaining during the day produce IL-10. There is also evidence that intracellular IL-6 expression exhibits distinct diurnal pattern, with increasing IL-6 levels during the night. [13]. These findings suggest that both immune cell number and function are subject to circadian regulation.
The core of the circadian clock is composed of two transcription factors, Clock and Bmal1 (Arntl; MOP3), which form a heterodimeric complex. Analyses of mice with genetic alterations in Bmal1 and Clock provided further insight into the relationship between immunity and the circadian clock. Bmal1-/- mice exhibit no locomotor rhytmicity and are significantly less active than normal mice [14]. In addition, Bmal1-/- mice exhibit increased segmented neutrophils and platelets numbers, and decreased B lymphocytes numbers [15]. The daily rhythmic variability in white blood cell populations, and the total number of white blood cells, red blood cells, and platelets were similarly reduced in mice that express a transcription-deficient mutant form of Clock [16].
Data suggest that the immune system is not only subject to circadian regulation, but may also provide inputs that alter the circadian activity of the clock(s) [17]. It is also known that activities such as sleep, movement, and food intake that are normally regulated by the circadian clock, are modified during periods of systemic inflammation [18, 19]. Tumor necrosis factor (TNFα, IL-1β, and IL-6 are among the prominent pro-inflammatory cytokines produced in human during the acute period of systemic inflammation. Cavadini et al [20] reported that infusion of TNFα in mice triggered a significant reduction in locomotor activity, prolonged rest time, and impaired expression of several clock-related genes in hepatic tissue. The authors proposed that TNFα-induced behavioral changes were triggered, at least in part, through alterations in clock gene expression [20]. These observations underscore the possibility that pro-inflammatory cytokines produced by activated immune cells have the capacity to reset the circadian clock in peripheral tissues during inflammation.
The status of circadian clock genes in humans during periods of systemic inflammation is largely undetermined. Okada et al [21] recently reported that the administration of endotoxin to rats impaired the expression of the clock genes Per1, Per2, and DBP in the liver, and Per2 and DBP in the SCN, with the expression nadir between 10-14 hours post-endotoxin challenge. The objective of this study was to determine whether clock gene expression is perturbed in human PBL during the acute period of systemic inflammation induced by an in vivo endotoxin (LPS) challenge [22, 23]. Our data revealed that in vivo endotoxin causes profound and concurrent suppression of many clock genes in PBL with a nadir between 3-6 hours post-infusion. These observations identify endotoxin as a potent and acute entrainer of the circadian clock gene network in PBL in humans.
Materials and Methods
Study population and procedures
Male and female subjects (age 18-29) were recruited by public advertisement to participate in a study approved by the Institutional Review Board of the Robert Wood Johnson Medical School. Subjects were administered a standard dose of endotoxin (2 ng/kg, NIH Clinical Center Reference Endotoxin, CC-RE, Lot 2, Bethesda, Maryland) [24] or saline (0.9% sodium chloride). Additional information is provided in the Supplementary Materials.
Gene expression analyses in PBL
Blood was drawn into Paxgene™ tubes (PreAnalytix). Total RNA was extracted as per PAXgene Blood RNA kit protocol (Qiagen), quantified using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), and reversed transcribed to cDNA using High capacity cDNA Archive kit (Applied Biosystems). Gene expression was analyzed in duplicates by quantitative real-time (qRT) PCR using inventoried TaqMan® gene expression assays (Applied Biosystems). A list of the gene expression assays is provided in the Supplementary Materials. The relative gene expression analysis was performed using the 2-ΔΔCT method [25]. The level of beta-2-microglobulin (B2M) expression was used as an internal reference, since the expression of B2M is not affected by endotoxin administration in vivo [24, 26, 27]. Data are expressed as fold change relative to time 0. For the sample at time 0, ΔΔCT equals zero and 20 equals one, so that the fold change in gene expression relative to the control equals one [25].
Gene expression analyses in leukocyte cell subpopulations
Detailed protocols can be found in the Supplement.
Cluster analysis
Details can be found in the Supplementary Materials. In brief, the data were clustered into a tree form using the standard hierarchical Unweighted Pair Group Method with Arithmetic Mean algorithm (UPGMA), with one minus the cross-correlation coefficient as a measure of similarity [28, 29]. All clustering operations were carried out in MatLab 2008a program.
TNFα, IL-6, and cortisol levels were determined as described [30, 31].
Melatonin
Plasma melatonin levels were determined using a direct Melatonin radioimmunoassay kit (Rocky Mountains Diagnostics, Colorado Springs, CO 08903). The assay range was 0-1000 pg/ml.
Statistics
Analysis of three groups or more was by one-way ANOVA with Newman-Keuls post-test. Two groups were compared by unpaired Student's t test. The operations were carried out using Prism 4 software Version 4.0b (GraphPad Software, Inc., La Jolla, CA). P values less than 0.05 were considered to be statistically significant.
Results
Day study
Subjects were administered endotoxin (n=4) or saline (control; n=2) at 09:00 h (time 0). The relative gene expression values determined on the day prior to the challenge are shown in Supplementary Figure 1. Of the 10 genes examined, only Rora and Rev-erb showed significant time-dependent expression differences prior to infusion (P <0.01; one way ANOVA) with a peak at 06:00 h (Supplementary Figure 1).
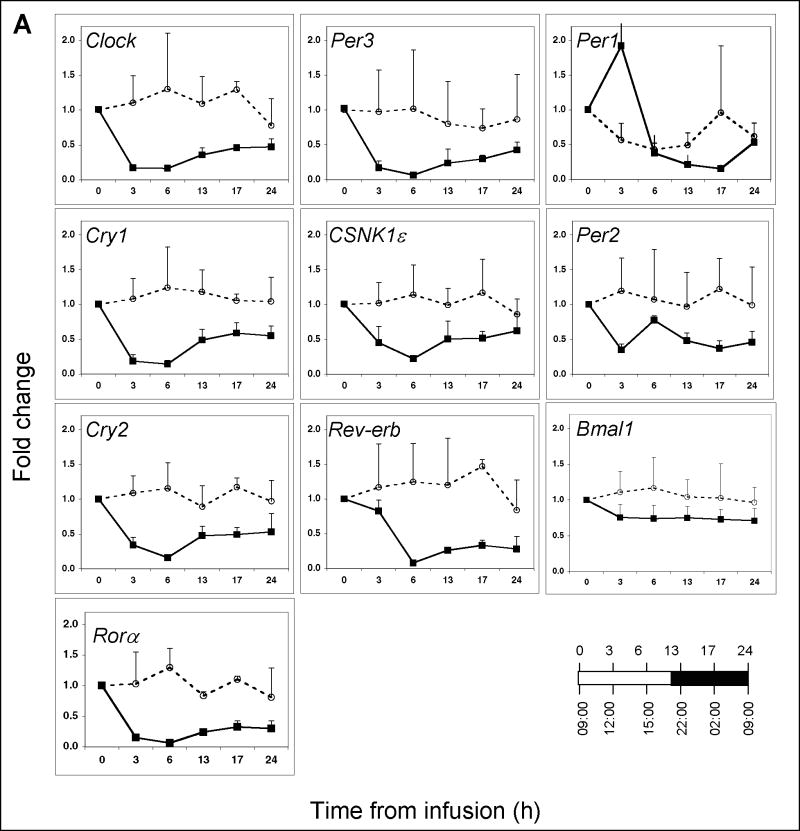
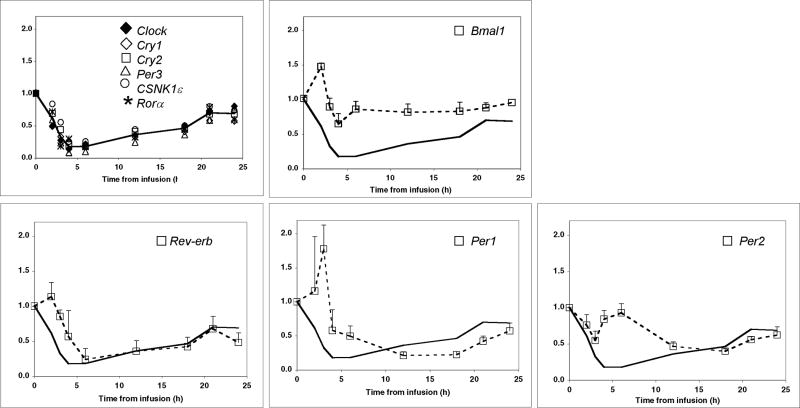
The infusion of endotoxin triggered a significant decline in Clock, Per3, Cry1, Cry2, Rora, CSNK1ε, and Rev-erb expression, reaching the nadir within 3-6 hours (Figure 1A) (P ≤ 0.0001 for all genes; one-way ANOVA). By 6 hours post-infusion, the expression values of these seven aforementioned genes had decreased by 80-90% relative to baseline. Bmal1 expression was not altered significantly post-endotoxin infusion (P>0.05, one-way ANOVA). The expression values of all genes, with the exception of Bmal1, differed significantly over the interval between 02:00 h (-7 h) pre-infusion and 02:00 h (17 h) post-infusion (Supplementary Table 1). These findings suggest that the expression levels of nine out of the ten genes examined remained suppressed for at least 17 hours post-endotoxin challenge.
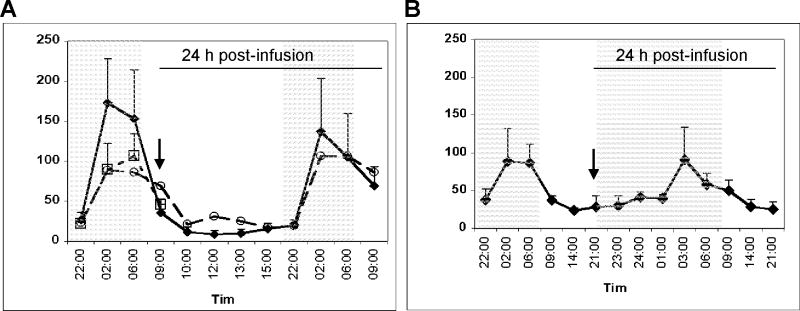
Figure 1. Clock gene expression in peripheral blood leukocytes obtained from human subjects challenged with endotoxin or saline at 09:00 h.
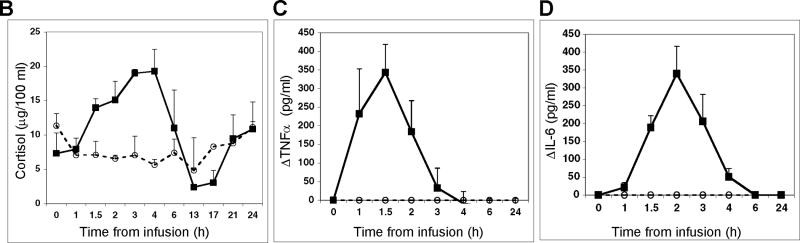
(A) Blood was collected at the indicated time points post-endotoxin (closed symbols; n=4 subjects) or post-saline (open symbols; n=2 subjects) infusion. Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 h infusion time, taken to be time 0. The relationship between the time from infusion and the time-of-day is illustrated in the lower right hand corner. Data are expressed as mean fold change ± SD. (B) Plasma cortisol levels (n=4) as a function of time post-endotoxin were monitored by direct radioimmunoassay. (C) TNFα (n=4) and (D) IL-6 (n=4) levels were determined by enzyme-linked immunoassay.
Plasma cortisol and pro-inflammatory cytokines levels in endotoxin- and saline-challenged subjects
Plasma cortisol levels in humans normally peak 3-4 hours after the end of the sleep/darkness period [9, 32]. Plasma concentrations of cortisol, as well as the pro-inflammatory cytokines TNFα and IL-6, increase significantly in response to an endotoxin challenge [22, 31, 33]. The anticipated increase in cortisol concentrations was detected in all endotoxin subjects by 1.5 hours post-challenge. The cortisol concentration peaked between 3-6 hours and returned to baseline levels at 24 hours (Figure 1C). As previously reported in this model system [22, 33], TNFα concentration peaked within 1-1.5 hours post-infusion and returned to baseline by 3 hours (Figure 1D), whereas IL-6 concentration peaked within 2 hours and returned to baseline within 6 hours (Figure 1E).
Nocturnal study
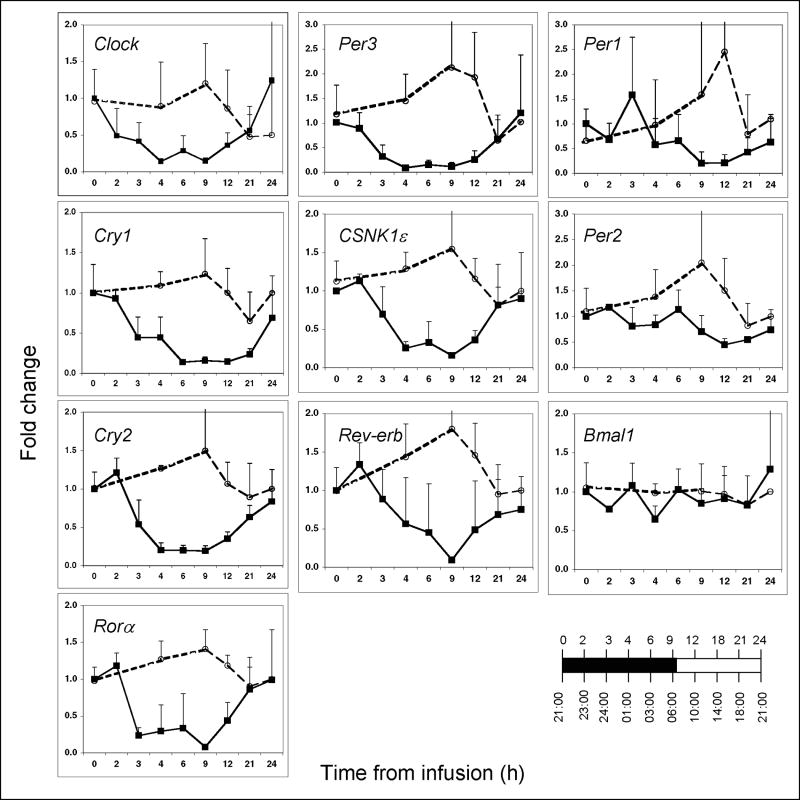
We considered the circadian/diurnal phase at the time of endotoxin challenge as a possible influence on the expression of clock genes in endotoxin challenged PBL. We therefore also studied volunteer subjects challenged with endotoxin at 21:00 h (n=3). Consistent with the data presented in Figure 1A for the day subjects, the relative gene expression levels of all ten genes were close to or above baseline during the 24 hours pre-infusion (Figure 2). The infusion of endotoxin at 21:00 h triggered profound temporal changes in clock gene expression (Figure 2). The changes were similar to those seen in subjects challenged with endotoxin at 09:00 h. Clock, Per3, Cry 1 and Cry2, Rev-erb, Rora, and CSNK1ε expression decreased by approximately 80-90% in response to the endotoxin challenge (p<0.001) reaching their nadir between 4 and 9 hours post-infusion. As was observed in the day subjects, the nadir in Per1 expression was reached several hours after the nadir of most other genes. Per2 and Bmal1 were the least affected by endotoxin. These observations establish that endotoxin suppresses clock gene expression when administered at various times during the diurnal cycle.
Figure 2. Clock gene expression in peripheral blood leukocytes obtained from human subjects challenged with endotoxin at 21:00 h.
Blood was collected at the indicated times prior to (open symbols) and post-endotoxin infusion (closed symbols) (n=3 subjects). Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 21:00 h infusion time, taken to be time 0. The relationship between the time from infusion and the time-of-day is illustrated in the lower right hand corner.
Circadian clock gene expression in PBL and leukocyte subpopulations
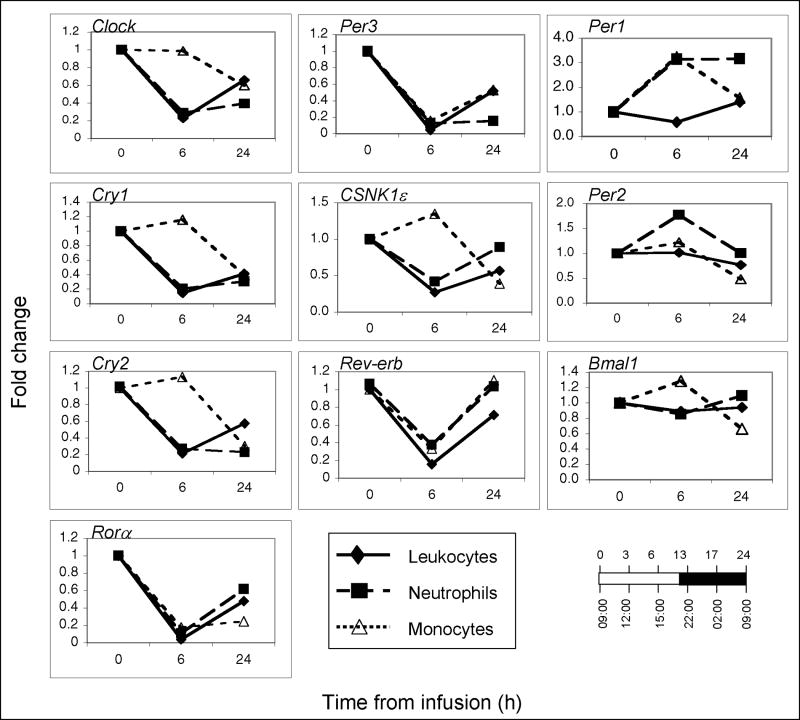
Circadian clock gene expression was also compared among PBL, monocyte- and neutrophil-subpopulations (n=3 for each). Blood was obtained prior to endotoxin-infusion (time 0; 09:00 h), and 6- and 24-hours post-infusion. As shown in Figure 3 (additional data are presented in Supplementary Figure 2), the expression patterns of Clock, Cry 1-2, Rora, Per3, CSKN1ε, Rev-erb and Bmal1 noted in PBL were similar to those observed in neutrophils. The changes in Clock, Cry1, Cry2, and CSKN1ε expression observed in monocytes were time-delayed relative to those observed in neutrophils and PBL, whereas Per3, Rora, and Rev-erb expression were reduced in monocytes by 6 h post-infusion. Per1 expression showed a great degree of variability within cell populations even when derived from a single donor (Figure 3 and Supplementary Figure 2). In contrast, as observed in PBL, Bmal1 expression was the least affected by endotoxin in neutrophils and monocytes. The data suggest that the temporal changes in clock gene expression observed in PBL reflect changes that unfold in neutrophils, and to a lesser degree in monocytes.
Figure 3. Clock gene expression in peripheral blood leukocytes, neutrophils and monocytes obtained from human subjects challenged with endotoxin at 09:00 h.
Blood was collected at time 0, 6 and 24 hours post-endotoxin infusion. Total peripheral blood leukocytes, neutrophils and monocytes subpopulations (n=3 for each) were obtained prior to RNA isolation. Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 h infusion time, taken to be time 0. The relationship between the time from infusion and the time-of-day is illustrated in the lower right hand corner. The data shown are averages (n=3) (all data sets are presented in Fig. 2S).
Endotoxin does not affect the melatonin secretory rhythm
The plasma melatonin levels in humans normally peak during the night [34, 35]. Analyses of plasma melatonin levels prior to infusion, and post-endotoxin or -saline infusion, revealed the anticipated increase in melatonin in the late part of the night (Figure 4). These observations suggest that endotoxin does not affect the apparent activity of the master clock during the acute period of systemic inflammation.
Figure 4. Endotoxin does not alter the melatonin secretory rhythm.
(A) Plasma melatonin concentrations were determined in blood samples obtained from control subjects (n=8 subjects) at 22:00 h, 02:00 h, 06:00 h, 09:00 h and 10:00 h (open squares). Samples were also obtained at the indicated times of the day pre- and post-saline infusion (open circles; n=2 subjects), and pre- and post-endotoxin infusion (closed diamonds; n=4 subjects). Arrow indicates the 09:00 h infusion time. (B) Samples were obtained at the indicated times of the day pre- and post-endotoxin infusion (closed diamonds; n=4 subjects). Arrow indicates the 21:00 h infusion time.
Circadian clock genes exhibit highly related responses to endotoxin
To better define the relationship among circadian clock gene expression in endotoxin-challenged PBL, the gene expression data presented in Figures 1 and 2 were clustered into a tree form (Supplementary Figure 3 A-D). Clustering analyses of the combined day- and night-placebo data suggested correlations between Per2 and Bmal1, and Cry1 and Cry2 (Supplementary Figure 3 A). In contrast with the results observed for the placebo subjects, the clustering analyses identified four genes, Clock, Per3, Cry1 and Cry2 that had highly related responses to endotoxin in both the day (Supplementary Figure 3 B) and night (Supplementary Figure 3 C) subjects. Per 1, Per 2, Per 3, and/or Bmal1, showed no correlated expression in subjects administered endotoxin (Supplementary Figure 3 B-D). Co-clustering of the day and night expression data indicated that CSNK1ε and Rora patterns correlate with Clock, Per3, Cry1 and Cry2 (Supplementary Figure 3 D). When the day and night endotoxin subjects data were analyzed as a group, the similar temporal decline in Clock, Cry1, Cry2, Per3, CSNK1ε, and Rora expression became apparent (Figure 5). The line featured in each of the panels in Figure 5 represents the mean of Clock, Cry1, Cry2 and Per3 expression values. Bmal1, Rev-erb, Per1, and Per2 displayed distinct expression patterns as compared to the common response pattern of the remaining six genes. The large number of clock genes exhibiting a similar endotoxin-induced response suggests that clock gene expression is both synchronized and suppressed during the acute period of systemic inflammation.
Figure 5. Clock gene expression in human peripheral blood leukocytes.
The relative gene expression values for each gene post-endotoxin infusion were combined for all day (n=4 subjects) and night subjects (n=3 subjects), and are presented as an average. Where shown, bars represent standard error of the mean. The line that is featured in each panel was drawn based on the mean expression values of Clock, Cry1, Cry2, and Per3, which exhibited correlated expression in both the day and night subjects.
Discussion
The expression of many circadian clock genes is suppressed in PBL during the acute period of systemic inflammation
We found that the expression of key genes implicated in the regulation of circadian clock function, including Clock, Per3, Cry1 and Cry2, Rora, and Rev-erb, is decreased in PBL by 80-90% within 3-6 hours post-endotoxin infusion. The Clock/Bmal1 complex regulates the transcription of genes with an E-box promoter region, which include Period (Per1, Per2, Per3), Cryptochrome (Cry1, Cry2) and Rev-erb [36]. Per/Cry protein complexes relocate from the cytosol to the nucleus, where they function as Clock/Bmal1 repressors, while Rev-erba and Rora bind to the promoter region of Bmal1 to either suppress or enhance its expression, respectively [3]. The repressor Per proteins are phosphorylated by Casein kinase 1ε (CSNK 1ε) [37]. The phosphorylated proteins are ubiquitinated and targeted for degradation by the proteosome. Once the Per proteins are degraded, the de-repressed Clock/Bmal complex reinitiates its activity cycle. The precipitous declines in expression of multiple genes that act at various points in the circadian clock network noted in this study provide a strong indication that clock activity in PBL is severely impaired during the acute period of systemic inflammation.
Per1, Per2, Per3, and Bmal1 gene oscillations have been observed in human peripheral blood mononucleated cells (PBMCs) as well as whole blood cells [9, 32, 38, 39]. In one study [32], Per1 and Per2 expression in PBMCs peaked in the early hours of the day, whereas Bmal1 peaked in the middle of the wake period. In another study, Per2 and Bmal1 cycled with a similar rhythm in PBMCs [39]. Recently, the expression of ten circadian clock genes, including Per 1-3, Cry1 and Cry2, Clock, and Bmal1, was examined in PBMCs [35]. Of the ten genes examined, only Per 1-3 showed rhythmic expression in most subjects, with no significant acrophase differences among the three genes. The goal of the present study was to determine the fate of clock gene expression in PBL during the acute period of systemic inflammation. Hence, in contrast with the entrainment protocols used by others [9, 32, 35, 39], our subjects were not entrained by an extended sleep/wake schedule prior to or during the study phase. These differences may explain the limited similarity among Per gene expression patterns noted among our placebo subjects.
Prior studies revealed a limited coordination among clock gene expression patterns in PBMCs obtained from sleep/wake entrained normal human subjects, and a significant inter-subjects variability [32, 35, 39]. In contrast, Cry1 and Cry2, as well as Clock, Per3, CSNK1ε, Rev-erb and Rora exhibited correlated expression in PBL obtained from endotoxin subjects. The correlated expression patterns observed in PBL, neutrophils and monocytes after an endotoxin challenge suggest that endotoxin is a potent entrainer of the circadian clock network in circulating inflammatory cells.
Melatonin, cortisol, and circadian clock gene expression in PBL during the acute period of systemic inflammation
In humans, the activity of the central master clock is generally correlated with the secretion of melatonin in the middle to late part of the night [40] [34, 35, 39]. We found that the melatonin rhythms remained intact in endotoxin-challenged subjects, peaking in the late part of the night. As previously reported [31], endotoxin triggered a surge in plasma cortisol levels with a peak between 3-4 hours. While cortisol levels peaked, many clock genes reached their expression nadir. These data suggest that centrally regulated plasma melatonin- and cortisol-rhythms, and PBL clock gene expression are independently regulated in response to endotoxin. These observations raise the possibility that the master clock and circadian clock gene expression in PBL become misaligned during the acute period of systemic inflammation induced by endotoxin.
Okada et al [21] recently observed transient changes in clock gene expression in SCN and liver of endotoxin challenged rats. In that study, endotoxin did not affect the rhythmicity of clock gene expression. Consistent these observations, our data also indicate that endotoxin transiently suppresses clock gene expression in peripheral tissue(s). Furthermore, our data establish that significant perturbations in clock gene expression in PBL unfold while the rhytmicity of the master clock, determined based on plasma melatonin levels, appears to remain intact in humans.
Despite our incomplete understanding of the mechanism(s) by which endotoxin influences circadian rhytmicity, a limited comparison between endotoxin and the two other well-characterized circadian entrainers, light and food, is possible. A short and abrupt period of bright light during the dark period is sufficient to reset the activity of the central clock in humans, as determined by changes in core body temperature and plasma cortisol production [41]. The magnitude of light-induced circadian phase resetting is dependent on the phase of the clock at the time of the input [41]. In contrast, endotoxin appears to trigger similar changes in circadian gene expression in PBL irrespective of the central clock phase. In addition, endotoxin disrupts the temporal relationship among plasma cortisol and melatonin, and circadian gene expression in PBL. These observations suggest that the mechanism by which systemic endotoxin affects clock gene expression differs from the process by which light entrains the clock. The effect of endotoxin on the circadian activity may be more similar to that of feeding. In rodents, feeding during the subjective night or the subjective day alters the rhythmic expression of Per1, Per2, Per3, and Cry1, in the liver within a few days. In contrast, the phase of Per1 and Per2 expression in the SCN was not affected by food entrainment [42]. Thus, both endotoxin and feeding appear to affect peripheral clock gene expression but not the activity of the central master clock.
Pro-inflammatory cytokines and clock gene expression in PBL
Pro-inflammatory cytokines, which are released in the early stages of systemic inflammation, have been implicated in the regulation of circadian activity in mice. TNFα- or IL-1β-infusion suppressed the expression of several clock genes, including Per2 and Per3, in mice livers by binding to the E-box motifs in their promoters [20]. Clock and Bmal1 expression was not affected these cytokines [20]. TNFα levels surge in response to endotoxin, reaching a zenith within 1.5-2 hours post-challenge, while the expression of most clock genes examined in this study remained suppressed for up to seventeen hours. Thus, while TNFα and/or IL-1β are likely to contribute to the regulation of several clock genes, additional factors appear to regulate clock gene expression in human PBL during the acute period of systemic inflammation.
Boivin et al. [9] were the first to examine circadian clock gene expression in human PBMCs. Analysis of PBL circumvents the need for cell-purification step(s), minimizes manipulation time, and hence is a practical approach for sample acquisition in the clinical setting. However, the use of a mixed cell population such as PBL introduces some level of uncertainty as the proportion of each immune cell type changes dynamically after endotoxin challenge, returning to baseline within 12 hours post-treatment [22, 43, 44]. To address this potential limitation, clock gene expression was compared among PBL, monocytes, and neutrophils at select time points post-infusion. The studies revealed that the changes in clock gene expression observed in PBL were replicated in neutrophils. Interestingly, by 6 hours post-endotoxin infusion, several but not all clock genes, were also suppressed in monocytes. These findings establish that the changes in circadian clock gene expression observed in PBL are primarily reflective of changes occurring in neutrophils, and to a lesser extent in monocytes.
In conclusion, our study defines a severe misalignment of central circadian entrainment cues from peripheral clock gene expression in PBL during acute systemic inflammation. Godin and Buchman [45] proposed that systemic inflammatory responses might trigger uncoupling between biological oscillators. Others have reported that the circadian rhytmicity of melatonin secretion is suppressed in severely ill patients [46, 47], while in rodents, absence of circadian cues during recovery from sepsis impairs survival [48]. These findings add a new element to the potential link between circadian regulation and disease.
Supplementary Material
Blood was collected at the indicated time. Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 h infusion time, taken to be time 0.
Blood was collected at time 0, 6 and 24 hours post-endotoxin infusion. Total peripheral blood leukocytes, neutrophils and monocytes subpopulations, were obtained as described in the Materials and Methods section. Where indicated, leukocytes, neutrophils, and monocytes were isolated from single donors. Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 infusion time, taken to be time 0.
The gene expression data for each subject was cross-correlated across all studied genes within a subject. The cross-correlation data were normalized to one, where one represents a perfect correlation between gene responses. The correlation matrices for (A) placebo day and night subjects (09:00 h plus 21:00 h saline infusion time; n=3), (B) endotoxin day subjects (09:00 h infusion; n=4), (C) endotoxin night subjects (21:00 h infusion; n=3), (D) endotoxin day and night subjects (09:00 h plus 21:00 h endotoxin infusion time; n=7), were clustered into a tree form.
Circadian gene expression values were examined in PBL obtained at 02:00 h pre- and post-infusion (n=4 subjects). Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 h infusion time, taken to be time 0. Data are expressed as Mean± SEM. Statistical analysis was performed using unpaired Student's t-test. Statistical significance was designated at the 95% confidence level
Acknowledgments
The authors thank Ashwini Kumar, Marie Macor, Michael Reddell, and Zhiyong Zhang for excellent technical assistance. We are grateful to Eric Mintz (Kent State University, Ohio) for confirming the clustering analysis.
This study was supported by the National Institutes of Health-NIGMS (grant no. GM36495)
Abbreviations
- IL
interleukin
- PBL
peripheral blood leukocytes
- PBMCs
peripheral blood mononucleated cells
- SCN
suprachiasmatic nucleus
- TNF
tumor necrosis factor
Footnotes
The authors have no potential conflicts of interest to disclose.
References
- 1.Turek FW. Are the suprachiasmatic nuclei the location of the biological clock in mammals? Nature. 1981;292(5821):289–290. doi: 10.1038/292289a0. [DOI] [PubMed] [Google Scholar]
- 2.Ishida N, Kaneko M, Allada R. Biological clocks. Proc Natl Acad Sci U S A. 1999;96(16):8819–8820. doi: 10.1073/pnas.96.16.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev. 2003;13(3):271–277. doi: 10.1016/s0959-437x(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12(7):540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnason GA, Jordan RC, Wood PA, Li Q, Lincoln DW, Sothern RB, Hrushesky WJ, Ben-David Y. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158(5):1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 9.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102(12):4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 10.Born J, Lange T, Hansen K, Molle M, Fehm HL. Effects of sleep and circadian rhythm on human circulating immune cells. J Immunol. 1997;158(9):4454–4464. [PubMed] [Google Scholar]
- 11.Dimitrov S, Benedict C, Heutling D, Westermann J, Born J, Lange T. Cortisol and epinephrine control opposing circadian rhythms in T cell subsets. Blood. 2009;113(21):5134–5143. doi: 10.1182/blood-2008-11-190769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange T, Dimitrov S, Fehm HL, Westermann J, Born J. Shift of monocyte function toward cellular immunity during sleep. Arch Intern Med. 2006;166(16):1695–1700. doi: 10.1001/archinte.166.16.1695. [DOI] [PubMed] [Google Scholar]
- 13.O'Connor MF, Motivala SJ, Valladares EM, Olmstead R, Irwin MR. Sex differences in monocyte expression of IL-6: role of autonomic mechanisms. Am J Physiol Regul Integr Comp Physiol. 2007;293(1):R145–151. doi: 10.1152/ajpregu.00752.2006. [DOI] [PubMed] [Google Scholar]
- 14.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Yang Z, Niu Z, Wang W, Peng J, Li Q, Ma MY, Zhao Y. The mortality of MOP3 deficient mice with a systemic functional failure. J Biomed Sci. 2006;13(6):845–851. doi: 10.1007/s11373-006-9108-4. [DOI] [PubMed] [Google Scholar]
- 16.Oishi K, Ohkura N, Kadota K, Kasamatsu M, Shibusawa K, Matsuda J, Machida K, Horie S, Ishida N. Clock mutation affects circadian regulation of circulating blood cells. J Circadian Rhythms. 2006;4:13. doi: 10.1186/1740-3391-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coogan AN, Wyse CA. Neuroimmunology of the circadian clock. Brain Res. 2008;1232:104–112. doi: 10.1016/j.brainres.2008.07.087. [DOI] [PubMed] [Google Scholar]
- 18.Kelley KW, Bluthe RM, Dantzer R, Zhou JH, Shen WH, Johnson RW, Broussard SR. Cytokine-induced sickness behavior. Brain Behav Immun. 2003;17 1:S112–118. doi: 10.1016/s0889-1591(02)00077-6. [DOI] [PubMed] [Google Scholar]
- 19.Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9(1):46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, Fontana A. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci U S A. 2007;104(31):12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada K, Yano M, Doki Y, Azama T, Iwanaga H, Miki H, Nakayama M, Miyata H, Takiguchi S, Fujiwara Y, et al. Injection of LPS causes transient suppression of biological clock genes in rats. J Surg Res. 2008;145(1):5–12. doi: 10.1016/j.jss.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Lowry SF. Human endotoxemia: a model for mechanistic insight and therapeutic targeting. Shock. 2005;24 1:94–100. doi: 10.1097/01.shk.0000191340.23907.a1. [DOI] [PubMed] [Google Scholar]
- 23.Andreasen AS, Krabbe KS, Krogh-Madsen R, Taudorf S, Pedersen BK, Moller K. Human endotoxemia as a model of systemic inflammation. Curr Med Chem. 2008;15(17):1697–1705. doi: 10.2174/092986708784872393. [DOI] [PubMed] [Google Scholar]
- 24.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–1037. doi: 10.1038/nature03985. Epub 2005 Aug 1031. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Spek CA, Verbon A, Aberson H, Pribble JP, McElgunn CJ, Turner T, Axtelle T, Schouten J, Van Der Poll T, Reitsma PH. Treatment with an anti-CD14 monoclonal antibody delays and inhibits lipopolysaccharide-induced gene expression in humans in vivo. J Clin Immunol. 2003;23(2):132–140. doi: 10.1023/a:1022528912387. [DOI] [PubMed] [Google Scholar]
- 27.Wiersinga WJ, Dessing MC, Kager PA, Cheng AC, Limmathurotsakul D, Day NP, Dondorp AM, van der Poll T, Peacock SJ. High-throughput mRNA profiling characterizes the expression of inflammatory molecules in sepsis caused by Burkholderia pseudomallei. Infect Immun. 2007;75(6):3074–3079. doi: 10.1128/IAI.01733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brazma A, Vilo J. Gene expression data analysis. FEBS Lett. 2000;480(1):17–24. doi: 10.1016/s0014-5793(00)01772-5. [DOI] [PubMed] [Google Scholar]
- 29.Datta S, Datta S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics. 2003;19(4):459–466. doi: 10.1093/bioinformatics/btg025. [DOI] [PubMed] [Google Scholar]
- 30.Van der Poll T, Lowry SF. Epinephrine inhibits endotoxin-induced IL-1 beta production: roles of tumor necrosis factor-alpha and IL-10. Am J Physiol. 1997;273(6 Pt 2):R1885–1890. doi: 10.1152/ajpregu.1997.273.6.R1885. [DOI] [PubMed] [Google Scholar]
- 31.Richardson RP, Rhyne CD, Fong Y, Hesse DG, Tracey KJ, Marano MA, Lowry SF, Antonacci AC, Calvano SE. Peripheral blood leukocyte kinetics following in vivo lipopolysaccharide (LPS) administration to normal human subjects. Influence of elicited hormones and cytokines Ann Surg. 1989;210(2):239–245. doi: 10.1097/00000658-198908000-00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James FO, Boivin DB, Charbonneau S, Belanger V, Cermakian N. Expression of clock genes in human peripheral blood mononuclear cells throughout the sleep/wake and circadian cycles. Chronobiol Int. 2007;24(6):1009–1034. doi: 10.1080/07420520701800736. [DOI] [PubMed] [Google Scholar]
- 33.van Deventer SJ, Buller HR, ten Cate JW, Aarden LA, Hack CE, Sturk A. Experimental endotoxemia in humans: analysis of cytokine release and coagulation, fibrinolytic, and complement pathways. Blood. 1990;76(12):2520–2526. [PubMed] [Google Scholar]
- 34.James FO, Cermakian N, Boivin DB. Circadian rhythms of melatonin, cortisol, and clock gene expression during simulated night shift work. Sleep. 2007;30(11):1427–1436. doi: 10.1093/sleep/30.11.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kusanagi H, Hida A, Satoh K, Echizenya M, Shimizu T, Pendergast JS, Yamazaki S, Mishima K. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neurosci Res. 2008;61(2):136–142. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278(42):41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 37.Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. 2004;24(2):584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takimoto M, Hamada A, Tomoda A, Ohdo S, Ohmura T, Sakato H, Kawatani J, Jodoi T, Nakagawa H, Terazono H, et al. Daily expression of clock genes in whole blood cells in healthy subjects and a patient with circadian rhythm sleep disorder. Am J Physiol Regul Integr Comp Physiol. 2005;289(5):R1273–1279. doi: 10.1152/ajpregu.00126.2005. [DOI] [PubMed] [Google Scholar]
- 39.Teboul M, Barrat-Petit MA, Li XM, Claustrat B, Formento JL, Delaunay F, Levi F, Milano G. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med. 2005;83(9):693–699. doi: 10.1007/s00109-005-0697-6. [DOI] [PubMed] [Google Scholar]
- 40.Arendt J. Melatonin: characteristics, concerns, and prospects. J Biol Rhythms. 2005;20(4):291–303. doi: 10.1177/0748730405277492. [DOI] [PubMed] [Google Scholar]
- 41.Czeisler CA, Kronauer RE, Allan JS, Duffy JF, Jewett ME, Brown EN, Ronda JM. Bright light induction of strong (type 0) resetting of the human circadian pacemaker. Science. 1989;244(4910):1328–1333. doi: 10.1126/science.2734611. [DOI] [PubMed] [Google Scholar]
- 42.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajkrt D, Camoglio L, Tiel-van Buul MC, de Bruin K, Cutler DL, Affrime MB, Rikken G, van der Poll T, ten Cate JW, van Deventer SJ. Attenuation of proinflammatory response by recombinant human IL-10 in human endotoxemia: effect of timing of recombinant human IL-10 administration. J Immunol. 1997;158(8):3971–3977. [PubMed] [Google Scholar]
- 44.Talwar S, Munson PJ, Barb J, Fiuza C, Cintron AP, Logun C, Tropea M, Khan S, Reda D, Shelhamer JH, et al. Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol Genomics. 2006;25(2):203–215. doi: 10.1152/physiolgenomics.00192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godin PJ, Buchman TG. Uncoupling of biological oscillators: a complementary hypothesis concerning the pathogenesis of multiple organ dysfunction syndrome. Crit Care Med. 1996;24(7):1107–1116. doi: 10.1097/00003246-199607000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Mundigler G, Delle-Karth G, Koreny M, Zehetgruber M, Steindl-Munda P, Marktl W, Ferti L, Siostrzonek P. Impaired circadian rhythm of melatonin secretion in sedated critically ill patients with severe sepsis. Crit Care Med. 2002;30(3):536–540. doi: 10.1097/00003246-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 47.Olofsson K, Alling C, Lundberg D, Malmros C. Abolished circadian rhythm of melatonin secretion in sedated and artificially ventilated intensive care patients. Acta Anaesthesiol Scand. 2004;48(6):679–684. doi: 10.1111/j.0001-5172.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- 48.Carlson DE, Chiu WC. The absence of circadian cues during recovery from sepsis modifies pituitary-adrenocortical function and impairs survival. Shock. 2008;29(1):127–132. doi: 10.1097/shk.0b013e318142c5a2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blood was collected at the indicated time. Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 h infusion time, taken to be time 0.
Blood was collected at time 0, 6 and 24 hours post-endotoxin infusion. Total peripheral blood leukocytes, neutrophils and monocytes subpopulations, were obtained as described in the Materials and Methods section. Where indicated, leukocytes, neutrophils, and monocytes were isolated from single donors. Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 infusion time, taken to be time 0.
The gene expression data for each subject was cross-correlated across all studied genes within a subject. The cross-correlation data were normalized to one, where one represents a perfect correlation between gene responses. The correlation matrices for (A) placebo day and night subjects (09:00 h plus 21:00 h saline infusion time; n=3), (B) endotoxin day subjects (09:00 h infusion; n=4), (C) endotoxin night subjects (21:00 h infusion; n=3), (D) endotoxin day and night subjects (09:00 h plus 21:00 h endotoxin infusion time; n=7), were clustered into a tree form.
Circadian gene expression values were examined in PBL obtained at 02:00 h pre- and post-infusion (n=4 subjects). Gene expression levels were analyzed by qRT-PCR and are expressed as fold change relative to the 09:00 h infusion time, taken to be time 0. Data are expressed as Mean± SEM. Statistical analysis was performed using unpaired Student's t-test. Statistical significance was designated at the 95% confidence level