Abstract
Primary cilia are evolutionarily conserved cellular organelles that organize diverse signaling pathways1,2. Defects in the formation or function of primary cilia are associated with a spectrum of human diseases and developmental abnormalities3. Genetic screens in model organisms have discovered core machineries of cilium assembly and maintenance4. However, regulatory molecules that coordinate the biogenesis of primary cilia with other cellular processes, including cytoskeletal organization, vesicle trafficking and cell-cell adhesion, remain to be identified. Here we report the results of a functional genomic screen using RNA interference (RNAi) to identify human genes involved in ciliogenesis control. The screen identified 36 positive and 13 negative ciliogenesis modulators, which include molecules involved in actin dynamics and vesicle trafficking. Further investigation demonstrated that blocking actin assembly facilitates ciliogenesis by stabilizing the pericentrosomal preciliary compartment (PPC), a previously uncharacterized compact vesiculotubular structure storing transmembrane proteins destined for cilia during the early phase of ciliogenesis. PPC was labeled by recycling endosome markers. Moreover, knockdown of modulators that are involved in the endocytic recycling pathway affected the formation of PPC as well as ciliogenesis. Our results uncover a critical regulatory step that couples actin dynamics and endocytic recycling with ciliogenesis, and also provide potential target molecules for future study.
Cilium assembly and disassembly are interconnected with several complex cellular processes such as cell cycle, cell polarization and cell migration, suggesting that there may be large numbers of ciliogenesis modulators1,5. In accordance with this, recent proteomic analyses together with comparative genomics and bioinformatics studies have identified over a thousand cilia- or basal body-associated proteins, referred to as the “ciliome”6,7. Although these approaches can provide a comprehensive list of candidates, the discovery of key regulators of ciliogenesis, which could reveal potential therapeutic targets, requires functional analysis. Thus we developed a high-throughput assay using small interfering RNA (siRNA) to evaluate the functional impact of 7784 therapeutically relevant genes across the human genome (Supplementary Table 1). The screen was based on an in vitro ciliogenesis model: serum starvation-induced ciliogenesis in telomerase-immortalized human retinal pigmented epithelial (htRPE) cells. EGFP-tagged Smoothed (Smo), a transmembrane protein that accumulates in primary cilia8, was used as a cilium marker for automated quantification of ciliated cells (Supplementary Fig. 1a,b,c). Sensitivity of the screen strategy was assessed by siRNAs targeting KIF3A, a critical protein for cilium assembly (Supplementary Fig. 1d). We selected a screen condition suitable for identifying both positive and negative modulators: siRNA transfection was performed at a reduced (sub-optimal) cell density in order to detect these dynamic ranges of ciliogenesis activity, and ciliation was assessed after 48 hr serum starvation (Supplementary Fig. 1e,f).
The primary screen identified 153 positive modulator hits and 79 negative modulator hits (Supplementary Table 1 and Supplementary Fig. 2a). We next performed a confirmation screen with the following modifications: (1) optimal cell density for ciliogenesis (confluent at the time of serum starvation) was used to minimize the selection of siRNAs which indirectly influence ciliogenesis through an effect on cell density, (2) cilium length was measured for validating negative modulators because length increase was a highly specific indicator for enhanced ciliogenesis, where optimal ciliogenesis condition was used (Supplementary Fig. 2b). The confirmation screen identified 40 positive and 13 negative modulators, which are associated with various molecular processes (Supplementary Table 2 and Supplementary Fig. 3). Remarkably, the screen hits include INPP5E, a gene recently identified as mutated in a human ciliopathy, Joubert syndrome 9. A positive modulator Agtpbp1 (formerly Nna1) is mutated in Purkinje cell degeneration (pcd) mice, which exhibit ciliopathy phenotypes: retinal degeneration and defective spermatogenesis10. Although the library was selected for therapeutically relevant genes and thus not highly representative of ciliome genes (tending to be structural), we did confirm 12 genes encoding ciliome proteins (Supplementary Table 3 and Supplementary Fig. 3).
We rescreened the confirmed positive modulators using anti-acetylated tubulin immunofluorescence as a cilium marker, and found 4 genes that did not affect cilium assembly (Supplementary Table 3 and Supplementary Fig. 3). The confirmed negative modulators were further classified according to silencing phenotypes observed in the absence of serum starvation. Silencing of 5 genes facilitated ciliogenesis even without serum-starvation (Supplementary Table 3 and Supplementary Fig. 3).
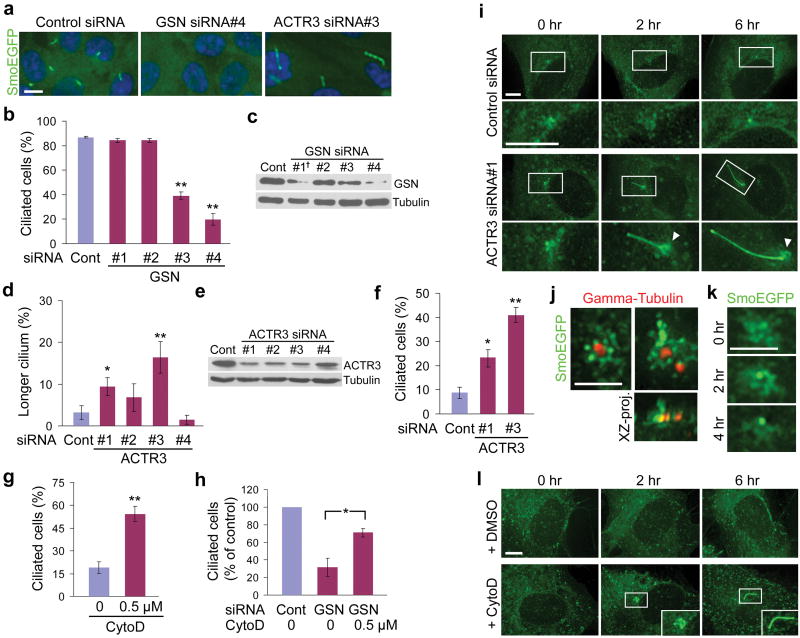
Among the ciliogenesis modulators with high confirmation screen scores (Supplementary Table 2) are two Gelsolin (GSN) family proteins GSN and AVIL, which regulate cytoskeletal actin organization by severing actin filaments11. Depletion of GSN proteins by two independent siRNAs significantly reduced ciliated cell numbers, suggesting that actin filament severing is involved in ciliogenesis (Fig. 1a,b,c). In contrast, silencing of actin-related protein ACTR3 (also called ARP3), which is a major constituent of the ARP2/3 complex that is necessary for nucleating actin polymerization at filament branches12, caused a significant increase in cilium length (Fig. 1a,d,e) and also facilitated ciliogenesis independent of serum starvation (Fig. 1f). These observations suggest an inhibitory role of branched actin network formation in ciliogenesis.
Figure 1. Regulators of actin dynamics modulate cilium assembly.
a, b, GSN knockdown reduced ciliated cell numbers. a, d, ACTR3 knockdown increased the number of cells with longer cilium (> 6 μm). c, e, Western blots showing protein levels (†GSN siRNA#1 was cytotoxic). f, ACTR3 knockdown induced ciliogenesis without serum starvation. g, CytochalasinD treatment for 8 hrs increased ciliated cell numbers without serum starvation. h, Ciliogenesis defect by GSN knockdown was rescued by 8 hr cytochalasinD treatment. i, Live imaging of htRPE-SmoEGFP cells after 60 hr siRNA transfection. Arrowheads, the pericentrosomal preciliary compartment (PPC) at the ciliary base. j, Morphology of PPC. Gamma-tubulin labels the centrosome. k, Live imaging of a PPC positive cell in serum-free medium. l, Live cell imaging in serum-free medium containing DMSO or cytochalasinD. Values, mean ± SD [n = 4 (b,d), 2 (f) and 3 (g,h)]. Student's t-test: *P < 0.05, **P < 0.01. Scale bars, 10 μm (a); 5 μm (i,l); 2.5 μm (j,k).
To examine the link between actin dynamics and ciliogenesis, we treated htRPE cells with actin polymerization inhibitor cytochalasinD, which facilitated ciliogenesis independent of serum starvation and promoted cilium elongation (Fig. 1g and Supplementary Fig. 4). Ciliogenic effect of cytochalasinD was also observed in HEK293T cells (Supplementary Fig. 5a,b). Moreover, cytochalasinD treatment significantly rescued ciliogenesis defect caused by GSN knockdown (Fig. 1h). CytochalasinD facilitated ciliogenesis at doses below that which affect stress fiber formation, excluding the possibility of global actin cytoskeleton rearrangement in ciliogenesis control (Supplementary Fig. 5c). Involvement of actin dynamics in ciliogenesis is further supported by the finding that α-PARVIN (PARVA), a component of the focal adhesion complex regulating actin cytoskeletal dynamics and cell signaling13, exhibits knockdown phenotypes similar to that of ACTR3 (Supplementary Fig. 6).
In order to identify the mechanism of altered ciliogenesis, we performed time-course live imaging. Remarkably, a number of non-ciliated htRPE cells transfected with ACTR3 siRNAs (14/30 cells) developed pronounced primary cilia (2.9±0.9 μm in length) within 2 hrs of serum starvation, whereas none of the control cells (0/30 cells) displayed cilia longer than 1.5 μm (Fig. 1i and Supplementary Fig. 7a). Furthermore, faster cilium elongation after the initiation of ciliogenesis was observed in ACTR3 depleted cells [mean cilium extension rate for first 6 hrs after serum starvation: ACTR3 depleted cells, 0.75±0.27 μm/hr (n=17); control cells, 0.11±0.06 μm/hr (n=6)]. These observations suggest that slow progression of ciliogenesis in htRPE cells is not the result of slow transduction of serum starvation-mediated ciliogenic signal or unavailability of core cilium assembly machineries, but is ascribed to the presence of an inhibitory regulation involving ACTR3.
Earlier electron microscopic studies found small vesicles tightly associated with the centriole initiating cilium assembly in fibroblasts and smooth muscle cells14,15. This suggests that ciliogenesis in non-polarized cells could be initiated by vesicle docking to the basal body, whereas basal body docking to the apical plasma membrane initiates ciliogenesis in polarized epithelial cells. More recent studies also have reported that the biogenesis of ciliary membrane that is coordinated with axoneme assembly may involve fusion of transport vesicles at the base of cilia16,17. SmoEGFP allowed us to visualize transport vesicles targeted to the ciliary base. Interestingly, a subset of htRPE-SmoEGFP cells (∼10 %: 42/434 interphase cells) cultured in the presence of serum exhibited compact SmoEGFP positive vesiculotubular structures (termed here PPC) tightly associated with the centrosome (Fig. 1J). In these cells, ciliogenesis occurred at PPC during 4 hr live imaging (Fig. 1k). Notably, an attenuation of SmoEGFP positive PPC was observed after the initiation of ciliogenesis (Fig. 2k and Supplementary Fig. 7b, control cells). After 4 hr serum starvation (when cilia are detected in ∼20 % of cells), 3.4 % of total cilia (17/500 cilia) were accompanied by PPC, suggesting that the transition of PPC to cilia occurs over the course of hours. PPC positive cilia were rarely observed after 24 hr serum starvation (2/500 cilia), confirming that PPC is a transient structure (Supplementary Fig. 8a). SmoEGFP revealed similar transient structures in IMCD cells (Supplementary Fig. 8b).
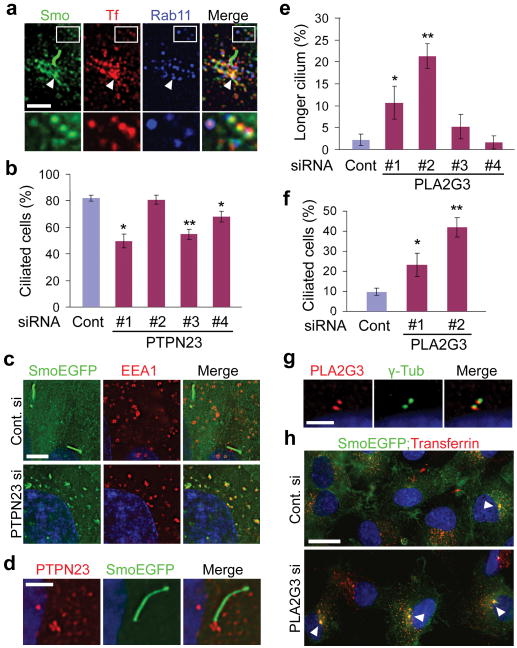
Figure 2. Endocytic recycling pathway is linked to ciliogenesis.
a, PPC overlapping with endocytosed transferrin-alexa594 and Rab11 (arrowhead). b, PTPN23 knockdown decreased the number of ciliated cells. c, PTPN23 knockdown caused an accumulation of SmoEGFP on EEA1-positive early endosomes. d, PTPN23 immunofluorescence was detected at the ciliary base. e, PLA2G3 knockdown increased the number of cells with longer cilia (> 6 μm). f, PLA2G3 knockdown induced ciliogenesis without serum starvation. g, Pericentriolar localization of PLA2G3. h, PLA2G3 knockdown increased the number of cells exhibiting recycling endosomes concentrated at high levels around the centrosome (arrowheads). Values, mean ± SD [n = 4 (b,e) and 3 (f)]. Student's t-test: *P < 0.05, **P < 0.01. Scale bars, 2.5 μm (a,d,g); 5 μm (c); 20 μm (h).
Remarkably, the majority of newly formed cilia (13/17 cilia) displaying mature lengths (4.5±1.9 μm) in ACTR3 depleted-live cells maintained pronounced PPC at the ciliary base (Fig. 1i and Supplementary Fig. 7b) although PPC eventually disappeared after 24 hr serum starvation as in control (Supplementary Fig. 8a). Moreover, facilitated ciliogenesis in cytochalasinD treated cells was preceded by a promotion of PPC formation [16 PPC positive cells/30 live-imaged cells treated with cytochalasinD for 2 hrs vs. 5 PPC positive cells/30 control cells (Fig. 1l)]. Short-term live imaging showed that cytochalasinD also affects the stability of the pre-existing PPC (Supplementary Fig. 9). These observations suggest that actin network formation may negatively modulate ciliogenesis by destabilizing the PPC, a potential temporary reservoir of lipid and membrane proteins for efficient ciliogenesis.
Recycling endosomes are a dynamic vesiculotubular compartment, exporting endocytosed membrane proteins and lipids to the cell surface via vesicular intermediates18. We found that PPC extensively overlaps with a subset of recycling endosomes concentrated around the centrosome: PPC was labeled by endocytosed transferrin and recycling endosome marker Rab11 (Fig. 2a and Supplementary Fig. 10). PTPN23, a nontransmembrane tyrosine phosphatase, has been implicated in cargo sorting at early endocytic compartments19, and is one of the high-scored hits. Silencing of PTPN23 significantly reduced the number of ciliated cells (Fig. 2b and Supplementary Fig.11). In addition, PTPN23 knockdown caused an accumulation of SmoEGFP on the early endosomes marked by EEA1 (Fig. 2c). PPC was not observed from the cells exhibiting early endosomal SmoEGFP accumulation (0/80 cells; Supplementary Fig. 12). Supporting the link between recycling endosomes and ciliogenesis, knockdown of ASAP1 (not included in the initial screen library), a gene required for pericentrosomal enrichment of recycling endosomes20, caused a decrease in the number of ciliated cells (Supplementary Fig. 13a,b,c). These observations suggest that a trafficking pathway connecting endocytic compartments to PPC is involved in cilium assembly. Pharmacological blocking of endocytic degradation pathway to lysosomes using concanamycinA did not block PPC formation or ciliogenesis, indicating that endocytic recycling pathway is specifically involved in ciliogenesis (Supplementary Fig. 13d). Detection of PTPN23 immunoreactivity at the basal bodies further supported a direct role for PTPN23 in ciliary vesicle targeting (Fig. 2d).
Among the unexpected screen hits was PLA2G3, a secreted phospholipase21. Involvement of cytoplasmic Phospholipase A2 enzymes in intracellular vesicle trafficking has been suggested based on their potential for introducing membrane curvature22. Depletion of PLA2G3 facilitated cilium extension and induced ciliogenesis independent of serum starvation, indicating that PLA2G3 is a negative ciliogenesis regulator (Fig. 2e,f and Supplementary Fig. 14). Confirming this result, application of oleyloxyethyl phosphorylcholine (OPC), an inhibitor for secreted phospholipase23, increased the number of ciliated cells (Supplementary Fig. 15a). Notably, PLA2G3 immunofluorescence was detected at the centriole pair (Fig. 2g). In accord with its localization, knockdown of PLA2G3 significantly increased the number of cells displaying recycling endosomes concentrated at high levels around the centrosome [17.4±1.4 % in control cells and 48.2±2.7 % in PLA2G3 depleted cells examined after 56 hr transfection without serum starvation; Student's t-test, P=0.006 (Fig. 2h)]. The majority of pericentrosomal recycling endosomes in PLA2G3 depleted cells (122/153) were associated with PPC or cilia. Overexpressed PLA2G3 exhibited an extensive co-localization with endocytosed transferrin and inhibited ciliogenesis (Supplementary Fig. 14d,e,f). These results further demonstrate the link between endocytic recycling pathway and ciliogenesis. Moreover, higher intensities of SmoEGFP fluorescence was observed in cilia from cells depleted of PLA2G3, demonstrating an inhibitory role for PLA2G3 in ciliary membrane protein targeting (Supplementary Fig. 15b,c).
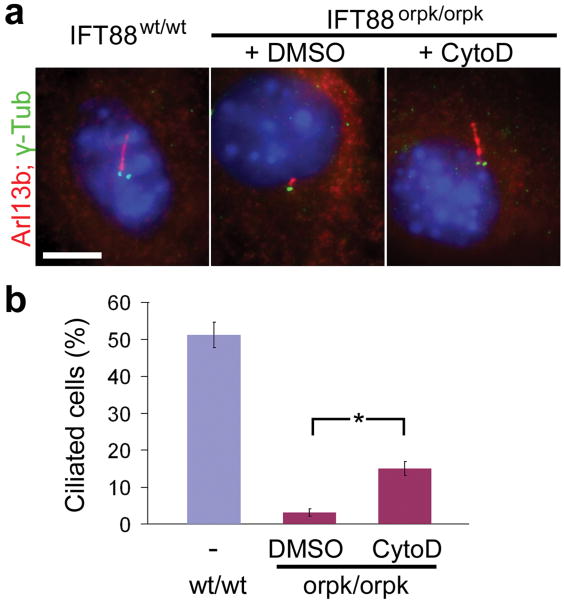
The fact that both axoneme assembly and ciliary membrane biogenesis were facilitated by inhibition of actin polymerization (Supplementary Fig. 16) prompted us to test if cytochalasinD treatment could rescue ciliogenesis defect caused by loss-of-function mutation in core ciliogenesis machinery. IFT88 transports molecules required for cilium assembly4, and cells carrying homozygous IFT88 hypomorphic mutation (orpk/orpk) fail to grow cilia with normal length24. Remarkably, 16 hr cytochalasinD treatment partially, but significantly, rescued ciliogenesis defect of IFT88 mutant cells (Fig. 3). A recent study showed that IFT particles are associated with recycling endosomes and are implicated in the recycling of T-cell receptors in lymphocytes25. Thus it is likely that cytochalasinD facilitates ciliary recruitment of IFT particles through the endocytic recycling pathway. CytochalasinD can interfere with many cellular processes involving actin polymerization, and thus is unlikely to be useful for ciliopathy treatment. However, small molecules specifically targeting actin modulators implicated in ciliogenesis might be good candidates for ciliopathy treatment.
Figure 3. Pharmacological rescue of ciliary defect on IFT88 mutant cells.
a, Cytochalasin D treatment for 16 hrs partially rescued cilium elongation defect caused by hypormorphic IFT88 mutation. b, Percentage of cells with the primary cilium longer than 1.5 μm. Values, mean ± SD (n = 3). Student's t-test: *P < 0.05. Scale bar, 5 μm.
In summary, our high-throughput functional screen identified modulators of ciliogenesis with diverse molecular functions, connecting ciliogenesis with other basic cellular processes. Furthermore, we discovered several proteins involved in the control of cilium length, a key determinant of normal cilium function. We anticipate that the development of small molecules that target these proteins may provide novel strategies for the treatment of the ciliopathies.
Method Summary
Cell culture
htRPE cells were maintained in DMEM:F12 supplemented with 10 % fetal bovine serum. Plasmid DNA harboring mouse SmoEGFP fusion gene was transfected to htRPE cells and a stable cell line (htRPE-SmoEGFP) was established by G418 selection. For ciliogenesis induction, culture medium was replaced with DMEM (without supplement) when cells are ∼90 % confluent, and cultured for 48 hrs before fixation. Overexpressed SmoEGFP was targeted to ciliary membrane independent of Shh.
siRNA library screen
An arrayed library containing 31111 unique siRNAs targeting 7784 human genes (Ambion; human druggable genome siRNA library V3.1; including siRNAs targeting 665 Kinases, 101 Kinase modulators, 231 Phosphatases, 307 Ligases, 57 Lipases, 444 Signaling molecules and 426 Cytoskeleton interacting proteins) was screened in duplicates. A high-throughput imaging was done using IC100 automated microscope system (Beckman Coulter), and the images were analyzed by Cytoshop software. For a confirmation screen, re-arrayed 913 siRNAs targeting 232 primary screen hits were tested in quadruplicates.
Supplementary Material
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Acknowledgments
The screen was performed at the Burnham Institute Functional Genomics Core (LJINCC grant P30 NS057096 to S.Lipton) with technical assistance from A.Cortez and E.Sisman and with imaging support from the UCSD Microscopy Core (P30 NS047101 and P30 CA23100). We thank P.A.Beachy for SmoEGFP plasmid and J.F.Reiter for IFT88orpk/orpk MEFs. We thank C.Kintner, V.M.Fowler, S.J.Field and S.Ferro-Novick for discussion. This work was supported by grants from NARSAD (Young Investigator Award to J.K.), NINDS (RO1 NS052455 to J.G.G) and the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper a www.nature.com/nature.
Author Contributions J.G.G. conceived and directed the project. J.K., S.H., and P.A. designed the screen and J.K., S.H. and E.S. performed the screen. J.K., K.O., K.L. and T.I. analyzed the screen data. J.K. designed the follow-up experiments and J.K. and J.L. performed the experiments. J.K. interpreted the results and J.K. and J.G.G. wrote the paper with contributions from S.H and P.A.
The authors declare competing financial interests (patent application).
References
- 1.Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum JL, Witman GB. Intraflagellar transport. Nature Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 5.Christensen ST, et al. The primary cilium coordinates signaling pathways in cell cycle control and migration during development and tissue repair. Curr Top Dev Biol. 2008;85:261–301. doi: 10.1016/S0070-2153(08)00810-7. [DOI] [PubMed] [Google Scholar]
- 6.Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nature Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 8.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 9.Bielas SL, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nature Genet. 2009;41:1032–1036. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Gonzalez A, et al. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295:1904–1906. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- 11.Silacci P, et al. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- 12.Cooper JA, Schafer DA. Control of actin assembly and disassembly at filament ends. Curr Opin Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 13.Sepulveda JL, Wu C. The parvins. Cell Mol Life Sci. 2006;63:25–35. doi: 10.1007/s00018-005-5355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archer FL, Wheatley DN. Cilia in cell-cultured fibroblasts. II. Incidence in mitotic and post-mitotic BHK 21-C13 fibroblasts. J Anat. 1971;109:277–292. [PMC free article] [PubMed] [Google Scholar]
- 16.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura S, et al. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxfield FR, McGraw TE. Endocytic recycling. Nature Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 19.Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue H, Ha VL, Prekeris R, Randazzo PA. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol Biol Cell. 2008;19:4224–4237. doi: 10.1091/mbc.E08-03-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valentin E, et al. Novel human secreted phospholipase A(2) with homology to the group III bee venom enzyme. J Biol Chem. 2000;275:7492–7496. doi: 10.1074/jbc.275.11.7492. [DOI] [PubMed] [Google Scholar]
- 22.Brown WJ, Chambers K, Doody A. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic. 2003;4:214–221. doi: 10.1034/j.1600-0854.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- 23.Hope WC, Chen T, Morgan DW. Secretory phospholipase A2 inhibitors and calmodulin antagonists as inhibitors of cytosolic phospholipase A2. Agents Actions. 1993;39:C39–42. doi: 10.1007/BF01972714. Spec No. [DOI] [PubMed] [Google Scholar]
- 24.Yoder BK, et al. Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am J Physiol Renal Physiol. 2002;282:F541–552. doi: 10.1152/ajprenal.00273.2001. [DOI] [PubMed] [Google Scholar]
- 25.Finetti F, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nature Cell Biol. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.