Abstract
Objective
CCL3L and CCL4L genes encode HIV-suppressive chemokines, colocalize on chromosome 17q12 and have copy number variation. Copy number variation of CCL3L associates with HIV-AIDS susceptibility. Here, we determined the influence of the combinatorial content of distinct CCL3L and CCL4L genes on HIV-AIDS susceptibility.
Methods
By designing gene-specific assays, the association between doses of all CCL3L or CCL4L genes or their individual duplicated components (CCL3La/b and CCL4La/b) with HIV-AIDS susceptibility was determined in 298 perinatally exposed Ukrainian children.
Results
The odds of transmission was increased in children with less than two copies of CCL3L or CCL4L, compared with those with at least two copies, and 10-fold higher when both mother and offspring had less than two CCL3L or CCL4L copies, compared with mother–child pairs with at least two copies. The extent of the pair-wise correlations between CCL3La, CCL3Lb, CCL4La and CCL4Lb copy number varied extensively, with an inverse correlation between CCL4L genes that transcribe a classical chemokine (CCL4La) versus aberrantly-spliced transcripts (CCL4Lb). Children possessing only CCL4Lb progressed four times faster to AIDS than those with only CCL4La. A lower content of CCL3L and CCL4L genes that transcribe classical chemokines was associated with enhanced HIV-AIDS susceptibility.
Conclusion
Transmission risk is greatest when mother and offspring both have low CCL3L or CCL4L gene doses. The impact on HIV-AIDS susceptibility of the chemokine gene-rich locus on 17q12 is dependent on the balance between the doses of genes conferring protective (CCL3La and CCL4La) versus detrimental (CCL4Lb) effects. Hence, the combinatorial genomic content of distinct genes within a copy number variable region may determine disease susceptibility.
Keywords: AIDS, CCL3L, CCL4L, HIV, transmission
Introduction
The results of the recent STEP trial [1] showing that a HIV-1 vaccine may have increased the risk of acquiring HIV prompted a call to define with greater precision the correlates of protection. A powerful approach to accomplish this is to identify host genetic determinants that alter risk of HIV acquisition and disease progression rates. Identification of such determinants in children exposed perinatally to HIV can be especially informative because approximately 70–90% of such children resist infection, despite high exposure to virus and absence of antiretroviral prophylaxis [2]. Here, we investigated such children to determine the impact on HIV-AIDS susceptibility of duplicated genomic regions that are enriched for chemokine genes for whom in-vitro studies [3–10] implicate a central role for their encoded products in HIV-AIDS pathogenesis.
Copy numbers of DNA segments [denoted as copy number variations (CNVs)] are implicated in disease susceptibility [11–16]. A CNV with relevance to HIV-AIDS susceptibility is for the segmental duplication that contains the gene encoding CC chemokine ligand 3-like 1 (CCL3L1). CCL3L1 [macrophage inflammatory protein 1 alpha P (MIP-1αP)], a nonallelic isoform of CCL3 (MIP-1αS), is the most potent agonist of CC chemokine receptor 5 (CCR5), the major HIV coreceptor, and among CCR5 chemokine ligands it exhibits maximal HIV-suppressive properties [5–8]. Association studies for the Δ32 and other polymorphisms in CCR5 [17–23] established a key role for CCR5 expression in HIV pathogenesis. By analogy, the genotype–phenotype associations for the copy number of the CCL3L1-containing segmental duplication suggest a role for this structural variation in HIV-AIDS pathogenesis. Prior studies indicate that a low copy number of CCL3L1 is associated with an increased risk of acquiring HIV infection [24–28], higher viral loads [24,26,29,30], a faster rate of decline of CD4+ T-cell counts or progression to AIDS [24,30,31], impaired recovery of CD4+ T-cell counts [31] and function [30] during antiretroviral therapy (ART), lower HIV-specific CD4+ and CD8+ T-cell responses [29], reduced CCL3 chemokine levels [24,25,32], higher numbers of HIV target cells (%CD4+/CCR5+ cells) [24,33] and reduced cell-mediated immune responses [30]. Extending this concept to nonhuman primates, a low copy number of CCL3L genes in Asian and Chinese macaques is associated with an accelerated progression to experimental AIDS in these animals [34].
However, these previous studies have not accounted for the full impact on HIV-AIDS susceptibility of CNV of other chemokine genes that along with CCL3L1 localize to chromosome 17q12, a hot spot for segmental duplications [11,12]. These genes include two other CCL3L (CCL3L2 and CCL3L3) genes and two CCL4L (MIP-1β-like) genes designated as CCL4L1 and CCL4L2 [3,4,9,10,32]. One possibility was that if the CNV of all the CCL3L and CCL4L genes reside on a single segmental duplication, then the associations of these CNVs with HIV-AIDS susceptibility should be identical. However, this is unlikely because this genomic region is replete with many potential breakpoints that may lead to nonidentical duplicated segments [36]. Underscoring this, although there is a high degree of correlation between the copy number of CCL3L and CCL4L genes, individuals contain more copies of CCL3L than CCL4L [32]. We therefore tested the hypothesis that the overall phenotypic impact of the chemokine gene-rich locus at chromosome 17q12 region on HIV-AIDS susceptibility will be influenced by the combinatorial content of different CCL3L and CCL4L-containing segmental duplications. To test this, we developed specific probes for the distinct CCL3L and CCL4L genes and determined their associations with HIV-AIDS susceptibility in a cohort of children exposed perinatally to HIV-1 from the Ukraine.
Methods
Cohort
We studied a cohort of 298 children from Ukraine who were exposed perinatally to HIV-1 between 1998 and 2006. Of these, 178 (59.7%) were infected. This distribution is not indicative of the perinatal transmission rate for HIV-1 infection, as the recruitment of children was based on HIV status. DNA was also available from the mothers of 89 children. To minimize potential confounding due to population stratification, only children and mothers of Slavic descent, the most common ethnic group found in Ukraine, were included in the study. These patients were recruited at the main outpatient HIV-1 reference centers (‘Municipal Centers for HIV Management and Prophylaxis’) in the Dnepropetrovsk region. All pregnant women within this region who test positive for HIV are referred to these centers. Additionally, all HIV-positive children born to these mothers also receive their medical care in these centers. Informed written consent was obtained from parents or legal guardians of all the children, and directly from the adults. The study was approved by the Institutional Review Boards of the participating institutions. The characteristics of the study individuals are given in Table 1.
Table 1. Characteristics of the children included in the study.
| Parameter | HIV infected | HIV uninfected | P |
|---|---|---|---|
| Number of children | 178 (59.7%) | 120 (40.3%) | – |
| Did not receive ZDV prophylaxis | 123 (69.1%) | 44 (36.7%) | 3.2 × 10−8 |
| Received breastfeeding [n (%)] | 17 (9.6%) | 5 (4.1%) | 0.077 |
| Hemoglobin (g/dl) [mean (SE)] | 10.8 (0.09) | 11.1 (0.11) | 0.030 |
| RBC count [×106/μl; mean (SE)] | 4.21 (0.08) | 4.36 (0.29) | 0.264 |
| Baseline CD4+ T-cell count (cells/μl) [mean (SE)] | 968.2 (62.2) | 2515.8 (11.2.9) | <1 × 10−22 |
| Baseline CD4 percentage [mean (SE)] | 22.9 (0.82) | 38.8 (0.75) | <1 × 10−22 |
| Average age at initiation of HAART [years, mean (SE)] | 5.00 (0.35) | – | – |
| Children developing AIDS [n (%)] | 56 (31.5%) | – | – |
| Total duration of follow-up (person years) | 735.33 | – | – |
| Duration of follow-up [years, median (IQR)] | 7.63 (5.29) | – | – |
ZDV, zidovudine; P, significance value; SE, standard error.
Study outcomes
The main outcome measures were HIV serostatus and progression to AIDS [1993 criteria of the Center for Disease Control and Prevention (CDC) classification for children].
Genotyping
Taq Man real-time assays similar to those described previously [24] and described in the Supplementary Materials and Fig. S1 were used to quantify the CNV of CCL3L and CCL4L genes. Primer and probe sequences are listed in Supplementary Table 1.
Statistical analysis
The association between gene copy number and risk of acquiring HIV infection was assessed by using logistic regression analyses, and rate of disease progression was investigated by using Kaplan–Meier survival curves and Cox proportional hazards modeling. Attributable fraction was estimated as described previously [24]. The pattern of correlations among the copy numbers of the different CCL3L and CCL4L genes was determined using principal components analyses. For this, a factor solution provided by using a minimum eigenvalue of one with the results optimized using the varimax rotation was used. Details of factor analyses are as described in the Supplementary Material. All statistical analyses were conducted using the Stata 7.0 (Stata Corporation, College Station, Texas, USA) software package.
Results
Nomenclature and PCR assay precision
In this study, we denoted CCL3L1 (MIM:601395), CCL3L2 (MIM:609467), CCL3L3 (MIM:609468), CCL4L2 (MIM:610757) and CCL4L1 (MIM:603782) as CCL3La, CCL3Lb, CCL3La, CCL4La and CCL4Lb, respectively (Fig. 1a and b), for the following reasons. In prior studies [24–32], because of the nature of the PCR primer probes used, we and others assessed the copy number of all CCL3L genes (CCL3L1, CCL3L2, CCL3L3) and had previously designated this composite as the CCL3L1-containing segmental duplication (Fig. 1a and b). CCL3L1 and CCL3L3 are separate genes, each having three identical exons that encode identical proteins [3,5], and therefore they are together denoted here as CCL3La (CCL3L1 + CCL3L3 = CCL3La; Fig. 1a–c). On the basis of current literature, CCL3L2, designated here as CCL3Lb, is thought to contain only two exons whose sequences are identical to those found in exons 2 and 3 in CCL3L1 and CCL3L3 [3,5]. Because CCL3Lb (CCL3L2) lacked the first exon found in CCL3La (CCL3L1 or CCL3L3), it has been considered as a pseudogene [3,5]. However, by bioinformatics and mRNA profiling, we identified novel 5′ exons for CCL3Lb, which give rise to two alternatively spliced transcripts (CCL3Lb-v1 and CCL3Lb-v2, Fig. 1c and data not shown). These alternatively transcribed mRNA species contain chemokine-like domains but are not predicted to encode classical chemokines (data not shown). Hence, CCL3La and CCL3Lb transcribe two distinct kinds of transcripts; the former encoding classical chemokines and the latter encoding alternatively spliced transcripts with novel 5′ exons (Fig. 1c).
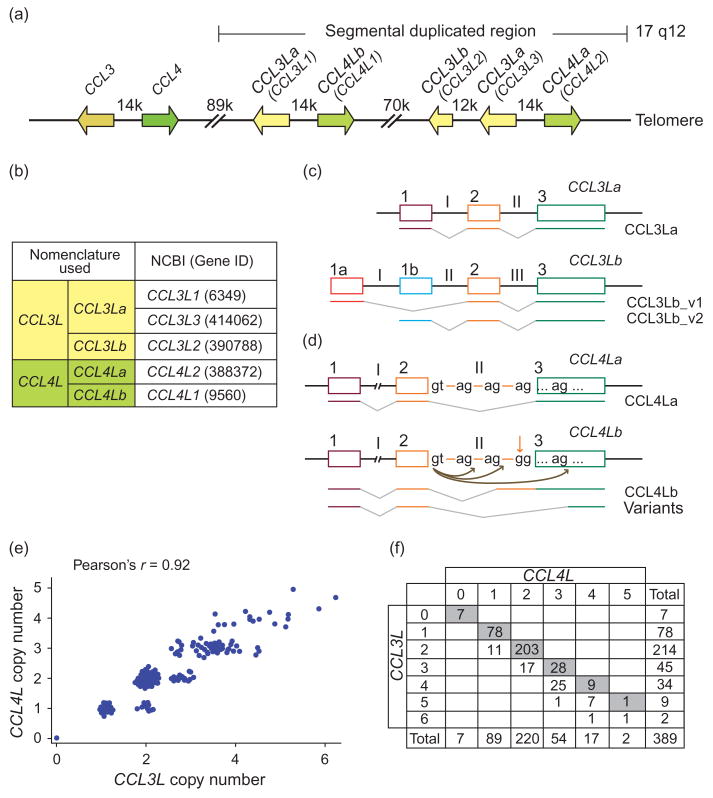
Fig. 1. Chromosome 17q12 segmental duplication, chemokine gene copy number variation nomenclature and correlation between copy number of CCL3L and CCL4L.
(a) Schematic representation of CCL3, CCL4, CCL3L and CCL4L genes. Arrows indicate the orientation of each gene. Shown on top is the distance between the indicated genes. Map is not to scale. (b) Nomenclature of CCL3L and CCL4L genes used here and previously [9,24,26,28,30,31]. (c) and (d) Schematic representation of the exon–intron structure of (c) CCL3L and (d) CCL4L genes. In (d), the downward pointing arrow indicates the A→G transition that leads to the generation of aberrantly spliced CCL4Lb transcripts, and the splicing patterns of these mRNA species are indicated by the curved arrows. Thus, a CCL4L copy, designated here as CCL4La, has the intact AG intron–exon splice sequence and is predicted to transcribe an intact full-length CCL4L transcript. By contrast, the other CCL4L copy, designated here as CCL4Lb, has a mutated intron–exon splice site sequence (GG), and this results in the formation of a set of aberrantly spliced transcripts that are predicted to produce CCL4L proteins that may not function as classical chemokines (as described by Colobran et al. [9]). Boxes represent exons, which are identified by Arabic numerals and the lines joining the boxes are the introns, which are identified by Roman numerals. Figure not to scale. (e) Scatter plot showing the correlation of the CCL3L and CCL4L genes copy numbers for all study individuals. (f) Distribution of CCL3L and CCL4L gene copy number among all study individuals. Grey represents number of individuals who have the same dose of CCL3L and CCL4L. The numbers below the grey boxes represent the number of individuals that have more copies of CCL3L than CCL4L.
Similarly, CCL4L represents a composite of CCL4Lb (CCL4L1) and CCL4La (CCL4L2), two distinct genes that have identical exonic sequences (Fig. 1a and b, [3,5]). Although these two genes have identical exonic sequences, a (A→G) transition in the splice acceptor site in the intron II of CCL4Lb relative to CCL4La leads to generation of aberrantly spliced CCL4Lb transcripts, that is, formation of transcripts with retention of portions of intron II or partial loss of exon 3 (Fig. 1d). Hence, CCL4La produces transcripts that are predicted to encode a classical chemokine, whereas CCL4Lb transcribes aberrantly spliced mRNA species (Fig. 1d).
On the basis of the aforementioned, we developed two separate assays to quantify the total copy number of all CCL3L or CCL4L genes, and separate assays each for the individual components of CCL3L (CCL3La and CCL3Lb) and CCL4L (CCL4La and CCL4Lb). The PCR assays we developed estimated the copy number of these individual chemokine gene CNVs with high precision (Fig. S1). The estimates of the sum of the copy number of CCL3La and CCL3Lb or CCL4La and CCL4Lb using these individual PCR assays were very similar to the estimate of the total copy number of CCL3L or CCL4L genes, respectively, with 100% concordance between the estimates for zero, one, two or at least two copies (data not shown).
CCL3L–CCL4L dose and HIV-AIDS risk
The distribution of CCL3L copy number in Ukrainian HIV-positive or HIV-negative children and HIV-positive mothers were similar to those observed in individuals of European descent [24], and the average copy number of CCL3L in this population was two (Fig. S2a). Although the copy number of CCL3L and CCL4L were highly correlated (92%, P < 0.0001), the gene dose of CCL3L was higher than that for CCL4L (Fig. 1e and f). Given this high correlation, predictably, the distribution of CCL4L and CCL3L were very similar (compare Fig. S2a and b), and therefore associations of the total CCL4L gene dose with HIV-AIDS susceptibility would be expected to be similar to those detected for the total CCL3L gene dose.
In a previous study [24], we found that in European–Americans or Argentinean children, possession of a copy number of CCL3L that was less than the average found in the overall population (two) was associated with an increased risk of acquiring HIV infection. Concordantly, among Ukrainian children exposed perinatally to HIV, possession of a copy number of CCL3L or CCL4L that was less than two was associated with a trend for a nearly 75% increased risk of acquiring HIV before or after accounting for receipt of zidovudine (ZDV) prophylaxis, respectively, compared with children who had at least two copies of CCL3L or CCL4L (Fig. 2a and b).
Fig. 2. Association of the CCL3L and CCL4L gene copy number with risk of acquiring HIV infection.
(a and b) The risk of HIV acquisition based on the (a) CCL3L and (b) CCL4L copy number in children. Left panel shows the distribution of CCL3L or CCL4L copy number categorized as less than two or at least two in the HIV-positive and HIV-negative children. Right panel shows results of logistic regression analyses unadjusted (shown in green) and adjusted (shown in red) for receipt of ZDV prophylaxis. The diamonds and error bars represent the OR and 95% CI, respectively. (c) Association of CCL3L copy number in the mother–child pairs with the risk of vertical transmission. The data on the left shows the distribution of the 89 mother–child pairs by gene copy number and the bar chart shows the proportion of children within each of the four categories who acquired HIV infection. The risk of HIV acquisition was estimated using multinomial logistic regression analysis which was unadjusted (left) and adjusted (right) for receipt of ZDV prophylaxis. ZDV, zidovudine; CI, confidence interval; OR, odds ratio; P, significance value.
We next determined whether risk of transmission differed based on whether a mother and her offspring had similar or dissimilar doses of CCL3L. Compared with mother–child pairs in which both the mother and her offspring had at least two CCL3L copies, the risk of transmission was significantly higher by a factor of approximately 10–13 only when both the mother and her offspring had a CCL3L gene dose of less than two (Fig. 2c). In these 89 mother–child pairs, identical associations were detected for the copy number of CCL4L. HIV-positive children with less than two CCL3L or CCL4L copies had experienced an approximately two times faster rate of progression to AIDS than those with a higher gene dose (Fig. 3a and b).
Fig. 3. Influence of the CCL3L copy number on HIV disease progression in children who did not receive perinatal zidovudine prophylaxis.
(a) Kaplan–Meier plots for time to AIDS stratified by CCL3L copy number in Ukrainian HIV-1-infected children (n = 123). Pink curve is for HIV-positive children with at least two CCL3L copy number, whereas green curve is for individuals with less than two copies of CCL3L. (b) Same as in (a), data are for CCL4L copy number. (c) Matrix of correlations among the indicated CCL3L and CCL4L genes in all study participants. The pair-wise comparisons revealed a highly statistically significant positive correlation (P < 0.001 for each pair-wise comparison) except for the frequencies of CCL4La and CCL4Lb, which displayed an inverse correlation. (d) Kaplan–Meier plots of subgroups based on CCL4La and CCL4Lb copy number and their effects on disease progression in infected children (n = 147). Zero (0) indicates that the gene is not present, 1 indicates the presence of one copy number of CCL4La or CCL4Lb. The red-colored Kaplan–Meier plot is for individuals who lack CCL4La, but have one or more copies of CCL4Lb; the green Kaplan–Meier plot is for individuals who have one copy each of CCL4La and CCL4Lb; and the blue Kaplan–Meier plot is for those who lack CCL4Lb and have one or more copies of CCL4La. The latter group represents the reference category. CI, confidence interval; N, number of children; P, significance value; r, Spearman's correlation coefficient; RH, relative hazard.
Relationships among CCL3L and CCL4L genes
We next investigated the phenotypic impact of the different components of CCL3L and CCL4L genes. Figure S2 (c) and (d) show the distribution of the copy numbers of the different CCL3L and CCL4L genes in the study individuals. The distribution patterns of CCL3La and CCL4La were very similar to those of the total copy numbers of CCL3L and CCL4L, respectively, whereas most study individuals lacked CCL3Lb and CCL4Lb (Fig. S2).
The pair-wise comparisons of the copy number of the different CCL3L and CCL4L genes revealed the following pattern. There was a highly statistically significant positive correlation between the copy numbers of CCL3La and CCL3Lb, CCL3La and CCL4La, CCL3La and CCL4Lb, CCL3Lb and CCL4La (Fig. 3c). In contrast, there was a highly statistically significant negative correlation between the copy number of CCL4La and CCL4Lb (r = −0.30, P < 0.0001; Fig. 3c).
On the basis of the observed inverse relationship between CCL4La and CCL4Lb (Fig. 3c) and because CCL4La but not CCL4Lb-derived transcripts are predicted to encode a classical chemokine (Fig. 1d), we surmised the following: those who had more ‘functional’ chemo-kine-encoding CCL4La copies than CCL4Lb copies might fare better in terms of their disease course than those who only possessed CCL4Lb copies. To test this possibility, we stratified HIV-infected children into three groups as shown in Fig. 3d. Consistent with our hypothesis, possession of only CCL4Lb was associated with a four-fold faster rate of disease progression than those who only possessed CCL4La (compare blue and red Kaplan–Meier plots, Fig. 3d). By contrast, those who had one copy of CCL4La and CCL4Lb displayed an intermediate disease phenotype (green Kaplan–Meier plot, Fig. 3d). The Kaplan–Meier plot of individuals who had higher copy numbers of CCL4La than CCL4Lb was similar to those who only possessed CCL4La (data not shown). These findings underscored the beneficial and negative associations of CCL4La and CCL4Lb copies on HIV disease course, respectively. There were too few individuals who were null for CCL3La to conduct similar analyses.
CCL3L–CCL4L components and HIV-AIDS susceptibility
The importance of accounting for the complexity imposed by the combinatorial gene content generated by variable CNV on the 17q12 was underscored by the observation that there was a positive correlation between the frequencies of each of the CCL3L and CCL4L genes except for the pair-wise comparison of CCL4La and CCL4Lb (Fig. 3c) and the contrasting disease-influencing effects associated with these two genes (Fig. 3d). This raised the possibility that the relative balance between the content of ‘protective’ versus ‘detrimental’ duplicated chemokine genes influences the HIV-AIDS-influencing phenotypic effect of the chemokine gene-rich 17q12 regions that have undergone segmental duplications. We used principal components analyses to address this possibility as well as to gain a greater understanding of the observed pattern of correlations between the chemokine gene copy numbers.
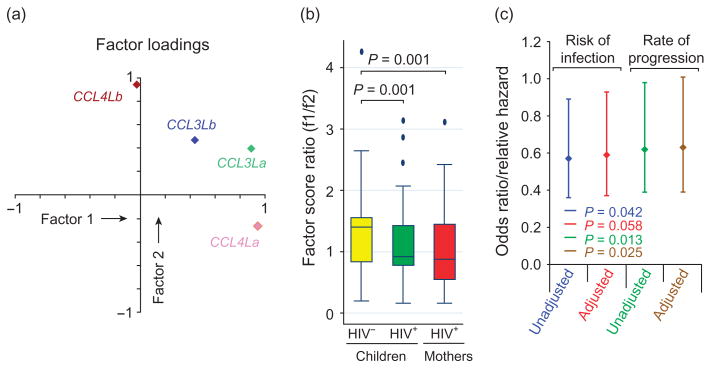
By factor analyses, we found that these four components loaded differentially onto a two-factor solution (Fig. 4a): CCL3La and CCL4La loaded heavily onto the first factor (factor 1), whereas the second factor (factor 2) was represented by CCL4Lb loading very highly onto it. Of note, CCL3Lb loaded almost equivocally on both the factors suggesting that this is a distinct component. The degree of uniqueness of CCL3La, CCL3Lb, CCL4La and CCL4Lb was 0.05, 0.59, 0.04 and 0.11, further emphasizing that with regards to the copy number distribution, the copy number of CCL3Lb was uniquely different from the distribution of CCL3La, CCL4La and CCL4Lb. Thus, factor 1 largely reflects CCL3La and CCL4La, whereas factor 2 largely reflects CCL4Lb.
Fig. 4. Factor analysis in study individuals.
(a) Results of principal components analyses. Two orthogonal factors (represented by abscissa and ordinate) were retained on the basis of an eigenvalue of more than one. The plot represents the varimax-rotated loadings for each of the components on the two factors (factors 1 and 2). Considering these loading patterns, the first factor represents mainly CCL3La and CCL4La, the two genes that transcribe functional chemokines, whereas factor 2 represents mainly CCL4Lb, which transcribes aberrantly spliced mRNA species. (b) Distribution of the factor 1/factor 2 (f1/f2) score ratio in study groups. HIV-positive children as well as their mothers had a significantly lower f1/f2 score ratio as compared with the HIV-negative children as assessed by Mann–Whitney test. For these analyses, we took the ratio of the scores for the first and second factors for a given study individual. As the factor scores were scattered over the interval (−2.21 and 4.27), we linearly transformed the raw factor scores by adding a constant of 2.5. This transformation converted all the factor scores into positive real numbers without affecting the original distribution of the factor scores. We then took the ratio of these positive factor scores. (c) Association of the log-transformed f1/f2 ratio with the risk of acquiring HIV (analyzed using logistic regression) and rate of disease progression (analyzed using Cox proportional hazards regression). The diamonds and error bars represent the point estimates and 95% CIs for OR (for risk of HIV acquisition) and relative hazards (for rate of disease progression). Results are from unadjusted and adjusted (for the receipt of prophylaxis) regression models. The numbers at the bottom show the significance values. CI, confidence interval; OR, odds ratio.
To assess whether biological interpretations made from the correlation patterns and principal components analyses were justifiable, we conducted the following analyses. Using the results from principal components analyses, we generated standardized scores for the first and second factor for each individual. On the basis of this, we surmised that a lower score for factor 1, which equates with a lower number of CCL3La and CCL4La copies would more likely associate with an HIV-positive status. Consistent with this, we observed that the median for the factor 1 score (in parenthesis) in HIV-negative children (0.25) was significantly higher than that found in HIV-positive children (0.13; P = 0.042 for comparison) and HIV-positive mothers (−0.43, P = 0.0067 for comparison). These observations suggested that a reduction in the overall gene content of CCL3La and CCL4La confers a differential risk of acquiring HIV.
We next examined whether the relative content of these two factors in a given individual was a determinant of the risk of acquiring HIV infection and progressing to AIDS. A ratio of the factors 1 (f1) and 2 (f2) scores (referred to here as f1/f2 score ratio) exceeding one indicates a relative enrichment of the first factor, whereas a value less than one indicates a relative enrichment of the second factor. We found that HIV-uninfected children had significantly higher values of the f1/f2 ratio compared with HIV-infected children and HIV-infected mothers (Fig. 4b). Using logistic regression analysis, we then determined if the risk of HIV acquisition is influenced by the f1/f2 score ratio. We found that each log-fold increase in the f1/f2 score ratio was associated with approximately 43 and 41% reduced likelihood of acquiring HIV before and after accounting for ZDV prophylaxis, respectively (Fig. 4c). Similarly and to a comparable extent, we found by using Cox proportional hazards regression analyses that a log-fold increase in the f1/f2 score ratio was associated with an approximately 38 and 37% decreased rate of progression to AIDS in the HIV-infected children before and after accounting for the receipt of prophylaxis, respectively (Fig. 4c).
Discussion
The present study has two major findings. First, they confirm and amplify the results of previous studies which showed that a low dose of CCL3L genes (referred previously as the CCL3L1-contianing segmental duplication) is associated with an increased risk of acquiring HIV and progressing rapidly to AIDS. Furthermore, our results demonstrate that a low CCL4L gene dose has similar associations. These results add credence to the notion that these genetic determinants may influence epidemic spread of HIV. Successful transmission of HIV-1 depends on both the infectiousness of the HIV-positive individual and susceptibility of the HIV-negative individual. Infectiousness is in part dependent on the viral load [37,38], and previous studies [24,26,29–31] indicate that a low copy number of CCL3L is associated with a progressive HIV disease course and higher viral load. Thus, it follows that mother–child dyads in which both members have a low copy number of CCL3L should have the highest probability of transmission, which is what we observed in this study. In 12.4% of mother–child pairs, both the mother and the child possessed less than two CCL3L (or CCL4L) copies and 91% of these children had acquired HIV infection. This translated to an odds ratio of more than 10 for the risk of transmission, which is among the highest odds for transmission that we have observed, and an attributable fraction of 18.4% (95% confidence interval 6.1–30.6%). These results suggest that a substantial proportion of vertical transmission in our study individuals may be explained by mother–child dyads in which each member has a low CCL3L/CCL4L dose. By analogy, we hypothesize that a similarly high proportion of transmission may occur among HIV-positive/HIV-negative sexual partner pairs when each member has a low CCL3L dose, a premise that is also supported by our modeling studies [39]. As there was complete concordance in the gene copy numbers of CCL3L and CCL4L in the mother–child pairs in which both mother and offspring had less than two CCL3L copies, these data also reflect the effects of the overall CCL4L gene copy number on transmission.
Second, our findings illustrate a chromosomal region whose genomic architecture is highly complex such that individuals do not inherit the same complement of CCL3L and CCL4L genes. However, we suggest that uncovering this complexity might be necessary to define the true phenotypic impact of these duplicated genes on HIV-AIDS susceptibility. This is exemplified by the finding that the copy numbers of CCL4La and CCL4Lb are inversely correlated and have opposing effects on AIDS progression rates, despite the fact that sequences of the coding exons of these two genes are identical [3,5]. Furthermore, we show that the balance between the copy numbers of the genes that transcribe classical versus aberrantly spliced CCL3L and CCL4L mRNA species influences HIV-AIDS susceptibility; a higher gene content of CCL4Lb (CCL4L1) or a lower content of CCL3La and CCL4La increased the risk of transmission and an accelerated disease course. Underscoring the negative influence of CCL4Lb on HIV acquisition, a previous study [9] also found that HIV-positive individuals were enriched for more copies of this gene compared with HIV-negative controls.
It is conceivable that there are additional tiers of genetic and mRNA complexity in this locus. We have identified novel 5′ exons for CCL3Lb, a gene that was previously thought to only have two exons, which are homologous to exons 2 and 3 of CCL3L1 or CCL3L3; for this reason, CCL3Lb was considered as a pseudogene [5]. We found that these novel 5′ exons give rise to two alternatively (CCL3Lb-v1 and CCL3Lb-v2) spliced transcripts. These transcripts contain chemokine-like domains, but are not predicted to encode classical chemokines, and additional studies will be required to uncover the functional properties of these CCL3L mRNA species.
The present findings have four implications. First, although CCL4La and CCL4Lb share 100% sequence identity in their coding sequences, the findings presented herein and those from our preliminary studies in other populations indicate that the pair-wise correlations among and between CCL3L and CCL4L genes, and the degree of the inverse correlation between the copy number of CCL4La and CCL4Lb genes may vary between populations. This suggests that genotype–phenotype association studies may need to account for interpopulation differences in the genomic architecture of this locus.
The second implication of our results is that the assessment of CCL4L dose is capturing the sum of two genes (CCL4La and CCL4Lb) whose copy number frequencies are inversely related and who have opposing effects on HIV disease course. Thus, the true phenotypic impact of CCL4La and CCL4Lb cannot be made using CCL3L copy number as a proxy for CCL4L or by evaluation of the composite CCL4L. This might explain in part why previous studies may not have found an association between CCL4L copy number and HIV disease [40]. Third, the CCL3Lb copy number loaded weakly and equivocally on factor 1 or 2, indicating that this gene may influence HIV-AIDS pathogenesis by as yet unidentified means. Consistent with this possibility, we have found that CCL3Lb (CCL3L2) generates alternatively transcribed transcripts whose 5′ exons differ from those of CCL3La (CCL3L1 and CCL3L3) (Fig. 1c). Fourth, greater insights into the immune correlates linked to a high gene dose of specific CCL3L and CCL4L chemokine genes that confer protective effects may provide novel insights into the correlates of protection. The importance of elucidating these correlates is also highlighted by the recent observation that CCL3L copy number may influence susceptibility to Simian-AIDS in rhesus macaques [34,41].
In summary, we confirm and extend significantly prior results indicating that a low copy number of CCL3L influences risk of HIV acquisition and disease progression in a cohort of children exposed perinatally to HIV. We demonstrate a role for CCL4L CNV in HIV-AIDS susceptibility. However, we show that dissecting the combinatorial genomic complexity posed by varying proportions of distinct CCL3L and CCL4L genes among individuals is required to elucidate the complete phenotypic impact of this locus in HIV, and suggest that the balance between the copy number of the different CCL3L and CCL4L genes influences HIV-AIDS susceptibility. From a broader perspective of relevance to the CNV field, it might be important to determine whether the combinatorial genomic content generated by distinct genes within a copy number variable region determines disease susceptibility.
Supplementary Material
Acknowledgments
Supported by award no. UKB2-2705-DP-05 of the U.S. Civilian Research and Development (CRDF) to L.S.K. and S.K.A., the Veterans Administration Center on AIDS and HIV infection at the South Texas Veterans Healthcare System and a MERITaward (R37046326) from the NIH to S.K.A. This work was also supported by a Elizabeth Glaser Scientist Award and Burroughs Wellcome Clinical Scientist Award in Translational Research to S.K.A.
We thank George Crawford for invaluable programmatic support at UTHSCSA; Vince Marconi for critical advice; staff members and patients at the Department of Pediatrics at Dnepropetrovsk State Medical Academy for logistic support and participation in the study, respectively; Birju Shah, Vivian Ahn, Lisa Beachy and Jill King for technical assistance; and A.S. Ahuja for forbearance. Any opinions, findings, conclusions or recommendations expressed in this material are those of the authors and do not reflect the views of CRDF.
L.S.K. and S.K.A. conceived the project, obtained funding for the study and provided expertise into different aspects of the study. L.S.K. and Z.A.C. enrolled patients, collected samples, conducted experiments and analyzed data. G.C. and W.H. designed the experimental protocols and analyzed data. R.S. helped establishing the PCR assays. G.G., S.M. and S.K.A. planned and performed the mRNA cloning and sequencing work. M.J.D., S.S.A. and R.A.C. provided critical conceptual input and analyzed data. S.K.A., H.K. and G.C. wrote the initial draft and all authors critically reviewed the manuscript and provided feedback. H.K. and S.K.A. conceived the statistical design.
Footnotes
There are no conflicts of interest.
References
- 1.Fauci AS, Johnston MI, Dieffenbach CW, Burton DR, Hammer SM, Hoxie JA, et al. HIV vaccine research: the way forward. Science. 2008;321:530–532. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- 2.Prendergast A, Tudor-Williams G, Jeena P, Burchett S, Goulder P. International perspectives, progress, and future challenges of paediatric HIV infection. Lancet. 2007;370:68–80. doi: 10.1016/S0140-6736(07)61051-4. [DOI] [PubMed] [Google Scholar]
- 3.Modi WS. CCL3L1 and CCL4L1 chemokine genes are located in a segmental duplication at chromosome 17q12. Genomics. 2004;83:735–738. doi: 10.1016/j.ygeno.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Modi WS, Bergeron J, Sanford M. The human MIP-1beta chemokine is encoded by two paralogous genes, ACT-2 and LAG-1. Immunogenetics. 2001;53:543–549. doi: 10.1007/s002510100366. [DOI] [PubMed] [Google Scholar]
- 5.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 6.Nibbs RJ, Yang J, Landau NR, Mao JH, Graham GJ. LD78beta, a nonallelic variant of human MIP-1alpha (LD78alpha), has enhanced receptor interactions and potent HIV suppressive activity. J Biol Chem. 1999;274:17478–17483. doi: 10.1074/jbc.274.25.17478. [DOI] [PubMed] [Google Scholar]
- 7.Struyf S, Menten P, Lenaerts JP, Put W, D'Haese A, De Clercq E, et al. Diverging binding capacities of natural LD78beta isoforms of macrophage inflammatory protein-1alpha to the CC chemokine receptors 1, 3 and 5 affect their anti-HIV-1 activity and chemotactic potencies for neutrophils and eosinophils. Eur J Immunol. 2001;31:2170–2178. doi: 10.1002/1521-4141(200107)31:7<2170::aid-immu2170>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 8.Xin X, Shioda T, Kato A, Liu H, Sakai Y, Nagai Y. Enhanced anti-HIV-1 activity of CC-chemokine LD78beta, a nonallelic variant of MIP-1alpha/LD78alpha. FEBS Lett. 1999;457:219–222. doi: 10.1016/s0014-5793(99)01035-2. [DOI] [PubMed] [Google Scholar]
- 9.Colobran R, Adreani P, Ashhab Y, Llano A, Este JA, Dominguez O, et al. Multiple products derived from two CCL4 loci: high incidence of a new polymorphism in HIV+ patients. J Immunol. 2005;174:5655–5664. doi: 10.4049/jimmunol.174.9.5655. [DOI] [PubMed] [Google Scholar]
- 10.Howard OM, Turpin JA, Goldman R, Modi WS. Functional redundancy of the human CCL4 and CCL4L1 chemokine genes. Biochem Biophys Res Commun. 2004;320:927–931. doi: 10.1016/j.bbrc.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 11.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 12.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 14.Sebat J. Major changes in our DNA lead to major changes in our thinking. Nat Genet. 2007;39:S3–S5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- 15.Conrad DF, Hurles ME. The population genetics of structural variation. Nat Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski JR. An evolution revolution provides further revelation. BioEssays. 2007;29:1182–1184. doi: 10.1002/bies.20686. [DOI] [PubMed] [Google Scholar]
- 17.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 18.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 19.Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 22.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191(Suppl 1):S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 23.Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, Begum K, et al. Evolution of human and nonhuman primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 25.Meddows-Taylor S, Donninger SL, Paximadis M, Schramm DB, Anthony FS, Gray GE, et al. Reduced ability of newborns to produce CCL3 is associated with increased susceptibility to perinatal human immunodeficiency virus 1 transmission. J Gen Virol. 2006;87:2055–2065. doi: 10.1099/vir.0.81709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn L, Schramm DB, Donninger S, Meddows-Taylor S, Coovadia AH, Sherman GG, et al. African infants' CCL3 gene copies influence perinatal HIV transmission in the absence of maternal nevirapine. AIDS. 2007;21:1753–1761. doi: 10.1097/QAD.0b013e3282ba553a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima T, Ohtani H, Naruse T, Shibata H, Mimaya JI, Terunuma H, Kimura A. Copy number variations of CCL3L1 and long-term prognosis of HIV-1 infection in asymptomatic HIV-infected Japanese with hemophilia. Immunogenetics. 2007;59:793–798. doi: 10.1007/s00251-007-0252-4. [DOI] [PubMed] [Google Scholar]
- 28.Sadam M, Karki T, Huik K, Avi R, Rüütel K, Uusküla A, Lutsar I. CCL3L1 variable gene copy number influence on the susceptibility to HIV-1/AIDS among Estonian intravenous drug user [abstract #296]. 15th Conference on Retroviruses and Opportunistic Infections; 2008. [Google Scholar]
- 29.Shalekoff S, Meddows-Taylor S, Schramm DB, Donninger SL, Gray GE, Sherman GG, et al. Host CCL3L1 gene copy number in relation to HIV-1-specific CD4+ and CD8+ T-cell responses and viral load in South African women. J Acquir Immune Defic Syndr. 2008;48:245–254. doi: 10.1097/QAI.0b013e31816fdc77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, Anaya JM, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja SK, Kulkarni H, Catano G, Agan BK, Camargo JF, He W, et al. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–3026. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 33.Ketas TJ, Kuhmann SE, Palmer A, Zurita J, He W, Ahuja SK, et al. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degenhardt JD, de Candia P, Chabot A, Schwartz S, Henderson L, Ling B, et al. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in rhesus macaques (Macaca mulatta) PLoS Genet. 2009;5:e1000346. doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colobran R, Comas D, Faner R, Pedrosa E, Anglada R, Pujol-Borrell R, et al. Population structure in copy number variation and SNPs in the CCL4L chemokine gene. Genes Immun. 2008;9:279–288. doi: 10.1038/gene.2008.15. [DOI] [PubMed] [Google Scholar]
- 36.Cardone MF, Jiang Z, D'Addabbo P, Archidiacono N, Rocchi M, Eichler EE, Ventura M. Hominoid chromosomal rearrangements on 17q map to complex regions of segmental duplication. Genome Biol. 2008;9:R28. doi: 10.1186/gb-2008-9-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wawer MJ, Gray RH, Sewankambo NK, Serwadda D, Li X, Laeyendecker O, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–1409. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 38.Cohen MS, Hellmann N, Levy JA, DeCock K, Lange J. The spread, treatment, and prevention of HIV-1: evolution of a global pandemic. J Clin Invest. 2008;118:1244–1254. doi: 10.1172/JCI34706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkarni H, Marconi VC, Agan BK, McArthur C, Crawford G, Clark RA, et al. Role of CCL3L1-CCR5 genotypes in the epidemic spread of HIV-1 and evaluation of vaccine efficacy. PLoS ONE. 2008;3:e3671. doi: 10.1371/journal.pone.0003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao W, Tang J, Song W, Wang C, Li Y, Wilson CM, Kaslow RA. CCL3L1 and CCL4L1: variable gene copy number in adolescents with and without human immunodeficiency virus type 1 (HIV-1) infection. Genes Immun. 2007;8:224–231. doi: 10.1038/sj.gene.6364378. [DOI] [PubMed] [Google Scholar]
- 41.Gornalusse G, Mummidi S, He W, Silvestri G, Bamshad M, Ahuja SK. CCL3L copy number variation and the co-evolution of primate and viral genomes. PLoS Genet. 2009;5:e1000359. doi: 10.1371/journal.pgen.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.