We burdened mice with intraperitoneal injection of the endoplasmic reticulum stress-inducing reagent tunicamycin, and found that wild-type mice were able to recover from the insult, whereas ATF6α-knockout mice exhibited liver dysfunction and steatosis. Our results establish links between endoplasmic reticulum stress, lipid metabolism and steatosis
Abstract
Accumulation of unfolded proteins in the endoplasmic reticulum (ER) activates homeostatic responses collectively termed the unfolded protein response. Among the three principal signaling pathways operating in mammals, activating transcription factor (ATF)6α plays a pivotal role in transcriptional induction of ER-localized molecular chaperones and folding enzymes as well as components of ER-associated degradation, and thereby mouse embryonic fibroblasts deficient in ATF6α are sensitive to ER stress. However, ATF6α-knockout mice show no apparent phenotype under normal growing conditions. In this report, we burdened mice with intraperitoneal injection of the ER stress-inducing reagent tunicamycin and found that wild-type mice were able to recover from the insult, whereas ATF6α-knockout mice exhibited liver dysfunction and steatosis. Thus, ATF6α-knockout mice accumulated neutral lipids in the liver such as triacylglycerol and cholesterol, which was ascribable to blockage of β-oxidation of fatty acids caused by decreased mRNA levels of the enzymes involved in the process, suppression of very-low-density lipoprotein formation due to destabilized apolipoprotein B-100, and stimulation of lipid droplet formation resulting from transcriptional induction of adipose differentiation-related protein. Accordingly, the hepatocytes of tunicamycin-injected knockout mice were filled with many lipid droplets. These results establish links among ER stress, lipid metabolism, and steatosis.
INTRODUCTION
Proteins must be properly folded to exert their respective functions assigned by genetic code. Newly synthesized secretory and transmembrane proteins gain tertiary and quaternary structures in the endoplasmic reticulum (ER), and only correctly folded proteins are transported to their final destinations. The quality of these proteins is tightly monitored by a system called ER quality control, which consists of two different mechanisms (Bukau et al., 2006). Proteins are folded with the assistance of a number of ER-localized molecular chaperones and folding enzymes (collectively termed ER chaperones hereafter). This mechanism is termed the productive folding mechanism and allows a majority of newly synthesized proteins to attain native conformations under normal conditions (Vabulas and Hartl, 2005). In contrast, proteins that are incompletely folded or misfolded, even after chaperone assistance, are retrotranslocated back to the cytosol to be degraded by the proteasome in a ubiquitin-dependent manner. This mechanism is termed ER-associated degradation (ERAD) and minimizes the amount of unfolded or misfolded proteins in the ER (Wilhovsky et al., 2000; Tsai et al., 2002).
Under a variety of conditions collectively termed ER stress, however, unfolded proteins are accumulated in the ER, which is detrimental to the cell. Eukaryotic cells from yeast to humans activate the unfolded protein response (UPR) to cope with ER stress (Mori, 2000; Schroder and Kaufman, 2005; Ron and Walter, 2007). In yeast cells, the UPR consists of transcriptional control only and is mediated by Ire1p, a transmembrane protein in the ER, whereas in metazoan cells it consists of translational and transcriptional control and is mediated by three ubiquitously expressed transmembrane proteins in the ER, namely, IRE1, PERK, and activating transcription factor (ATF)6. The major targets of transcriptional control conserved from yeast to humans are ER chaperones and ERAD components, induction of which leads to maintenance of the homeostasis of the ER. It has been shown that transcriptional induction of these ER quality control proteins is achieved with the Ire1p or Ire-1 pathway in Saccharomyces cerevisiae (Travers et al., 2000), Caenorhabditis elegans (Shen et al., 2005), and Drosophila melanogaster (Hollien and Weissman, 2006). In contrast, our lab and Kaufman's lab have shown that the ATF6 pathway is required for transcriptional induction of these ER quality control proteins in mouse embryonic fibroblasts (MEFs) (Wu et al., 2007; Yamamoto et al., 2007), thus revealing a switch in principal regulator from IRE1 to ATF6 during evolution.
ATF6 is not encoded by the yeast genome but is present as a single gene product in worm and fly cells. In contrast, ATF6 consists of the two closely related ATF6α and ATF6β in mammalian cells. Both ATF6α and ATF6β are ER membrane-bound transcription factors activated via ER stress-induced regulated intramembrane proteolysis (Haze et al., 1999, 2001; Ye et al., 2000). They are synthesized as type II transmembrane proteins in the ER, designated pATF6α(P) and pATF6β(P). On ER stress, these precursor forms relocate to the Golgi apparatus via COP II vesicles, where they are cleaved by sequential action of site-1 and site-2 proteases (Shen et al., 2002; Okada et al., 2003; Nadanaka et al., 2004). The ATF6α/β N-terminal fragments released from the membrane, designated pATF6α(N) and pATF6β(N), enter the nucleus and activate transcription of their target genes, because they contain all domains necessary for an active transcription factor (Yoshida et al., 2000; Yoshida et al., 2001). Ectopic expression of pATF6α(N) results in transcriptional induction of various ER chaperone genes (Okada et al., 2002).
ATF6α and ATF6β genes have been knocked out in mice (Wu et al., 2007; Yamamoto et al., 2007). Analysis of MEFs revealed that ATF6α is the main mediator of transcriptional induction of ER chaperones and that ATF6α heterodimerizes with XBP1, the downstream transcription factor of the IRE1 pathway, to mediate transcriptional induction of ERAD components. Based on the results of microarray analyses, which were subsequently confirmed by Northern blot hybridization and functional analysis, ATF6α is considered to be a transcription factor that is specialized in the control of ER quality control proteins (Adachi et al., 2008). Accordingly, MEFs deficient in ATF6α are sensitive to ER stress. In contrast, deletion of the ATF6β gene has almost no effect on the induction of ER quality control proteins. Nonetheless, ATF6β probably has overlapping functions with ATF6α and contributes to the UPR, because double knockout of ATF6α and ATF6β genes causes embryonic lethality even though mice lacking either ATF6α or ATF6β alone seem normal (Yamamoto et al., 2007).
Here, to define the role of ATF6α-mediated regulation of ER quality control proteins in vivo, we subjected ATF6α +/+ and −/− mice to pharmacological ER stress. This was found to cause liver steatosis. During the preparation of this article, Kaufman and coworkers conducted similar and extensive experiments and obtained similar conclusions (Rutkowski et al., 2008). However, we obtained results that connect the deficiency in ATF6α-mediated regulation of ER quality control proteins to liver steatosis more directly; namely, the level of apolipoprotein B-100 (apoB-100), a major protein component of very-low-density lipoprotein (VLDL), was markedly affected by the presence or absence of ATF6α.
MATERIALS AND METHODS
Production of ATF6α+/+, ATF6α−/−, ATF6β+/+, and ATF6β−/− Mice and Preparation of Tunicamycin and Lipopolysaccharide (LPS) Solutions
ATF6α or ATF6β gene was disrupted in the embryonic stem cell line J1 and male chimeras thereby obtained were crossed with C57BL/6J females to generate heterozygotes as described previously (Yamamoto et al., 2007). Male heterozygotes were backcrossed to C57BL/6J females five times for use in the experiments shown in Figures 1B, 2, 3A, and 6A, and more than seven times for use in the rest of the experiments, before the production of ATF6α+/+, ATF6α−/−, ATF6β+/+, and ATF6β−/− mice. Genotyping was conducted by polymerase chain reaction (PCR) as described previously (Yamamoto et al., 2007). Mice were housed in a temperature-controlled room with a 12-h light/dark cycle and had free access to normal chow diet. Tunicamycin and LPS were dissolved in 150 mM sucrose and phosphate-buffered saline (PBS), respectively. Livers taken from mice before or after injection were immediately frozen in liquid nitrogen, unless otherwise indicated.
Figure 1.
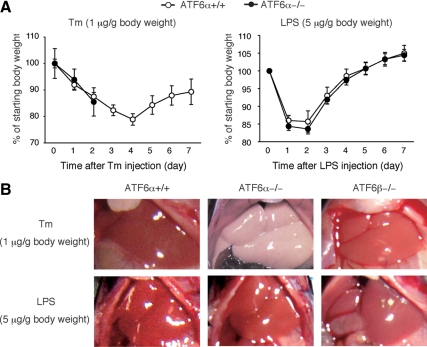
Effect of tunicamycin injection on body weight and liver appearance of ATF6α +/+ and −/− mice. (A) Tunicamycin (Tm) or LPS was intraperitoneally injected into ATF6α +/+ and −/− mice each at a dose of 1 or 5 μg/g body weight, respectively. Body weights of injected mice were determined daily for 7 d and are presented after normalization to that at day 0 of injection as the means ± SEM (n = 4 for Tm-injected ATF6α −/− mice and n = 5 for others). All ATF6α −/− mice injected with tunicamycin died at day 3. (B) Tm or LPS was injected into ATF6α +/+ and −/− mice as well as ATF6β −/− mice as described in A. The abdomen of injected mice was dissected and photographed after 48 h.
Figure 2.
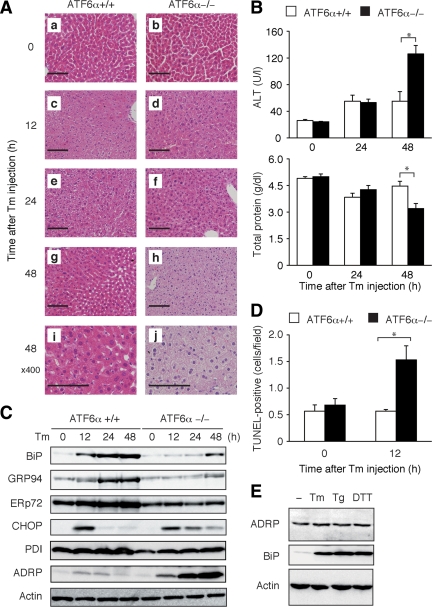
Effect of tunicamycin injection on liver function of ATF6α +/+ and −/− mice. (A) Tunicamycin (Tm) was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Liver tissue sections were prepared at the indicated time points after injection and stained with hematoxylin and eosin. Bar, 100 μm (a–h) and 50 μm (i and j). (B) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Levels of ALT and total protein in the serum of injected mice were determined at the indicated time points and are presented as the means ± SEM (n = 3). *p < 0.05. (C) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Liver lysates were prepared at the indicated time points after injection and analyzed by immunoblotting using antibody to BiP, GRP94, ERp72, CHOP, PDI, ADRP, or actin. (D) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. TUNEL-positive cells were determined in liver tissue sections prepared 12 h after injection and are presented as the means ± SEM (n = 3). *p < 0.05. (E) HEK293T cells were untreated (−) or treated for 12 h with 2 μg/ml Tm, 300 nM thapsigargin (Tg), or 2 mM dithiothreitol (DTT). The cell lysates were prepared and analyzed by immunoblotting using antibody to ADRP, BiP, or actin.
Figure 3.
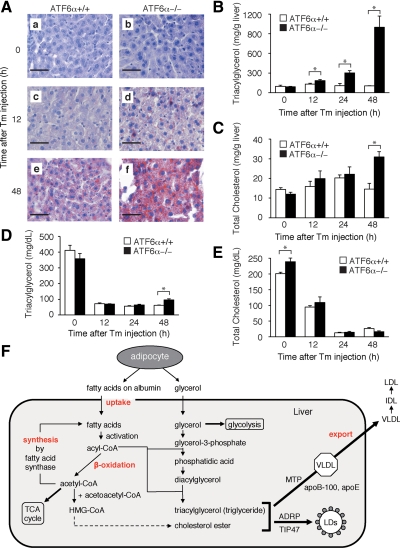
Effect of tunicamycin injection on lipid contents in the liver of ATF6α +/+ and −/− mice. (A) Tunicamycin (Tm) was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Liver tissue sections were prepared at the indicated time points after injection and stained with oil red O. Bar, 50 μm. (B) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Levels of triacylglycerol (triglyceride) in the liver were determined at the indicated time points after injection and are presented as the means ± SEM (n = 3). *p < 0.05. (C) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Levels of total cholesterol in the liver were determined at the indicated time points after injection and are presented as the means ± SEM (n = 3). *p < 0.05. (D) Tm was injected into ATF6α +/+ and −/− mice as in Figure 1A. Levels of triacylglycerol (triglyceride) in plasma were determined at the indicated time points after injection and are presented as the means ± SEM (n = 4). *p < 0.05. (E) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Levels of total cholesterol in plasma were determined at the indicated time points after injection and are presented as the means ± SEM (n = 4). *p < 0.05. (F) Schematic presentation of metabolism of fatty acids and cholesterol.
Figure 6.
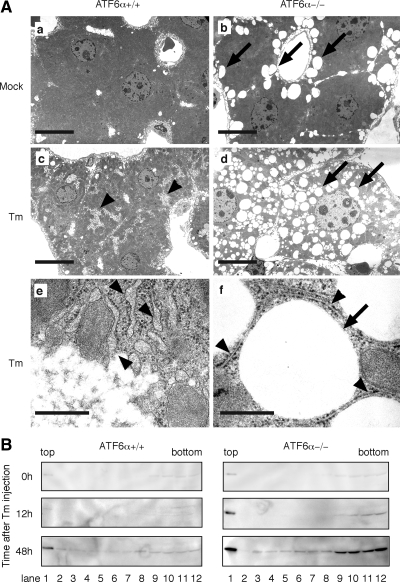
Induction of lipid droplet formation in ATF6α −/− mice in response to tunicamycin injection. (A) Tunicamycin (Tm) was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Liver tissue sections were prepared from untreated or tunicamycin-injected mice after 48 h and analyzed with electron microscopy. Bar, 10 μm (a–d) and 500 nm (e and f). (B) Tm was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Whole liver obtained at the indicated time points after injection was homogenized and then subjected to sucrose gradient (5–39% from top to bottom) centrifugation. An aliquot of 12 collected fractions was analyzed by immunoblotting using anti-ADRP antibody.
Histological Analysis
Unfrozen livers were fixed in 4% paraformaldehyde in PBS, embedded in paraffin, sectioned to 5 μm in thickness, and stained with hematoxylin and eosin. To visualize neutral lipids, frozen liver sections (10 μm in thickness) were stained with oil red o according to the standard procedure. Apoptosis was detected by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay by using an In Situ Cell Death Detection kit, alkaline phosphatase (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions. For electron micrographic analysis, mice were perfused with 0.1 M cacodylate buffer, pH 7.4, containing 2% paraformaldehyde and 2.5% glutaraldehyde. Livers were dissected and further fixed for 2 h at room temperature. Sections were processed as described previously (Harada et al., 1990) and examined with an electron microscope (model 1010; JEOL, Tokyo, Japan) at 100 kV.
Biochemical Determinations
Serum alanine aminotransferase and total protein levels were measured with blood samples obtained from a tail vein by using VetScan (Abaxis, Union City, CA). Total lipids were extracted from ∼30 mg of frozen liver according to a previously published method (Bligh and Dyer, 1959). Hepatic triacylglycerol (triglyceride) and cholesterol levels as well as those in plasma were measured using a Triglyceride Test kit (Wako Pure Chemicals, Osaka, Japan) and LabAssay Cholesterol kit (Wako Pure Chemicals), respectively.
Immunoblotting
Total liver lysates were prepared by homogenizing frozen liver with a Teflon pestle in 10 volumes of 80 mM Tris-HCl buffer, pH 6.8, containing 2% SDS, 50 μM MG132, and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan), followed by boiling, sonication, and clarification by centrifugation. Liver microsomes were prepared according to the published procedure (Uchiyama et al., 2006). In brief, frozen liver was homogenized with a Teflon pestle in 10 volumes of 20 mM Tris-HCl buffer, pH 7.5, containing 0.25 M sucrose, 50 μM MG132, and protease inhibitor cocktail. Homogenates were centrifuged at 8000 × g for 10 min, and the resulting supernatant was further centrifuged at 134,000 × g for 1 h. Microsomal pellets were suspended in the homogenizing buffer. Protein content was determined using a BCA Protein Assay kit (Pierce Chemical, Rockford, IL). These samples were analyzed by standard procedures (Sambrook et al., 1989) using Western Blotting Luminol Reagent obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Chemiluminescence was visualized using an LAS-3000 LuminoImage analyzer (Fujifilm, Tokyo Japan). Anti-ATF6β antibody was as described previously (Haze et al., 2001). Anti-KDEL (anti-BiP), anti-GRP94, anti-ERp72, anti-protein disulfide isomerase (PDI), and anti-HSP47 antibodies were purchased from Stressgen (Ann Arbor, MI). Anti-CHOP (anti-GADD153) and anti-apoB-100 antibodies were obtained from Santa Cruz Biotechnology. Anti-actin and anti-adipose differentiation-related protein (ADRP) antibodies were from Millipore (Billerica, MA) and Progen (Heidelberg, Germany), respectively.
Microarray Analysis and Northern Blot Hybridization
Total RNA was extracted from frozen livers by using an RNeasy mini kit (QIAGEN, Venlo, The Netherlands). Microarray analysis was performed essentially as described previously (Adachi et al., 2008). Total RNA extracted from livers of mice 48 h after intraperitoneal injection of vehicle (150 mM sucrose) alone, or tunicamycin was converted to cDNA, which was then labeled with cyanine 3-cytidine 5′-triphosphate (CTP) or cyanine 5-CTP, respectively. These labeled cRNA probes were mixed and hybridized with a 4 × 44K Agilent oligo microarray (Whole Mouse Genome; Agilent Technologies, Santa Clara, CA).
Northern blot hybridization was performed according to standard procedures published previously (Sambrook et al., 1989). Digoxigenin-labeled cDNA probes were prepared using PCR according to the manufacturer's instructions (Roche Diagnostics) and hybridized with RNA electrophoresed and blotted on a membrane. Subsequent reaction with anti-digoxigenin antibody (Roche Diagnostics) and treatment with the chemiluminescent detection reagent CDP-star (GE Healthcare, Chalfont St. Giles, United Kingdom) were performed according to the manufacturers' specifications. Chemiluminescence was visualized using an LAS-3000 LuminoImage analyzer (Fujifilm).
Sucrose Gradient Centrifugation to Separate Lipid Droplets (LDs)
LDs were separated by sucrose gradient centrifugation according to a previously published procedure (Pol et al., 2004). Approximately 50 mg of frozen liver was homogenized with a Teflon pestle in 10 volumes of 20 mM Tris-HCl buffer, pH 7.5, containing 8% sucrose, 50 μM MG132, and protease inhibitor cocktail, and then the homogenate was centrifuged at 1000 × g for 10 min. The resulting supernatant was mixed with an equal volume of 20 mM Tris-HCl buffer, pH 7.5, containing 70% sucrose. This mixture (350 μl; 39% sucrose) was placed at the bottom of a centrifugation tube onto which 350 μl each of 20 mM Tris-HCl buffer, pH 7.5, containing 20, 10, and 5% sucrose was layered in a stepwise manner. The loaded tubes were centrifuged at 50,000 rpm (120,000–214,000 × g) for 3 h at 4°C.
Cell Culture and Transfection
Human embryonic kidney (HEK)293T and HepG2 cells were cultured in DMEM (glucose at 4.5 g/l) supplemented with 10% fetal bovine serum, 2 mM glutamine, and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C in a humidified 5% CO2, 95% air atmosphere. Transfection was carried out using MicroPorator MP-100 (Digital Bio, Seoul, Korea) according to the manufacturer's instructions. pcDNA-ATF6α(171-373) to express a dominant-negative form of ATF6α was constructed previously (Haze et al., 1999).
RESULTS
Induction of Liver Dysfunction in ATF6α −/− Mice after Tunicamycin Injection
We injected tunicamycin intraperitoneally into ATF6α +/+ and −/− male mice (8–10 wk) at a dose of 1 μg/g body weight, which was previously used in analyses of CHOP and caspase-12 knockout mice (Zinszner et al., 1998; Nakagawa et al., 2000). Tunicamycin causes ER stress by inhibiting protein N-glycosylation (Kaufman, 1999). Results showed that both types of mice lost weight immediately after tunicamycin injection (Figure 1A, left). After a continuous decrease for 4 d, the body weight of ATF6α +/+ mice started to increase from day 5, indicating recovery from the tunicamycin insult. In marked contrast, all ATF6α −/− mice died at day 3, demonstrating that tunicamycin injection conferred fatal damage to ATF6α −/− mice. Importantly, ATF6α +/+ and −/− mice showed no significant difference in changes in body weight after injection of LPS, an endotoxin derived from Gram-negative bacterial outer membrane that is known to cause inflammation (Trent et al., 2006) (Figure 1A, right). It should be noted that our results are consistent with those previously published by Kaufman et al. (Wu et al., 2007; Rutkowski et al., 2008), although ATF6α −/− mice died much earlier in our experiments than in their experiments, despite the injection of the same amount of tunicamycin.
Dissection of the abdomen revealed that the liver of ATF6α −/− but not ATF6α +/+ mice turned pale at 48 h after tunicamycin injection (Figure 1B). No such marked difference was observed when ATF6α +/+ and −/− mice were challenged with LPS or when tunicamycin was injected into ATF6β +/+ or −/− mice (Figure 1B). Histological analysis using hematoxylin and eosin staining confirmed a marked difference between the liver of ATF6α +/+ and −/− mice, particularly at 48 h after tunicamycin injection (Figure 2A). Accordingly, the level of alanine aminotransferase (ALT), an index of hepatocyte injury, was increased more than twofold in the serum of ATF6α −/− mice compared with that of ATF6α +/+ mice, whereas the level of total protein, an index of liver function, as well as albumin was decreased, all three from 48 h after tunicamycin injection (Figure 2B and Supplemental Figure 1). We observed no significant difference in the morphology of the kidney or other organs between ATF6α +/+ and −/− mice after tunicamycin injection (our unpublished observation). These results indicate that tunicamycin-induced injury of the liver is specific to ATF6α −/− mice.
Immunoblotting analysis of liver lysates showed that the induction of four major ER chaperones, namely, BiP, GRP94, ERp72, and PDI, observed in tunicamycin-injected ATF6α +/+ mice was indeed lost or greatly mitigated in ATF6α −/− mice (Figure 2C), as expected from the results obtained with MEFs (Yamamoto et al., 2007). The liver damage in ATF6α −/− mice could be explained, at least in part, by tunicamycin-induced apoptosis of hepatocytes; TUNEL-positive cells were significantly increased in the liver of ATF6α −/− mice as early as 12 h after tunicamycin injection compared with ATF6α +/+ mice (Figure 2D). This apoptosis may be independent of CHOP, which is implicated in ER stress-induced cell death (Zinszner et al., 1998), because CHOP was induced at the protein level in the liver of both ATF6α +/+ and −/− mice at 12 h after tunicamycin injection (Figure 2C). Regardless, these results suggest the importance of ATF6α-mediated upregulation of the levels of ER quality control proteins in protecting the mouse liver from ER stress-induced damage.
Induction of Liver Steatosis in ATF6α −/− Mice after Tunicamycin Injection
Consistent with the discolored appearance of the liver (Figure 1B), we found that the liver of ATF6α −/− mice injected with tunicamycin was much more positive to oil red O staining than that of ATF6α +/+ mice (Figure 3A). Indeed, the level of triacylglycerol (triglyceride) in the liver of ATF6α −/− mice was markedly increased compared with that of ATF6α +/+ mice 48 h after tunicamycin injection (Figure 3B). Furthermore, the level of total cholesterol in the liver of ATF6α −/− mice was significantly increased compared with that of ATF6α +/+ mice at this time (Figure 3C). In contrast, levels of triacylglycerol and total cholesterol in the plasma decreased similarly in both ATF6α +/+ and −/− mice after tunicamycin injection (Figure 3, D and E). Thus, tunicamycin injection caused the specific accumulation of neutral lipids in the liver of ATF6α −/− mice, leading to liver steatosis.
Triacylglycerol is synthesized mainly in liver via three cycles acylation of glycerol-3-phosphate by using acyl-CoA, which is produced through the activation of fatty acids (Figure 3F). Fatty acid levels in the liver are regulated by the balance between uptake, synthesis, catabolism (β-oxidation), and export (Bradbury, 2006; Reddy and Rao, 2006; Duval et al., 2007). To determine which of theses processes was affected by tunicamycin injection, we performed microarray analysis by using RNA independently prepared three times from liver of ATF6α +/+ and −/− mice 48 h after the injection of tunicamycin or vehicle (150 mM sucrose) alone. Fluorescent intensities obtained with 9656 of 41,534 genes mounted on slides were at least twofold higher than background intensities in all 12 determinations (3 each of vehicle and tunicamycin injection in ATF6α +/+ and −/− mice). As shown in Table 1, genes involved in fatty acid metabolism were generally down-regulated in ATF6α −/− mice, probably due to tunicamycin-induced liver dysfunction (see ratio [%] that is change in knockout [KO] divided by change in wild type [WT]). We first checked genes involved in the uptake of fatty acids, namely, the plasma membrane fatty acid binding protein (FABPpm)/mitochondrial aspartate amino transferase (AST), fatty acid translocase/CD36, and fatty acid transport protein (FATP)/solute carrier family 27 a1 (Slc27a1) genes (Bradbury, 2006; Duval et al., 2007). As shown in Table 1, expression levels of these genes were unchanged or lower in the liver of ATF6α −/− mice than in that of ATF6α +/+ mice 48 h after tunicamycin injection, suggesting that fatty acid uptake was not a key event in the accumulation of triacylglycerol in ATF6α −/− mice.
Table 1.
Determination of expression levels of genes involved in lipid metabolism in tunicamycin- or vehicle-injected ATF6α +/+ and −/− mice by microarray analysisa
| Gene name | Basal (%) KO vehicle WT vehicle | Change in WT (%) Tm Vehicle | Change in KO (%) Tm Vehicle | Ratio (%) Change in KO Change in WT | GenBank |
|---|---|---|---|---|---|
| FA uptake | |||||
| FABPm/AST | 92 | 88 ± 7.1 | 90 ± 6.1 | 100 | NM_010325 |
| Fatty acid translocase/CD36 | 90 | 93 ± 31 | 30 ± 3.7 | 32 | L23108 |
| FATP/Slc27al | 75 | 52 ± 17 | 32 ± 7.0 | 62 | NM_011977 |
| FA activation | |||||
| ACSL1 | 110 | 61 ± 2.6 | 7.6 ± 1.6 | 12 | NM_007981 |
| FABP1 | 170 | 123 ± 57 | 0.35 ± 0.17 | 0.28 | NM_017399 |
| FATP2/Slc27a2 | 120 | 101 ± 5.2 | 5.7 ± 2.6 | 5.6 | NM_011978 |
| FATP5/Slc27a5 | 110 | 82 ± 32 | 6.3 ± 1.0 | 7.7 | NM_009512 |
| FA β-oxidation | |||||
| PPARα | 50 | 56 ± 12 | 14 ± 3.8 | 25 | NM_011144 |
| CPT-Ia | 83 | 101 ± 11 | 42 ± 4.2 | 42 | NM_013495 |
| CPT-II | 110 | 60 ± 9.3 | 16 ± 5.6 | 27 | NM_009949 |
| ACOX1 | 130 | 215 ± 29 | 32 ± 10 | 15 | NM_015729 |
| FA synthesis | |||||
| SREBP-1 | 64 | 30 ± 14 | 6.8 ± 2.9 | 23 | NM_011480 |
| Fatty acid synthase | 97 | 12 ± 2.2 | 4.4 ± 1.9 | 37 | NM_007988 |
| Stearoyl-CoA desaturase1 | 70 | 9.6 ± 2.9 | 7.8 ± 1.6 | 81 | NM_009127 |
| ChREBP | 83 | 72 ± 23 | 8.9 ± 3.3 | 12 | NM_021455 |
| PPARγ | 70 | 93 ± 11 | 112 ± 15 | 120 | NM_011146 |
| PPRC1 | 61 | 107 ± 24 | 454 ± 61 | 420 | BC066048 |
| LD formation | |||||
| ADRP | 45 | 132 ± 44 | 966 ± 442 | 730 | NM_007408 |
| TIP47 | 81 | 89 ± 7.1 | 450 ± 280 | 510 | NM_025836 |
| FA export | |||||
| MTP | 95 | 82 ± 2.6 | 30 ± 9.0 | 37 | NM_008642 |
| Apolipoprotein | |||||
| Apolipoprotein A-I | 73 | 73 ± 33 | 3.0 ± 5.9 | 4.1 | NM_009692 |
| Apolipoprotein A-II | 193 | 125 ± 46 | 6.9 ± 1.9 | 5.5 | NM_013474 |
| Apolipoprotein A-V | 300 | 98 ± 21 | 3.6 ± 0.40 | 3.7 | NM_080434 |
| Apolipoprotein B | 120 | 122 ± 30 | 15 ± 8.0 | 12 | AK147540 |
| Apolipoprotein C-I | 120 | 93 ± 18 | 19 ± 10 | 20 | NM_007469 |
| Apolipoprotein C-II | 110 | 120 ± 29 | 17 ± 2.2 | 14 | NM_009695 |
| Apolipoprotein C-III | 73 | 38 ± 4.2 | 6.8 ± 1.1 | 18 | NM_023114 |
| Apolipoprotein E | 110 | 104 ± 15 | 22 ± 9.4 | 21 | NM_009696 |
| Cholesterol metabolism | |||||
| SREBP-2 | 91 | 41 ± 8.9 | 11 ± 2.5 | 27 | AK172219 |
| HMG-CoA synthase1 | 100 | 22 ± 7.9 | 13 ± 7.7 | 59 | NM_145942 |
| HMG-CoA synthase2 | 72 | 77 ± 16 | 11 ± 3.7 | 14 | NM_008256 |
| HMG-CoA reductase | 86 | 14 ± 7.3 | 6.1 ± 1.3 | 44 | AK159899 |
| LDL receptor | 75 | 26 ± 8.1 | 3.8 ± 6.9 | 15 | BC053041 |
| LRP1 | 96 | 72 ± 11 | 8.8 ± 2.3 | 12 | NM_008512 |
a Aray analysis was conducted using total RNA independently prepared three times from livers 48 h after intraperitoneal injection of tunicamycin or vehicle (150 mM sucrose) alone into ATF6α +/+ (WT) and −/− (KO) mice. Basal (%), change in WT (%), change in KO (%), and ratio (%) were determined as indicated at the top. Gene accession numbers also are shown.
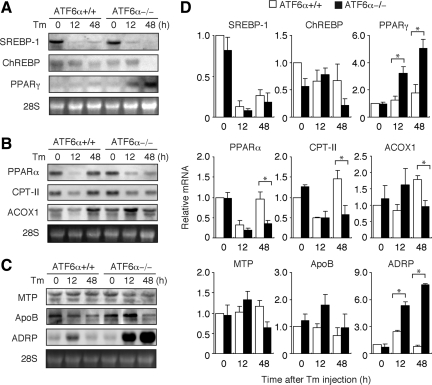
We next checked genes involved in the synthesis of fatty acids. De novo synthesis of fatty acids is regulated by three transcription factors, namely, sterol regulatory element-binding protein (SREBP)-1, carbohydrate response element-binding protein (ChREBP), and peroxisome proliferator-activated receptor (PPAR)γ (Reddy and Rao, 2006). Fatty acid synthase and stearoyl-CoA desaturase are targets of SREBP-1. Microarray and Northern blot hybridization analyses showed that the level of SREBP-1 mRNA was severely decreased in the liver of both ATF6α +/+ and −/− mice after tunicamycin injection (Table 1 and Figure 4A). The level of ChREBP mRNA also was decreased in the liver of ATF6α −/− mice after tunicamycin injection. Although both microarray and Northern blot hybridization showed an increase in the level of PPARγ mRNA in the liver of ATF6α −/− compared with ATF6α +/+ mice at 12 and 48 h after tunicamycin injection (Table 1 and Figure 4A), this enhancement effect might be canceled by a marked decrease in the level of SREBP-1 mRNA. These results suggest that the synthesis of fatty acids is generally suppressed after tunicamycin injection regardless of genotype.
Figure 4.
Effect of tunicamycin injection on expression levels of various genes involved in fatty acid metabolism in the liver of ATF6α +/+ and −/− mice. (A) Tunicamycin (Tm) was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Total RNA was prepared from livers at the indicated time points after injection and analyzed by Northern blot hybridization by using a digoxigenin-labeled probe specific to SREBP-1, ChREBP, or PPARγ. (B) Total RNA was prepared and analyzed as described in A by using a probe specific to PPARα, CPT-II, or Acox1. (C) Total RNA was prepared and analyzed as described in A by using a probe specific to MTP, ApoB, or ADRP. (D) Intensities of each band obtained in A–C were quantified and are presented as the means ± SEM (n = 3) after normalization to the value obtained for uninjected ATF6α +/+ mice. *p < 0.05.
Fatty acids are catabolized to acetyl-CoA for use in energy production mainly by β-oxidization in mitochondria and peroxisomes (Figure 3F). Carnitine palmitoyl transferases (CPTs)-I and -II and acyl-CoA oxidase (Acox1) are key enzymes in mitochondrial and peroxisomal β-oxidization, respectively. PPARα is a master transcriptional regulator of genes involved in β-oxidization in mitochondria and peroxisomes (Reddy and Rao, 2006; Duval et al., 2007). Expressed mRNA levels of these proteins in liver of ATF6α −/− mice were significantly lower than those in ATF6α +/+ mice 48 h after tunicamycin injection when determined by microarray (Table 1). These observations were confirmed by Northern blot hybridization (Figure 4B). PPARα mRNA and CPT-II mRNA levels were decreased at 12 h after tunicamycin injection in ATF6α +/+ and −/− mice and then recovered at 48 h after tunicamycin injection in ATF6α +/+ but not in ATF6α −/− mice. The level of Acox1 mRNA in ATF6α −/− mice was significantly lower than that in ATF6α +/+ mice 48 h after tunicamycin injection. These results suggest that a sustained decrease in the catabolism of fatty acids in the liver of ATF6α −/− mice contributes to the accumulation of triacylglycerol in response to tunicamycin injection.
Sustained Decrease in apoB-100 in ATF6α −/− Mice after Tunicamycin Injection
Amounts of triacylglycerol and cholesterol excess to those required by the liver are incorporated into VLDL particles and secreted into the plasma (Figure 3F). Two proteins are essential to the formation of VLDL. apoB-100 is a major protein component of VLDL, which is present on the surface of VLDL, whereas microsomal triglyceride transfer protein (MTP), a luminal ER protein, mediates the assembly of apoB-100 with neutral lipids. We therefore checked the mRNA levels of these proteins in the microarray analysis data. The level of MTP mRNA in the liver of tunicamycin-injected ATF6α −/− mice was lower than that in similarly treated ATF6α +/+ mice (Table 1), which was confirmed by Northern blot hybridization analysis of mRNA prepared from livers at 48 h after tunicamycin injection (Figure 4C). PDI is an essential component of the microsomal triglyceride transfer enzyme (Noiva, 1999). We found that the protein level of PDI was lower in the liver of tunicamycin-injected ATF6α −/− mice than in similarly treated ATF6α +/+ mice (Figure 2C). These observations suggested that the microsomal triglyceride transfer activity might be decreased in the absence of ATF6α-mediated transcriptional control. Given that the levels of SREBP-1 mRNA and ChREBP mRNA involved in the synthesis of fatty acids were decreased after tunicamycin injection (Figure 4A), apoB-100 might not be lipidated in the liver of tunicamycin-injected ATF6α −/− mice as efficiently as in similarly treated ATF6α +/+ mice. This leads to degradation of apoB-100 (Oyadomari et al., 2006), thereby contributing to lower level of apoB-100 observed in the liver of tunicamycin-injected ATF6α −/− mice than in similarly treated ATF6α +/+ mice (see below), although the difference in SREBP-1 mRNA, MTP mRNA, and PDI protein levels between ATF6α +/+ and −/− mice was not striking.
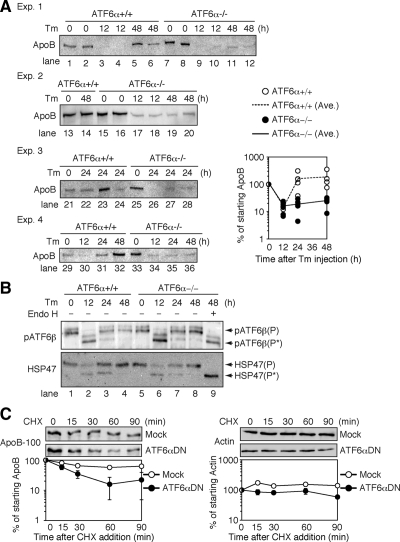
The level of apoB mRNA in the liver of tunicamycin-injected ATF6α −/− mice was severely decreased compared with that in similarly treated ATF6α +/+ mice in microarray analysis (Table 1). In contrast, Northern blot analysis showed that apoB mRNA levels in the liver of tunicamycin-injected ATF6α +/+ and −/− mice were similar at all time points (Figure 4C). Because apoB mRNA is very long (see below), it could not be adequately hybridized to short oligonucleotide probes used in microarray analysis, giving rise to erroneous underestimation. Because apoB-100 consists of 4536 amino acids, and thus represents one of the biggest proteins in the cell, and contains 25 cysteines and 20 N-linked glycosylation sites (Fisher and Ginsberg, 2002), we considered that the folding of apoB-100 would be difficult under ER stress conditions. We therefore examined the effect of tunicamycin injection on the level of apoB-100 in the liver of ATF6α +/+ and −/− mice. As shown in Figure 5A, levels in uninjected ATF6α +/+ and −/− mice were similar (lanes 1, 2, 7, 8, 13, 15, 16, 21, 25, 29, and 33) and were similarly markedly decreased 12 h after tunicamycin injection (Figure 5A, lanes 3, 4, 9, 10, 17, 18, 30, and 34). Importantly, the level of apoB-100 in the liver of ATF6α +/+ mice was restored at 48 h after tunicamycin injection (Figure 5A, lanes 5, 6, 14, and 32). In marked contrast, the level of apoB-100 in the liver of ATF6α −/− mice remained low at 48 h after tunicamycin injection (Figure 5A, lanes 11, 12, 19, 20, and 36).
Figure 5.
Effect of tunicamycin injection on the level of apoB-100 in the liver of ATF6α +/+ and −/− mice. (A) Tunicamycin (Tm) was injected into ATF6α +/+ and −/− mice as described in Figure 1A. Microsomes were prepared from livers at the indicated time points after injection. Aliquots (100 μg) of microsomal lysates were analyzed by immunoblotting using anti-apoB-100 antibody. The results of four independent experiments are shown. Bottom right, intensity of each band was quantified, normalized with the respective value at 0 h, and plotted against time after tunicamycin injection. (B) Top, 100-μg aliquots of microsomal lysates prepared as described in A were digested with (+) or without (−) endo H and then analyzed by immunoblotting using anti-ATF6β antibody. The migration positions of pATF6β(P) and pATF6β(P*), the unglycosylated form of pATF6β(P), are indicated. Bottom, 100-μg aliquots of microsomal lysates prepared as described in A were analyzed by immunoblotting using anti-HSP47 antibody. (C) HepG2 cells transfected with vector alone (Mock) or vector to express a dominant-negative form of ATF6α (ATF6αDN) were treated with cycloheximide (CHX) for the indicated periods. Cell lysates were prepared and analyzed by immunoblotting using antibody to ApoB-100 or actin. Intensity of each band was quantified, normalized with the respective value at 0 h, and plotted against time of cycloheximide treatment. Values are presented as the mean ± SEM (n = 3).
To provide an explanation for the above-mentioned observation and to exclude the unfavorable possibility that tunicamycin was effective in ATF6α −/− mice for a longer time than in ATF6α +/+ mice, we checked the glycosylation status of proteins in the ER before and after tunicamycin injection by using pATF6β(P), a glycoprotein spanning the ER membrane once. For this purpose, liver lysates were digested with or without endoglycosidase H (endo H) and analyzed by immunoblotting with anti-ATF6β antibody. The mobility of pATF6β(P) differed before and after endo H treatment (Figure 5B, compare lane 8 with lane 9), indicating glycosylation of ATF6β with a high mannose-type oligosaccharide [faster migrating band representing unglycosylated ATF6β was designated pATF6β(P*)]. ATF6β was detected as glycosylated pATF6β(P) in the liver of uninjected ATF6α +/+ and −/− mice, as expected (Figure 5B, lanes 1 and 5). In contrast, ATF6β was detected as unglycosylated pATF6β(P*) even without endo H treatment 12 h after tunicamycin injection in ATF6α +/+ and −/− mice (Figure 5B, lanes 2 and 6), indicating that tunicamycin indeed blocked protein N-glycosylation. Analysis of ATF6β showed that ATF6β started to be glycosylated again from 24 h after tunicamycin injection (Figure 5B, lanes 3 and 7) and was fully glycosylated after 48 h (Figure 5B, lanes 4 and 8), probably due to the catabolism of tunicamycin. We also checked the glycosylation status of HSP47, an ER-resident molecular chaperone specific to collagen (Nagata, 2003). Results showed that the unglycosylated form of HSP47 (representing newly synthesized molecules) was produced after tunicamycin injection in the livers of ATF6α +/+ and −/− mice (Figure 5B, compare lane 2 with lane 1 and lane 6 with lane 5) and disappeared from the liver lysate of ATF6α −/− mice no later than it did from that of ATF6α +/+ mice (Figure 5B, lanes 3, 4, 7, and 8). Thus, analyses of ATF6β and HSP47 revealed no significant difference in the duration of tunicamycin action in ATF6α +/+ and −/− mice.
The above-mentioned results suggested the possibility that tunicamycin-induced unglycosylation caused misfolding of apoB-100, which was then degraded by the proteasome, because it was previously shown that apoB-100 was ubiquitinated and degraded by the proteasome when assembly of apoB-100 with lipids was blocked (Zhou et al., 1998). apoB-100 levels in the liver of ATF6α +/+ mice were recovered at 24 h albeit that the extent varied among individuals (Figure 5A, lanes 22, 23 24, and 31), probably due to differences in the rate of catabolism of tunicamycin, whereas apoB-100 levels all remained low in the liver of ATF6α −/− mice (Figure 5A, lanes 26, 27, 28, and 35). To test this notion further, we transfected HepG2 cells with vector alone or vector expressing a dominant negative form of ATF6α (containing the basic leucine zipper region but lacking the transcriptional activation domain) and determined the stability of apoB-100 by cycloheximide chase experiments. As shown in Figure 5C, expression of dominant-negative ATF6α destabilized apoB-100 without affecting the stability of actin, suggesting that ATF6α-mediated transcriptional control is required for the productive folding of apoB-100. Together, we considered that apoB-100 in the liver of tunicamycin-injected mice could be properly folded again when the tunicamycin insult was dismissed only if ER chaperones were induced by the action of ATF6α but remained misfolded and degraded if the induction of ER chaperones was compromised due to the absence of ATF6α.
Induction of Lipid Droplet Formation in ATF6α −/− Mice after Tunicamycin Injection
Among genes involved in fatty acid metabolism, we observed striking differences in the results of microarray analysis on the two genes; changes in ADRP mRNA and tail-interacting protein of 47 kDa (TIP47) mRNA levels were 7.3- and 5.1-fold higher in liver of ATF6α −/− mice than that of ATF6α +/+ mice (Table 1). This marked increase in ADRP mRNA after tunicamycin injection was confirmed by Northern blot hybridization (Figure 4C). These mRNA encode two of several proteins that cover LDs, namely, perilipin, ADRP, and TIP47, collectively termed PAT proteins (Martin and Parton, 2006). We found that signals for perilipin obtained in microarray analysis were too low to be analyzed. Immunoblotting analysis also revealed marked induction of ADRP in the liver of ATF6α −/− mice but not of ATF6α +/+ mice after tunicamycin injection (Figure 2C). Interestingly, analysis of HEK293T cells showed that ADPR was not a target of the UPR, because it was not induced by any of three ER stress inducers, namely, tunicamycin, thapsigargin, and dithiothreitol (Figure 2E). Because ADRP was induced only after tunicamycin injection in the liver of ATF6α −/− mice (Figure 2C), it is unlikely that ATF6α suppresses the expression of ADRP constitutively. The mechanism of ADRP induction remains to be determined.
LDs, the site of lipid storage in the cell, contain neutral lipids such as triacylglycerol and cholesterol ester in their core, which are surrounded by a monolayer of phospholipids (Tauchi-Sato et al., 2002; Martin and Parton, 2006). We therefore analyzed liver sections by electron microscopy. As shown in Figure 6A, hepatocytes of ATF6α +/+ and −/− mice before tunicamycin injection were quite similar except that hepatocytes of −/− mice contained a few vacuole-like structures (compare Figure 6A, a and b, vacuoles are indicated by arrows). Interestingly, ribosome-bound rough ER were markedly enlarged 48 h after tunicamycin injection in hepatocytes of ATF6α +/+ mice (Figure 6A, c and e [with higher magnification], rough ER are indicated by arrowheads). In marked contrast, the rough ER in hepatocytes of ATF6α −/− mice seemed normal, but the hepatocytes instead contained an extremely large number of vacuole-like structures (Figure 6Ad), which probably represented LDs, because the structures were completely round and not surrounded by lipid bilayers (Figure 6Af, with higher magnification). The presence of ADRP in the structure was confirmed as follows. Liver taken from ATF6α +/+ and −/− mice 0, 12, and 48 h after tunicamycin injection was homogenized, and the homogenates were separated by sucrose gradient centrifugation. LDs are known to be recovered in the top fraction; we found that ADRP was indeed present in the top fraction and that levels were markedly increased after tunicamycin injection into ATF6α −/− mice (Figure 6B). Based on these results, we concluded that tunicamycin injection caused liver steatosis by blocking β-oxidation of fatty acids, suppressing VLDL formation, and facilitating LD formation in ATF6α −/− mice.
DISCUSSION
The liver is a major site for the production of proteins as well as lipoprotein particles circulating in plasma, and the ER is a key place for their synthesis and maturation. The protein quality control system in the ER is tightly and dynamically regulated by the UPR, an intracellular signaling from the ER. Among three ubiquitous signaling pathways operating in mammalian UPR, namely, the IRE1α, PERK, and ATF6α pathways, ATF6α plays a pivotal role in up-regulating various ER quality control proteins in response to ER stress. However, ATF6α-deficient mice show no apparent phenotype under normal growing conditions (Wu et al., 2007; Yamamoto et al., 2007). In this report, we subjected ATF6α-deficient mice to ER stress pharmacologically and observed the induction of liver steatosis, which progressed from the blockage of β-oxidation of fatty acids, suppression of VLDL formation, and facilitation of LD formation.
Kaufman and coworkers conducted similar experiments with ATF6α-deficient mice and found that tunicamycin injection caused liver steatosis, consistent with our results (Rutkowski et al., 2008). Furthermore, they created mice harboring liver-specific deletion of IRE1α as well as mice exhibiting liver-specific blockage of ER stress-induced phosphorylation of eukaryotic initiation factor 2α, an event immediately downstream of PERK activation. Tunicamycin injection into these two types of mice also caused fatty liver. Thus, defects in the three principal UPR signaling pathways all lead to hepatic steatosis. They found prolonged induction of the proapoptotic transcription factor CHOP as a consequence of tunicamycin injection into ATF6α-deficient mice and mice harboring liver-specific deletion of IRE1α, and they also found suppression of CCAAT/enhancer-binding protein (C/EBP)α as a consequence of tunicamycin injection into all three types of mice. They postulated that defective UPR signaling resulted in sustained ER stress, leading to the sustained induction of CHOP even during the period in which the hepatocytes of wild-type mice recovered from the insult via activation of the UPR. This notion was supported by formation of fatty liver, prolonged induction of CHOP, and sustained suppression of C/EBPα after tunicamycin injection into mice deficient in p58IPK, a DnaJ domain-containing cochaperone of the DnaK protein BiP present in the ER (Rutkowski et al., 2007). CHOP expression suppresses C/EBPα because CHOP can function as a dominant-negative regulator of C/EBP family members (Ron and Habener, 1992). This in turn leads to the suppression of metabolic gene expression through the suppressed expression of their master regulators, such as SREBP-1 and PPARα, via possible transcriptional control: it was shown previously that C/EBPα binding sites seem to exist on their promoter regions (Villacorta et al., 2007). Because PPARα is a master transcriptional regulator of genes involved in β-oxidization in mitochondria and peroxisomes (Reddy and Rao, 2006; Duval et al., 2007), suppression of its expression contributes to the accumulation of fatty acids. Our results obtained with ATF6α-deficient mice are generally consistent with those by Kaufman and coworkers. We indeed observed a more sustained induction of CHOP in ATF6α-deficient mice than in wild-type mice (Figure 2C) as well as suppression of PPARα expression (Figure 4B).
We sought to correlate the induction of liver steatosis and LD formation in hepatocytes more directly with the failure of ATF6α-deficient mice to up-regulate ER quality control proteins in response to ER stress. We therefore checked expression of apoB-100 at a protein level, which was not conducted by Kaufman and coworkers. apoB-100, a huge (4536-amino acid) and heavily-glycosylated (20 potential sites) protein, is an essential component of VLDL, accounting for approximately one third of total lipoproteins present in VLDL (Fisher and Ginsberg, 2002). Previous results showed that the half-life of apoB mRNA is quite long (∼16 h) (Pullinger et al., 1989) but that only 30–60% of newly synthesized apoB-100 is secreted into the extracellular milieu (Borchardt and Davis, 1987; Sato et al., 1990), indicating that the level of apoB-100 is regulated posttranslationally and that the proper folding of apoB-100 is difficult even under normal conditions. We suspected that the folding of apoB-100 is made even more difficult under ER stress conditions. Indeed, we found that apoB-100 levels were greatly mitigated when protein N-glycosylation in the liver was blocked by tunicamycin injection, as evidenced from the analysis of ATF6β and HSP47, regardless of the genotype of mice (Figure 5A). Importantly, apoB-100 levels were restored in wild-type mice when the effects of tunicamycin disappeared but remained low in ATF6α-deficient mice (Figure 5A). We cannot formally rule out the possibility that decreased level of apoB-100 mRNA in tunicamycin-treated ATF6α-deficient mice contributed to sustained decrease in apoB-100 protein level due to the discrepancy between the data of microarray analysis (Table 1) and Northern blot hybridization analysis (Figure 4C). Nonetheless, our results suggest that the levels of ER quality control proteins can directly affect the productive folding process of apoB-100. Resulting down-regulation of apoB-100 favors the accumulation of triacylglycerol and cholesterol, leading to the formation of LDs in hepatocytes and then to the development of liver steatosis.
Although tunicamycin injection-induced liver steatosis represents an extreme condition, such that ATF6α-deficient mice die within 3 d after this pharmacological challenge, our and Kaufman's results suggest the possibility that chronic ER stress is involved in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). NAFLD encompasses a wide spectrum of liver diseases ranging from simple steatosis, characterized by the accumulation of triglyceride within hepatocytes, to nonalcoholic steatohepatitis, which may lead to liver fibrosis or cirrhosis (Day, 2002; Harrison et al., 2002; McClain et al., 2004; Adams et al., 2005; Sanal, 2008). The development of NAFLD is affected by genetic and environmental factors (Day, 2002) and occurs more frequently among people with diabetes (50%) and obesity (76%) (Adams et al., 2005). The study of NAFLD in humans is hampered by the genetic heterogeneity within populations, and no universally applicable noninvasive diagnostic method is available. Animal models therefore provide valuable tools to investigation of the pathogenesis of NAFLD (Anstee and Goldin, 2006). One currently favored hypothesis for the development of NAFLD is the “two-hit theory” that was originally proposed for nonalcoholic steatohepatitis (Day and James, 1998). The first hit increases the vulnerability of the liver to the second hit through the accumulation of neutral lipids within hepatocytes (hepatic steatosis), and the second hit in turn leads to inflammation and hepatocyte injury (fibrosis or cirrhosis). Several factors have been suggested to play a role in the development of NAFLD, including insulin resistance, oxidative stress, mitochondrial dysfunction, dysregulated cytokine metabolism, drugs, toxins, and abnormal lipid metabolism (Browning and Horton, 2004; Adams et al., 2005; Wei et al., 2008). However, the mechanisms by which neutral lipids accumulate within hepatocytes and how steatosis turns into steatohepatitis remain elusive.
Interestingly, several recent studies have revealed an unexpected link between ER stress and liver steatosis or the involvement of UPR mediators in hepatic lipogenesis or glycolysis. Homocysteine-induced ER stress caused the activation of SREBPs and a subsequent increase in the levels of triglycerides and cholesterol in the liver (Werstuck et al., 2001). Selective deletion in the liver of XBP1, a downstream transcription factor of IRE1α, caused a decrease in lipid production, leading to severe hypocholesterolemia and hypotriglycemia (Lee et al., 2008). Selective blockage in the liver of ER stress-induced phosphorylation of eukaryotic initiation factor 2α, a downstream event of PERK activation, resulted in a decrease in hepatic glycogen levels and susceptibility to fasting hypoglycemia in lean mice, and enhanced glucose tolerance and diminished hepatosteatosis in mice fed a high-fat diet (Oyadomari et al., 2008).
Here, we provide a new linkage between ER stress and liver steatosis by revealing a new intersection at which the ATF6α branch of the UPR can directly affect lipid metabolism. In addition to the effects on β-oxidation of fatty acids and the synthesis of LD components, we showed that ATF6α-mediated regulation of ER quality control proteins is critical to facilitating the folding of apoB-100 and thereby maintain its level under ER stress conditions, which in turn minimizes intracellular levels of triglycerides and cholesterols in the liver by encapsulating these lipids in VLDL for secretion into plasma. Together with the results of Kaufman, our results suggest that chronic ER stress or chronic failure of ER quality control can act as a first hit in the development of NAFLD. Chronic ER stress might occur in the liver of individuals when the function of ER quality control proteins is compromised or when the homeostatic UPR is not activated efficiently. By subjecting ATF6α-deficient mice to continuous milder ER stress, we may be able to gain new insights into the pathogenesis of NAFLD. These studies are now in progress.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Toyoshi Fujimoto (Nagoya University Graduate School of Medicine) for valuable advice. We thank Ms. Kaoru Miyagawa for technical and secretarial assistance. This work was supported, in part, by Ministry of Education, Culture, Sports, Science and Technology of Japan grants 15GS0310 and 20247026 (to K. M.) and by a grant from the Ono Medical Research Foundation.
Abbreviations used:
- Acox
acyl-CoA oxidase
- ALT
alanine aminotransferase
- apoB-100
apolipoprotein B-100
- ChREBP
carbohydrate response element-binding protein
- CPT
carnitine palmitoyl transferase
- endo H
endoglycosidase H
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- LD
lipid droplet
- LPS
lipopolysaccharide
- MEF
mouse embryonic fibroblast
- NAFLD
nonalcoholic fatty liver disease
- PDI
protein disulfide isomerase
- PPAR
peroxisome proliferator-activated receptor
- SREBP
sterol regulatory element-binding protein
- TIP
tail-interacting protein
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- UPR
unfolded protein response
- VLDL
very-low-density lipoprotein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0133) on July 14, 2010.
REFERENCES
- Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. ATF6 is a Transcription Factor Specializing in the Regulation of Quality Control Proteins in the Endoplasmic Reticulum. Cell Struct. Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Adams L. A., Angulo P., Lindor K. D. Nonalcoholic fatty liver disease. CMAJ. 2005;172:899–905. doi: 10.1503/cmaj.045232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstee Q. M., Goldin R. D. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int. J. Exp. Pathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Borchardt R. A., Davis R. A. Intrahepatic assembly of very low density lipoproteins. Rate of transport out of the endoplasmic reticulum determines rate of secretion. J. Biol. Chem. 1987;262:16394–16402. [PubMed] [Google Scholar]
- Bradbury M. W. Lipid metabolism and liver inflammation. I. Hepatic fatty acid uptake: possible role in steatosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G194–G198. doi: 10.1152/ajpgi.00413.2005. [DOI] [PubMed] [Google Scholar]
- Browning J. D., Horton J. D. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Day C. P. Pathogenesis of steatohepatitis. Best Pract. Res. Clin. Gastroenterol. 2002;16:663–678. doi: 10.1053/bega.2002.0333. [DOI] [PubMed] [Google Scholar]
- Day C. P., James O. F. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- Duval C., Muller M., Kersten S. PPARalpha and dyslipidemia. Biochim. Biophys. Acta. 2007;1771:961–971. doi: 10.1016/j.bbalip.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Fisher E. A., Ginsberg H. N. Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J. Biol. Chem. 2002;277:17377–17380. doi: 10.1074/jbc.R100068200. [DOI] [PubMed] [Google Scholar]
- Harada A., Sobue K., Hirokawa N. Developmental changes of synapsin I subcellular localization in rat cerebellar neurons. Cell Struct. Funct. 1990;15:329–342. doi: 10.1247/csf.15.329. [DOI] [PubMed] [Google Scholar]
- Harrison S. A., Kadakia S., Lang K. A., Schenker S. Nonalcoholic steatohepatitis: what we know in the new millennium. Am. J. Gastroenterol. 2002;97:2714–2724. doi: 10.1111/j.1572-0241.2002.07069.x. [DOI] [PubMed] [Google Scholar]
- Haze K., Okada T., Yoshida H., Yanagi H., Yura T., Negishi M., Mori K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J., Weissman J. S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Parton R. G. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- McClain C. J., Mokshagundam S. P., Barve S. S., Song Z., Hill D. B., Chen T., Deaciuc I. Mechanisms of non-alcoholic steatohepatitis. Alcohol. 2004;34:67–79. doi: 10.1016/j.alcohol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Nadanaka S., Yoshida H., Kano F., Murata M., Mori K. Activation of mammalian unfolded protein response is compatible with the quality control system operating in the endoplasmic reticulum. Mol. Biol. Cell. 2004;15:2537–2548. doi: 10.1091/mbc.E03-09-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K. HSP47 as a collagen-specific molecular chaperone: function and expression in normal mouse development. Semin. Cell. Dev. Biol. 2003;14:275–282. doi: 10.1016/j.semcdb.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B. A., Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- Noiva R. Protein disulfide isomerase: the multifunctional redox chaperone of the endoplasmic reticulum. Semin. Cell. Dev. Biol. 1999;10:481–493. doi: 10.1006/scdb.1999.0319. [DOI] [PubMed] [Google Scholar]
- Okada T., Haze K., Nadanaka S., Yoshida H., Seidah N. G., Hirano Y., Sato R., Negishi M., Mori K. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J. Biol. Chem. 2003;278:31024–31032. doi: 10.1074/jbc.M300923200. [DOI] [PubMed] [Google Scholar]
- Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S., et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 2004;15:99–110. doi: 10.1091/mbc.E03-06-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullinger C. R., North J. D., Teng B. B., Rifici V. A., Ronhild de Brito A. E., Scott J. The apolipoprotein B gene is constitutively expressed in HepG2 cells: regulation of secretion by oleic acid, albumin, and insulin, and measurement of the mRNA half-life. J. Lipid Res. 1989;30:1065–1077. [PubMed] [Google Scholar]
- Reddy J. K., Rao M. S. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G852–G858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- Ron D., Habener J. F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant- negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski D. T., Kang S. W., Goodman A. G., Garrison J. L., Taunton J., Katze M. G., Kaufman R. J., Hegde R. S. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., et al. UPR Pathways Combine to Prevent Hepatic Steatosis Caused by ER Stress-Mediated Suppression of Transcriptional Master Regulators. Dev. Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Sanal M. G. The blind men ‘see’ the elephant-the many faces of fatty liver disease. World J. Gastroenterol. 2008;14:831–844. doi: 10.3748/wjg.14.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R., Imanaka T., Takatsuki A., Takano T. Degradation of newly synthesized apolipoprotein B-100 in a pre-Golgi compartment. J. Biol. Chem. 1990;265:11880–11884. [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shen X., Ellis R. E., Sakaki K., Kaufman R. J. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in. C. elegans. PLoS Genet. 2005;1:355–368. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J. Biol. Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trent M. S., Stead C. M., Tran A. X., Hankins J. V. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- Uchiyama S., Shimizu T., Shirasawa T. CuZn-SOD deficiency causes ApoB degradation and induces hepatic lipid accumulation by impaired lipoprotein secretion in mice. J. Biol. Chem. 2006;281:31713–31719. doi: 10.1074/jbc.M603422200. [DOI] [PubMed] [Google Scholar]
- Vabulas R. M., Hartl F. U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Villacorta L., Garcia-Barrio M. T., Chen Y. E. Transcriptional regulation of peroxisome proliferator-activated receptors and liver X receptors. Curr. Atheroscler. Rep. 2007;9:230–237. doi: 10.1007/s11883-007-0024-5. [DOI] [PubMed] [Google Scholar]
- Wei Y., Rector R. S., Thyfault J. P., Ibdah J. A. Nonalcoholic fatty liver disease and mitochondrial dysfunction. World J. Gastroenterol. 2008;14:193–199. doi: 10.3748/wjg.14.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werstuck G. H., et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J. Clin. Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhovsky S., Gardner R., Hampton R. HRD gene dependence of endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 2000;11:1697–1708. doi: 10.1091/mbc.11.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson R. B., Komuro R., Chen X., Dave U. P., Prywes R., Brown M. S., Goldstein J. L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Mori K. ATF6 activated by proteolysis directly binds in the presence of NF-Y (CBF) to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol. Cell. Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Fisher E. A., Ginsberg H. N. Regulated Co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J. Biol. Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.