We identify new mRNA targets for the QKI-6 RNA binding proteins using an unbiased approach. We show that AIP-1 mRNA is bound by QKI-6 within its 3′-UTR. This regulation is observed in oligodendrocytes and it is essential for oligodendrocyte process outgrowth.
Abstract
The quaking viable (qkv) mice represent an animal model of dysmyelination. The absence of expression of the QKI-6 and QKI-7 cytoplasmic isoforms in oligodendrocytes (OLs) during CNS myelination causes the qkv mouse phenotype. The QKI RNA-binding proteins are known to regulate RNA metabolism of cell cycle proteins and myelin components in OLs; however, little is known of their role in reorganizing the cytoskeleton or process outgrowth during OL maturation and differentiation. Here, we identify the actin-interacting protein (AIP)-1 mRNA as a target of QKI-6 by using two-dimensional differential gel electrophoresis. The AIP-1 mRNA contains a consensus QKI response element within its 3′-untranslated region that, when bound by QKI-6, decreases the half-life of the AIP-1 mRNA. Although the expression of QKI-6 is known to increase during OL differentiation and CNS myelination, we show that this increase is paralleled with a corresponding decrease in AIP-1 expression in rat brains. Furthermore, qkv/qkv mice that lack QKI-6 and QKI-7 within its OLs had an increased level of AIP-1 in OLs. Moreover, primary rat OL precursors harboring an AIP-1 small interfering RNA display defects in OL process outgrowth. Our findings suggest that the QKI RNA-binding proteins regulate OL differentiation by modulating the expression of AIP-1.
INTRODUCTION
The quaking viable (qkv) mice harbor an autosomal recessive deletion encompassing part of the qkI locus (Ebersole et al., 1996b). These mice represent an animal model of CNS dysmyelination, because they display rapid tremors that are especially pronounced in their hind limbs. The oligodendrocytes (OLs) of the qkv mice display a maturation defect and as a result do not properly compact myelin (Hogan and Greenfield, 1984). The exact reason that OLs from the qkv mice fail to mature is unknown; however, it is likely that there is a premature cessation of the OL differentiation due to defective RNA regulation (Chenard and Richard, 2008).
The qkI gene expresses three major alternatively spliced mRNAs (5, 6, and 7 kb) encoding QKI-5, QKI-6, and QKI-7 that differ in their C-terminal 30 amino acids (Ebersole et al., 1996b). The QKI-5 unique sequence contains a nuclear localization signal that renders this isoform exclusively nuclear (Wu et al., 2002). QKI-6 and QKI-7 are cytoplasmic (Pilotte et al., 2001) and in mice, their expression coincides with the process of myelination during the first two postnatal weeks (Ebersole et al., 1996a). The genetic deletion in qkv mice prevents the expression of the QKI-6 and QKI-7 isoforms in OLs, resulting in a dysmyelination phenotype (Ebersole et al., 1996b; Hardy et al., 1996a) and implying that the QKI-6 and QKI-7 isoforms play a key role in myelination. Indeed, the forced expression of QKI-6 and QKI-7 induces OL differentiation and maturation (Larocque et al., 2005). Moreover, a transgenic allele that expresses QKI-6 specifically in the OL lineage rescues the dysmyelination phenotype of qkv mice (Zhao et al., 2006b).
The QKI proteins belong to the heteronuclear ribonucleoprotein particle K homology domain family of RNA-binding proteins that bind RNA with sequence specificity and high affinity (Lee and Schedl, 2006; Lukong et al., 2008; Volk et al., 2008). The QKI proteins selectively interact with a short sequence termed the QKI response element [QRE; ACUAAY (N1-20) UAAY] (Galarneau and Richard, 2005) frequently located in the 3′-untranslated region (UTR) of the target mRNAs. Functional QREs have been identified in the mRNAs of myelin basic protein (MBP) (Li et al., 2000; Larocque et al., 2002), Krox-20 (Egr-2) (Nabel-Rosen et al., 2002; Galarneau and Richard, 2005), microtubule-associated protein 1B (Zhao et al., 2006a), and the cyclin-dependent kinase inhibitor p27KIP1 (Larocque et al., 2005). Many other mRNAs are predicted to be regulated by QKI, because a bioinformatics analysis led to the identification of 1433 mRNAs that contain at least one QRE (Galarneau and Richard, 2005). Gene ontology annotation identified development as a major category of putative QKI RNA targets, consistent with the dysmyelination phenotype of the qkv mice (Galarneau and Richard, 2005). Another category that was identified was cell adhesion (Galarneau and Richard, 2005), but a functional link between the QKI proteins and mRNAs encoding proteins regulating cell adhesion remains undefined.
To identify QKI-6–regulated targets, we introduced QKI-6 in CRL2020, a QKI-deficient glioblastoma multiforme cell line. Cellular extracts were prepared from QKI-6 expressing CRL2020 and a control green fluorescent protein (GFP) expressing CRL2020. Each extract was labeled, and the differentially expressed proteins were identified by two-dimensional difference gel electrophoresis (2D-DIGE), followed by mass spectrometry. We identified several cytoskeletal proteins, including AIP-1, tropomyosin (TPM)1, Actin, γ1 (ACTG1), and lamin B1 that are regulated by QKI-6. These findings are consistent with cell adhesion as being a major annotation category of the QKI RNA targets that we identified by using SELEX and bioinformatics (Galarneau and Richard, 2005). We confirmed that the AIP-1 mRNA is an RNA target for QKI-6. Moreover, we show that the AIP-1 mRNA levels are regulated in purified OLs and during CNS myelination. These findings define a new regulatory network in OLs controlled by the QKI proteins.
MATERIALS AND METHODS
Antibodies
The anti-myc (9E10) and A2B5 mouse monoclonal antibodies were obtained from the American Type Culture Collection (Manassas, VA). Antibodies against α-tubulin and β-actin were purchased from Sigma-Aldrich (St. Louis, MO). The QKI-6 antibodies were raised in rabbits by using the peptide KEYPIEPSGVLGMAFPTKG. Anti-AIP-1 was purchased from Aviva Systems Biology (San Diego, CA). Anti-voltage-dependent anion channel 2 (VDAC2) polyclonal and nucleophosmin 1 (NPM1) were from Abcam (Cambridge, MA). Anti-TPM3 was purchased from Abnova (Walnut, CA).
2D-DIGE
The patient derived glioblastoma multiforme CRL2020 cell line was purchased from American Type Culture Collection (Mulholland et al., 2006). These cells were transduced for 2 d in triplicates with either AdGFP or AdQKI-6, as described previously (Larocque et al., 2002). Differentially expressed protein spots were selected, picked, and identified by mass spectrometry by Applied Biomics (Hayward, CA).
QKI Knockdown in U343
U343 glioblastoma cell lines were transfected with siGENOME SMARTpool small interfering RNA (siRNA) targeting the mRNA for the QKI isoforms or siGFP (5′-GCT GAC CCT GAA GTT CAT CTG CGT-3′) as control for 2 d, followed by cell harvesting in Laemmli buffer. Lipofectamine RNAiMAX transfection reagent was used according to manufacturer's protocol (Invitrogen, Carlsbad, CA). siQKI (M-104651-00-0010) was purchased from Dharmacon RNA Technologies (Lafayette, CO).
Electrophoretic Mobility Shift Assay
Synthetic RNAs were prepared by runoff in vitro transcription using T7 MegaShortscript (Ambion, Austin, TX) according to the manufacturer's protocols. A constant concentration of 32P-labeled RNA (100 pmol) was incubated alone, with 1 μM glutathione transferase (GST), or with increasing concentrations (100 nM, 300 nM, and 1 μM) of GST-QKI-6 or GST-QKI-6:V157E mutant in the following buffer: 20 mM HEPES, pH 7.4, 330 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mg/ml heparin, and 0.01% IGEPAL CA630 [Sigma-Aldrich]). The 30-μl reactions were incubated at room temperature for 1 h, and then 3.3 μl of RNA loading dye was added to each sample. Next, 20 μl of each sample was separated on native Tris-glycine 8% acrylamide gels. The gels were dried and the bound, and unbound RNAs were analyzed using a Storm PhosphorImager (GE Healthcare, Baie-d'Urfe, QC, Canada).
Luciferase (Luc) Assays
The 3′-UTR of AIP-1 was amplified using the following primers (5′-AAT CAA GCT TAC GCA AAA ACA CTC CCA ATC-3′ and 5′-AAT CAC TAG TCT CTG GAT GGA CCG AAT CAG-3′) from the human AIP-1 full-length clone (Origene, Rockville, MD) and cloned into the pMIR-REPORT luciferase vector (Ambion). Human embryonic kidney (HEK)293 cells were cotransfected with either empty pMIR luciferase, the pLuc:AIP-1 3′-UTR, or the pLuc:AIP-1 3′-UTR with the mutated QRE along with either pcDNA (control), pcDNA-myc-QKI-6, or pcDNA-myc-QKI-6:V157E. pRLTK (Promega, Madison, WI) encoding Renilla luciferase was used to control transfection efficiency. The cell extracts were harvested 48 h after transfection, and luciferase activity was assayed using the Dual-Luciferase Reporter Assay kit (Promega) and measured using the GloMax 20/20 luminometer (Promega).
In Vivo RNA Binding Assay
CRL2020 cells transduced with AdQKI-6 were harvested in lysis buffer (0.1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 1 mg/ml heparin, and 0.5 U/μl RNasin). The lysates were immunoprecipitated with anti-myc antibody or immunoglobulin (Ig)G. RNA was isolated using TRIzol isolation reagent (Invitrogen) according to the manufacturer's protocol. Reverse transcription assays were performed using SuperScript II Reverse Transcriptase (Invitrogen). The sequences of the primers used were as follows: AIP-1, 5′-CAT TCC TTT GAA ATA AGG TT-3′ and 5′-AAA TAT GTA CTA CGG AAT TA-3′; p27KIP1, 5′-CCT TCC CCA AAA TTG CTT CT-3′ and 5′-CCG GCT AAC TCT GAG GAC AC-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-AGG GGT CTA CAT GGC AAC TG-3′ and 5′-GTC AGT GGT GGA CCT GAC CT-3′.
Real-Time Polymerase Chain Reaction (PCR)
Real-time PCR reactions were performed on the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) using SYBR Green PCR Master Mix (QIAGEN, Valencia, CA). Data analysis was performed using Real-Time PCR software 7500 version 2.0.1 (Applied Biosystems). The relative concentrations of the genes of interest were determined using the comparative Ct method after normalizing to the endogenous control (GAPDH). Primers from QIAGEN were as follows: TPM3, Hs_TPM3_2_SG; VDAC2, Hs_VDAC2_2_SG; AIP-1, Hs_WDR1_2_SG; NPM1, Hs_NPM1_2_SG; Actin, Hs_ACTB_2_SG; p27KIP1, Hs_CDKN1B_2_SG; and GAPDH, Hs_GAPDH_2_SG.
mRNA Half-Life Analysis
Total RNA was isolated from CRL2020 cell lines transduced with either AdGFP or AdQKI-6 for 24 h and treated with 5 μg/ml actinomycin D from 0 to 24 h. mRNA levels of AIP-1, p27KIP1, Actin, and GAPDH were examined by real-time PCR with the following primers from QIAGEN: AIP-1, Hs_WDR1_2_SG; Actin, Hs_ACTB_2_SG; p27KIP1, Hs_CDKN1B_2_SG; and GAPDH, Hs_GAPDH_2_SG.
OL AIP-1 Localization and Actin Filament Localization
Primary OL precursor cells were isolated, purified, and differentiated as described previously (Almazan et al., 1993). For nonphalloidin staining, 4% paraformaldehyde fixation was used followed by permeabilization in 0.5% Triton X-100 and blocking in 10% goat serum. For actin filament staining, cells were rinsed in a cytoskeletal buffer containing 137 mM NaCl, 5 mM KCl, 1 mM Na2HPO4, 0.4 mM KH2PO4, 5.5 mM glucose, 4 mM NaHCO3, 2 mM MgCl2, 2 mM EGTA, and 10 mM MES, pH 6.1 (Webb et al., 1995). The cells were then fixed and permeabilized in the cytoskeletal buffer, supplemented with 0.5% Triton X-100 and 0.25% glutaraldehyde for 10 min at room temperature, followed by a more stringent fixation in cytoskeletal buffer with 1% glutaraldehyde for 10 min at room temperature. After fixation, cells were blocked with 10% goat serum for 15 min at room temperature before incubation with phalloidin conjugated to Alexa 488 (500 nM) for 30 min at room temperature. Images were acquired using AxioImager 2 upright microscope (Carl Zeiss, Jena, Germany) and prepared using AxioVision 4.8.1 software (Carl Zeiss).
OL Total mRNA Analysis and mRNA Half-Life
Total RNA was isolated using the TRIzol (Invitrogen) from rat primary oligodendrocyte precursor cells (OPCs). mRNA half-life analysis was performed on purified rat primary OPCs differentiated for two time points (0 and 4 d) and treated with 5 μg/ml actinomycin D for 0–24 h. First-strand cDNA synthesis and real-time PCR was performed as described previously (Larocque et al., 2005). The following oligonucleotide primers used for purified OLs total and half-life mRNA assay were ordered from QIAGEN: Actin, Rn_Actg1_1_SG; GAPDH, Rn_Gapd_1_SG; AIP-1, Rn_Wdr1_1_SG; p27KIP1, Rn_Cdkn1b_2_SG; and MBP, Rn_Mbp_1_SG. Id2 primers were ordered from Invitrogen: 5′-GGA CAG AAC CAA ACG TCC AG-3′ and 5′-TAA GCT CAG AAG GGA ATT CAG-3′.
AIP-1 Knockdown in OLs
Purified OPCs were transfected with siGLO and siGENOME SMARTpool siAIP-1 or siGLO and sipGL2 (5′-CGT ACG CGG AAT ACT TCG A-3′) for 2 d, followed by a second siRNA treatment. siIMPORTER Transfection reagent (Millipore, Billerica, MA) was used according to the protocol provided for adherent cells. Twelve hours after the second siRNA treatment, the culture media were changed to either serum-free media (SFM) with growth factors or into SFM supplemented with T3 to promote differentiation. Quantification of cell types in sipGL2- or siAIP-1–transfected cells was determined by morphology and immunostaining using A2B5 antibodies. Approximately 50 cells from each triplicate of siAIP-1 and sipGL2 transfection were assessed for the presence of secondary branching. siGENOME SMARTpool siAIP-1 (M-085115-00-0010) and siGLO (D-001600-01-05) were purchased from Dharmacon RNA Technologies. Images were acquired using AxioImager 2 upright microscope (Carl Zeiss) and prepared using AxioVision 4.8.1 software (Carl Zeiss).
Animals
Animals were killed in accordance with a protocol approved by the Animal Care Committee at McGill University. qkv/qkv mice were obtained from JAX laboratories (Bar Harbor, ME) and maintained on a C57BL/6 background. The qkv mutation was amplified using primers directed against the breakpoint (5′-TCT AAA GAG CAT TTT CGA AGT-3′) and (5′-TTG CTA ACT GAA TAT TAC T-3′). Rat brains were weighed and solubilized in ice-cold lysis buffer (1:5, wt/vol) containing 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail (Roche, Laval, QC, Canada). Brains were homogenized by pulse sonication at 15-s intervals on ice, followed by centrifugation at 16,000 × g for 20 min. Supernatant protein concentrations were determined by bicinchoninic acid assay according to manufacturer's instructions (Thermo Fisher Scientific, Waltham, MA).
Immunocytochemistry
Mice were anesthetized with isoflurane and perfused with ice-cold phosphate-buffered saline followed by 4% paraformaldehyde. Brains were cryoprotected in 30% sucrose overnight at 4°C and embedded in OCT compound (Tissue-Tek, Markham, ON, Canada) over dry ice in acetone. Tissues were cryostat sectioned at 6 μm and collected on +/+ slides (Fisher, Ottawa, ON, Canada). Tissue sections were blocked in 10% goat serum in Tris-buffered saline + 0.5% Triton X-100 for 1 h followed by incubation with primary antibodies overnight at room temperature. Slides were incubated with Alexa-Flour 488 or 546 IgG at a dilution of 1:400 for 4 h.
RESULTS
Identification of Cellular Proteins Modulated by QKI-6 Expression Using 2D-DIGE
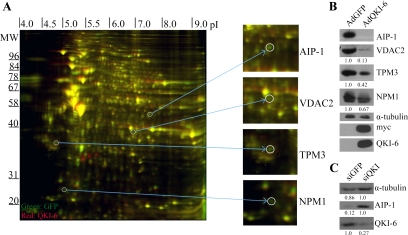
The QKI isoforms are sequence-specific RNA-binding proteins (Ryder and Williamson, 2004; Galarneau and Richard, 2005) known to modulate various aspects of RNA metabolism in myelinating cells (Chenard and Richard, 2008); however, few physiological QKI RNA targets are known. To identify QKI RNA targets, we reasoned that the mRNA and protein levels of certain targets would be modulated by the expression of the QKI isoforms. The QKI-deficient glioblastoma multiforme cell line CRL2020 (Mulholland et al., 2006) was transduced with a control adenovirus expressing GFP (AdGFP) or an adenovirus expressing myc-epitope–tagged QKI-6 and GFP (AdQKI-6). We focused on the QKI-6 isoform, because it has been shown to rescue the phenotype of the qkv mice (Zhao et al., 2006b). Approximately 95% of the cells infected with AdGFP or AdQKI-6 expressed GFP, a marker of infection. The cellular extracts were differentially labeled with fluorescent dyes and analyzed by 2D-DIGE (Figure 1A). The analysis revealed 21 putative QKI-6-regulated targets, 12 of which were up-regulated and nine of which were down-regulated. The putative QKI-6 targets were analyzed by mass spectrometry (Table 1). By using immunoblotting analysis, we confirmed the decrease in AIP-1, VDAC2, TPM3, and NPM1 with the ectopic expression of the QKI-6 isoform (Figure 1B). The expression of the myc-epitope–tagged QKI-6 was confirmed by immunoblotting with anti-myc and -QKI-6 antibodies, whereas anti-α-tubulin antibodies were used to demonstrate equivalent loading (Figure 1B).
Figure 1.
Two-dimensional DIGE identifies QKI-6–regulated targets in glioblastoma cells. (A) The differential protein expression between AdGFP-infected and AdQKI-6–infected CRL2020 cells was identified by 2D-DIGE. The unique protein spots (green or red, not yellow) were picked and identified by mass spectrometry. The molecular mass markers are shown on the ordinate in kilodaltons, and the isoelectric point is shown on the abscissa. Four representative unique spots were enlarged on the right, which were identified via mass spectrometry as AIP-1, VDAC2, TPM3, and NPM1. (B) Protein extracts from AdGFP and AdQKI-6 were analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting with anti-AIP-1, -VDAC2, -TPM3, and -NPM1 antibodies. Anti-α-tubulin antibodies were used to confirm equivalent protein expression. Anti-myc and -QKI-6 antibodies confirmed the expression of the myc-epitope–tagged QKI-6. The data shown are representative of three independent experiments and normalized to α-tubulin protein levels. The relative amounts are presented underneath each blot and were performed using Scion-Image (Scion, Frederick, MD). The standard deviations for VDAC2, TPM3, and NPM1 are ±0.1, ±0.12, and ±0.09, respectively. (C) Knockdown of QKI-6 was performed using siRNA against QKI or control siRNA against GFP. Protein extracts from siQKI and siGFP were analyzed by SDS-PAGE followed by immunoblotting with anti-AIP-1 and -QKI-6 antibodies. Anti-α-tubulin antibodies were used to confirm equivalent protein expression. Absolute quantified levels of protein bands are presented underneath each blot and were performed using Scion-Image.
Table 1.
Putative targets of QKI-6 identified by 2D-DIGE
| Gene | Gene name | Volume ratio QKI-6:GFP |
|---|---|---|
| ACO2 | Aconitase 2, mitochondrial | 1.5 |
| ACTB | A chain structure of crystalline profilin-β-actin | 1.8 |
| ACTG1 | Actin, γ1 | −1.9 |
| AIP-1 | Actin-interacting protein | −1.6 |
| ANXA2 | Annexin 2A | 1.9 |
| CAPZB | Capping protein (actin filament) muscle Z-line, β | −3.1 |
| CCT6A | Chaperonin containing TCP1, subunit 6A (ζ1) | −1.6 |
| Eef2 | Eukaryotic translation elongation factor 2 | 1.9 |
| GANAB | Glucosidase α neutral AB | 1.9 |
| GSN | Gelsolin | 1.7 |
| HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 isoform A | 2.3 |
| HSPA2/8 | Heat-shock 70-kDa protein 2 and 8 | 2.1 |
| HSPD1 | Heat-shock 60-kDa protein 1 (chaperonin) | 1.8 |
| Lamin B1 | Lamin B1 | −2.5 |
| NPM1 | Nucleophosmin | −1.7 |
| PKM2 | Pyruvate kinase | 2 |
| PYGB | Phosphorylase brain glycogen | 2.3 |
| TPI | Triosephosphate isomerase 1 | 1.8 |
| TPM3 | Tropomyosin 3 | −1.6 |
| VDAC2 | Voltage-dependent anion channel 2 | −6.1 |
| XRCC2 | XRCC2 DNA repair protein | −1.7 |
As an additional control for our 2D-DIGE analysis, we also assessed whether knockdown of the QKI-6 isoform in the U343 glioma cell line would affect expression of AIP-1 as this target was found to be most highly regulated in CRL2020 by exogenous QKI-6. Immunoblotting analysis of control knockdown, siGFP, compared with siQKI showed a threefold decrease in QKI-6 levels and a sevenfold increase in levels of AIP-1 after QKI knockdown normalized to α-tubulin levels, further confirming our initial 2D-DIGE analysis (Figure 1C).
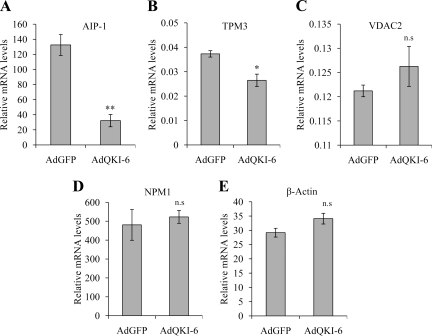
We next assessed whether QKI-6 also decreased the expression levels of the AIP-1, TPM3, VDAC2, and NPM1 mRNAs by using quantitative real-time reverse transcription (RT)-PCR. The mRNA levels of AIP-1 and TPM3 were significantly reduced with the expression of QKI-6 (Figure 2, A and B), whereas the mRNA levels of the VDAC2, NPM1, and the control β-actin were not significantly (n.s.) altered by QKI-6 expression (Figure 2, C–E). These findings suggest that QKI-6 may modulate the posttranscriptional regulation of AIP-1 and TPM3 mRNAs. We selected AIP-1 for further study, because it was the target that seemed to be most sensitive to QKI-6 expression while also harboring the consensus QRE sequences within its 3′-UTR.
Figure 2.
The AIP-1 mRNA is reduced with QKI-6 expression. Total RNA was harvested after infection of AdGFP or AdQKI-6 into CRL2020 cells. mRNA expression of AIP-1 (A), TPM3 (B), VDAC2 (C), NPM1 (D), and actin (E) was assayed by real-time PCR. The results shown represent triplicate analyses of each target. GAPDH was used as an internal control for RNA level normalization. One-way analysis of variance was performed; single asterisks denotes a p < 0.05 and double asterisks denote a p < 0.005.
QKI-6 Leads to AIP-1 mRNA Destabilization
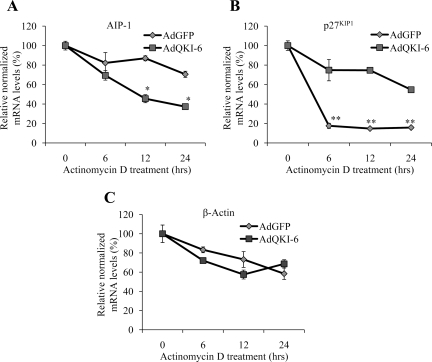
We examined whether QKI-6 regulates the half-life of the AIP-1 mRNA by treating AdGFP- and AdQKI-6–transduced CRL2020 cells with actinomycin D and measuring the half-life of the AIP-1 mRNA by real-time PCR. We showed previously that the QKI-6 isoform stabilizes the mRNA encoding p27 KIP1 (Larocque et al., 2005), and we monitored the half-life of mRNAs encoding p27KIP1 and β-actin, as positive and negative controls, respectively. The half-life of AIP-1 mRNA was observed to be >24 h in the AdGFP-transduced cells, whereas its half-life was decreased to ∼12 h in AdQKI-6–transduced cells (Figure 3A). The p27KIP1 mRNA stability increased with the expression of QKI-6 (Figure 3B), as observed previously (Larocque et al., 2005), whereas QKI-6 expression did not significantly affect the half-life of the β-actin mRNA (Figure 3C). These findings suggest that expression of the QKI-6 isoform decreases the half-life of the AIP-1 mRNA.
Figure 3.
QKI-6 associates with the AIP-1 mRNA and decreases its half-life. Total RNA was harvested after infection of AdGFP or AdQKI-6 into CRL2020 cells treated with actinomycin D for the indicated times. The levels of AIP-1 (A), p27KIP1 (B), and actin (C) mRNA normalized to levels of GAPDH were examined by real-time PCR. Two-way analysis of variance was performed in comparison with AdGFP; single asterisks denotes a p < 0.05 and double asterisks denote a p < 0.005.
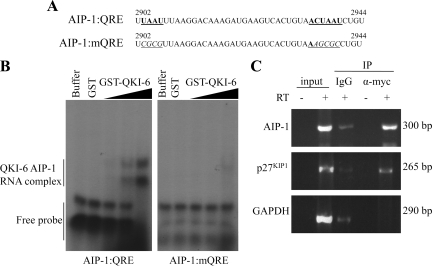
We showed previously that the QKI proteins bind a bipartite QRE with the sequence UAAY-(N1-20)-ACUAAY, where N represents any nucleotide and Y is a pyrimidine (Galarneau and Richard, 2005). We identified from nucleotides 2902–2944 in the human and mouse AIP-1 mRNA the sequence UAAU-(N26)-ACUAAU, reminiscent of a QRE. To assess that this putative QRE is bound by QKI-6, we performed electrophoretic mobility shift assays using in vitro-transcribed RNA and recombinant GST QKI-6 (GST-QKI-6) protein. As RNA probes, we used the wild-type AIP-1 3′-UTR QRE from nucleotides 2902–2944 and a mutant AIP-1 3′-UTR QRE (AIP-1:mQRE) that harbors nucleotide substitutions within the QRE that should prevent QKI-6 RNA binding (Figure 4A). GST-QKI-6 bound in a dose-dependent manner to the AIP-1:QRE but not to the AIP-1:mQRE (Figure 4B). These findings show that the AIP-1 mRNA harbors a QRE that is bound by QKI-6.
Figure 4.
The AIP-1 3′-UTR harbors a QKI response element (QRE). (A) The sequence of the human AIP-1 3′-UTR from nucleotides 2902–2944 is shown. The underlined and bold sequences denote the half-site and core QKI binding sites from left to right. The mutant AIP-1 sequences with mutations at the QRE sites are shown underlined and italicized. (B) Radioactive labeled RNA probes of the sequences shown in A were incubated with protein-RNA binding buffer alone (buffer), with GST, or with increasing amounts of GST-QKI-6. The samples were incubated at room temperature for 1 h and then separated by 8% Tris borate-EDTA-polyacrylamide gel electrophoresis gel. The migration of the QKI-6/RNA complex and free RNA probe is indicated. (C) CRL2020 cells transduced with AdQKI-6 were lysed and protein–RNA interactions examined by coimmunoprecipitation. Cell extracts were lysed and incubated with IgG as a control and anti-myc antibodies to immunoprecipitate myc-epitope–tagged QKI-6. The QKI-6 bound RNA and input RNA were analyzed by semiquantitative RT-PCR, and the DNA fragments separated on a 2% agarose gel and visualized by ethidium bromide staining.
As a control for viewing physiological interaction of QKI-6 and AIP-1 mRNA, we examined whether QKI-6 can coimmunoprecipitate with the AIP-1 mRNA in vitro. Anti-QKI-6 and control (IgG) immunoprecipitations were performed using AdQKI-6–transduced CRL2020 cell extracts. The coimmunoprecipitating RNAs were isolated, and the presence of AIP-1, p27KIP1, and GAPDH mRNAs was monitored by semiquantitative RT-PCR. We observed that QKI-6 associated with both the mRNAs of AIP-1 and p27KIP1 significantly above background, but it did not associate with the mRNA of the housekeeping gene GAPDH (Figure 4C). These data show that the AIP-1 mRNA associates with QKI-6.
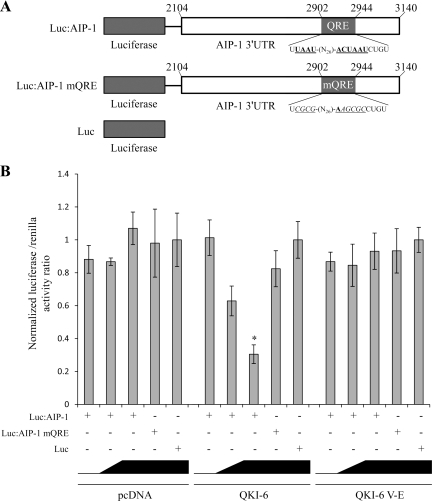
We next examined whether QKI-6 could regulate the expression of a heterologous reporter gene harboring the AIP-1 3′-UTR. Expression vectors encoding wild-type QKI-6 or a known RNA binding defective mutant (QKI-6:V157E; Larocque et al., 2002) were transfected in HEK293 cells with a luciferase reporter plasmid with or without the AIP-1 3′-UTR, as well as with an AIP-1 3′-UTR harboring a mutated QRE (Figure 5A). We observed that luciferase expression of the luciferase AIP-1 3′-UTR reporter plasmid decreased in a dose-dependent manner with the addition of QKI-6 but not with pcDNA or QKI-6:V157E (Figure 5B). Moreover, the luciferase expression was not modulated by QKI-6 in the absence of the AIP-1 3′-UTR or in presence of a mutated QRE in AIP-1 3′-UTR (Luc and Luc:AIP-1 mQRE; Figure 5B). These findings show the requirement for the QRE within the AIP-1 3′-UTR and the QKI-6 RNA binding activity to mediate the QKI-6–dependent AIP-1 repression.
Figure 5.
The 3′-UTR of AIP-1 confers QKI-6 sensitivity to a heterologous RNA. (A) Schematic illustration of pMIR-reporter vectors with or without the 3′-UTR of AIP-1, as well as with a mutated QRE version of the AIP-1 3′-UTR. (B) HEK293 cells were cotransfected with pcDNA, pcDNA-QKI-6, or pcDNA-QKI-6:V157E, along with the indicated pMIR-reporter vectors and pTKrenilla to control for transfection efficiency. The luciferase activity was expressed as a normalized ratio of the activity of luciferase/Renilla (± represents the SE of the mean). Each bar represents three separate experiments. One-way analysis of variance was performed in comparison with absence of QKI-6 transfection. Single asterisk denotes a p < 0.005.
AIP-1 Levels Decrease during CNS Myelination and OL Differentiation
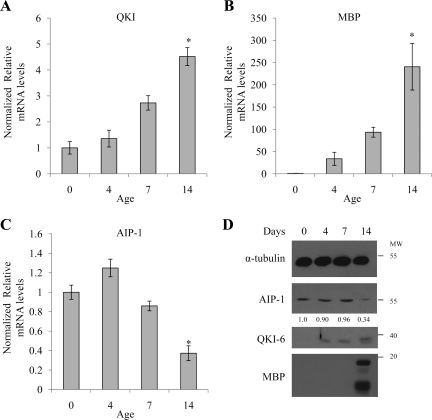
To elucidate whether AIP-1 is regulated during in vivo OL differentiation and CNS myelination, we investigated changes in AIP-1 levels in the brain of postnatal day 0 (P0), P7, and P14 wild-type rats. Total RNA was purified and the mRNA levels of QKI, MBP (marker of OL differentiation), and AIP-1 were analyzed by real-time RT-PCR and normalized to GAPDH. During peak in vivo OL differentiation and CNS myelination at P7 and P14, we confirmed that the QKI and MBP mRNA levels were significantly increased (Figure 6, A and B). AIP-1 mRNA decreased gradually at P7 and P14 (Figure 6C). The decrease in AIP-1 protein level also was observed at P14 by immunoblotting (Figure 6D). Moreover, the AIP-1 reduction at P14 also was observed by immunostaining O4-positive OLs in rat brains (Supplemental Figure 1). These findings show that AIP-1 mRNA and protein levels decrease in rat brains during in vivo OL differentiation and CNS myelination.
Figure 6.
The decrease in AIP-1 coincides with CNS myelination in rats. Rat pup brains taken at postnatal day 0, day 7, or day 14 were homogenized in lysis buffer, and the RNA was extracted using TRIzol or Laemmli buffer for Western blotting. RNA was analyzed by real-time PCR with primers for QKI (A), MBP (B), and AIP-1 (C). One-way analysis of variance was performed; single asterisk denotes a p < 0.05. Protein samples were analyzed by Western blotting (D) for α-tubulin, AIP-1, QKI-6, and MBP; α-tubulin levels were used to confirm equivalent loading, and MBP expression was used as a marker for OL differentiation. The anti-QKI-6 antibody also recognizes QKI-7 and hence the doublet observed at day 14. Changes in AIP-1 levels are shown and represent α-tubulin–normalized values in comparison with day 0 OLs; quantifications were performed using Scion-Image.
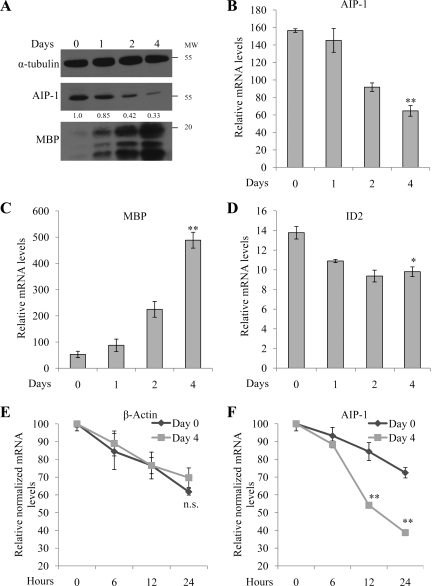
We next examined whether the level of AIP-1 mRNA and protein was regulated during in vitro OL differentiation. AIP-1 was monitored by immunoblotting protein extracts isolated from 0, 1, 2, and 4 d differentiated OLs. We observed an approximately threefold reduction in AIP-1 protein levels after 4 d of OL differentiation by using anti-α-tubulin immunoblotting as a loading control and anti-MBP antibodies were used as a marker of OL maturation (Figure 7A). In addition, purified primary rat OLs were differentiated for 0, 1, 2, and 4 d and the AIP-1 mRNA was quantified by real-time RT-PCR and normalized to GAPDH. We observed a gradual decrease in AIP-1 mRNA expression level reaching an ∼2.5-fold decrease at day 4 of OL differentiation (Figure 7B). The mRNA levels of MBP and the transcriptional repressor Id2 increased and decreased, respectively, confirming the proper differentiation of the OLs (Figure 7, C and D). Together, these findings show that AIP-1 mRNA and protein levels are significantly diminished during OL differentiation, suggesting that the reduction in AIP-1 protein and mRNA expression may be a prerequisite for OL branching and maturation.
Figure 7.
AIP-1 expression decreases during primary rat OL differentiation. (A) Primary rat OPCs were induced to differentiate for the indicated days, and the cells were lysed and protein extracts were separated by SDS-polyacrylamide gel electrophoresis. The presence of α-tubulin, AIP-1, and MBP isoforms were analyzed by immunoblotting. α-Tubulin levels were used to confirm equivalent loading, and MBP expression was used as a marker for OL differentiation. Total RNA levels were isolated from OLs differentiated for different time points (day 0–4) and analyzed by real-time PCR for changes in mRNA levels of AIP-1 (B), MBP (C), and Id2 (D). Real-time data represent three separate experiments. One-way analysis of variance was performed in comparison with day 0 samples; single asterisks denote a p < 0.05 and double asterisks denote a p < 0.005. RNA half-life for Actin (E) and AIP-1(F) was determined by actinomycin D treatment for 0–24 h and compared between different time points of OL differentiation (day 0 and day 4). Real-time data represent three separate experiments. Two-way analysis of variance was performed in comparison with day 0 OPCs; double asterisks denote a p < 0.005.
We monitored the half-life of the endogenous AIP-1 mRNA in the OL precursor cells (day 0) and in the mature OLs (day 4), in which the levels of AIP-1 are elevated and low, respectively (Figure 7A). OLs were incubated for up to 24 h with actinomycin D, and the levels of β-actin or AIP-1 mRNA were monitored by real-time PCR and normalized to GAPDH. We observed that the β-actin mRNA half-life was unaffected with OL differentiation (Figure 7E). However, the AIP-1 mRNA half-life was shorter (∼12 h) in day 4 mature OLs compared with >24 h in day 0 OL precursor cultures (Figure 7F). These findings show that the AIP-1 half-life decreases during OL differentiation.
AIP-1 Localization with Cofilin and Actin Filaments in OLs
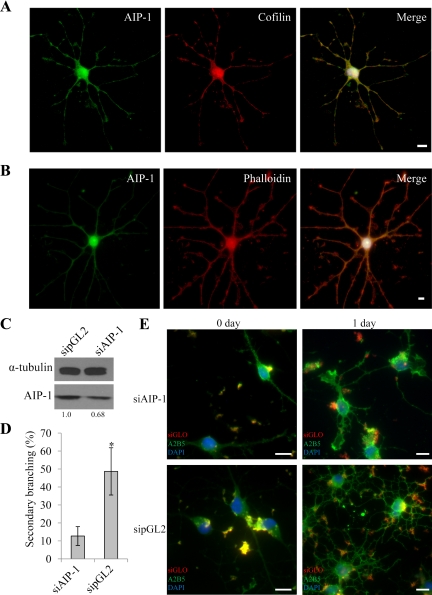
AIP-1 is known to colocalize with cofilin and actin filaments (Ono, 2003). To examine whether AIP-1 is localized with cofilin in OLs, we coimmunostained differentiated OLs with anti-AIP-1 and anti-cofilin antibodies, and the cells were visualized by fluorescence microscopy. Indeed, as expected, both AIP-1 and cofilin localized diffusely throughout OLs and the staining was enriched at branch points (Figure 8A). Moreover, similar staining was visualized with actin filaments by using phalloidin costained with AIP-1 (Figure 8B). These finding show that AIP-1 localization in OLs is similar to what has been observed in neurons stained with actin-depolymerizing factor (ADF)/cofilin (Sarmiere and Bamburg, 2004), consistent with AIP-1 being a regulator cofactor for actin remodeling with ADF/cofilin in OLs.
Figure 8.
AIP-1 depleted OLs exhibit defects in process extension. Primary rat OPCs were differentiated for 1 d. The cells were then immunostained for AIP-1 and cofilin (A) or AIP-1 and phalloidin (B); the nuclei were visualized with 4,6-diamidino-2-phenylindole (DAPI) staining, and the images were merged on the right. Bars, 50 μm for the full images and 25 μm for the branch point images. (C) Knockdown efficiency of AIP-1 was verified using Western blotting from total cell lysates extracted after 24 h of differentiation, and α-tubulin was used as a loading control. Changes in AIP-1 levels are shown and represent α-tubulin normalized values; quantifications were performed using Scion-Image. (D) Day 1 differentiated cells transfected with either siAIP-1 or sipGL2 were quantified for their differentiation progress by counting the number of secondary branching positive cells in five fields of view per triplicate. One-way analysis of variance was performed; single asterisk denotes a p < 0.05. (E) Primary rat OPCs were cotransfected with siRNAs targeting AIP-1 or an irrelevant gene (luciferase; sipGL2) and siGLO, a marker of transfected cells. The siRNA-incubated cells were left in proliferation media (day 0) or induced to differentiate for 24 h, and the cells were then immunostained with anti-A2B5 antibodies to monitor process outgrowth. Bars, 50 μm.
AIP-1 Is Required for OL Process Extension
We next investigated whether the presence of AIP-1 in the immature OL precursor cells was required for OL process extension. Immature OL precursor cells were transfected with siRNA targeting AIP-1 (siAIP-1) or a control siRNA targeting an irrelevant gene (luciferase; sipGL2). siGLO also was used as a marker to visualize transfected cells, which showed staining similar to previously reported results (Xian et al., 2005). We observed an ∼30% knockdown of AIP-1 in the entire immature primary OL precursor cell culture (Figure 8C). The number of secondary branching was assessed from undifferentiated and differentiated siGLO-positive OLs. We observed that siAIP-1–transfected OLs were significantly impaired in process extension compared with the control OL cultures (Figure 8, D and E). These findings suggest that AIP-1 is required for OL process extension.
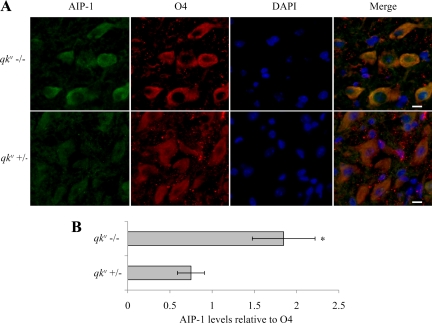
We next analyzed whether AIP-1 levels are affected in qkv/qkv mice, because these mice have been characterized previously to lack the expression of the QKI-6 and QKI-7 isoforms in OLs (Hardy et al., 1996b). Thus, we examined whether the AIP-1 expression was affected in the OLs of qkv/qkv mice. Because of mosaic expression patterns of the QKI isoforms in the brains of qkv/qkv mice (Hardy et al., 1996b), we resorted to immunofluorescence to examine the expression of AIP-1 in OLs. We observed that the O4-positive OLs of 14-d-old qkv/qkv mice mouse pups had an approximately twofold increase in AIP-1 protein level compared with OLs from qkv/+ mice (Figure 9, A and B). These findings suggest that the QKI isoforms regulate AIP-1 in mouse OLs.
Figure 9.
Increased AIP-1 expression in qkv/qkv mice. Brains were harvested from qkv/qkv mice and qkv/+ mice at postnatal day 14 and stained for AIP-1, O4, and 4,6-diamidino-2-phenylindole (DAPI); merged images are presented on the right (A). Images were taken in the cerebral cortex. Bars, 50 μm. (B) Immunostaining was quantified using AxioVision 4.8.1 software by standardizing levels of AIP-1 signal to O4 signal; five fields of view were used for each genotype. One-way analysis of variance was performed; single asterisk denotes a p < 0.05.
DISCUSSION
Here, we identified the mRNA of AIP-1, a regulatory protein for cofilin during actin filament severing and depolymerization (Ono, 2003), as an RNA target for the QKI-6 RNA-binding protein. The AIP-1 mRNA was bound by QKI-6 in the 3′-UTR, promoting AIP-1 mRNA destabilization and resulting in lower AIP-1 protein levels. We show the requirement for the QRE within the AIP-1 3′-UTR residing from nucleotides 2902–2944 by using a luciferase reporter gene, and we also show that QKI-6 RNA binding activity is required to mediate the QKI-6–dependent AIP-1 repression. QKI-6 and QKI-7 are up-regulated during murine CNS myelination and OL differentiation (Ebersole et al., 1996b). We observed that AIP-1 mRNA and protein levels decrease in rat brains during in vivo OL differentiation and CNS myelination, consistent with AIP-1 mRNA being an in vivo target of QKI-6. Furthermore, qkv/qkv mice that lack QKI-6 and QKI-7 within its OLs (Hardy et al., 1996b), had an increased level of AIP-1 in OLs. Moreover, we observe a decrease in AIP-1 mRNA and protein levels during in vitro OL differentiation. AIP-1 reduction with siRNA in the immature OL precursor cells impaired OL process outgrowth, as seen by the reduction in secondary branching. Together, these findings suggest that the QKI RNA-binding proteins regulate OL differentiation by modulating the expression of AIP-1.
The QKI proteins exhibit multiple RNA functions including mRNA stability, mRNA export, and alternative splicing (Chenard and Richard, 2008). We have shown previously that the p27KIP1 mRNA is stabilized with QKI-6/7 expression (Larocque et al., 2005), and we now show that QKI-6 also functions in reducing the half-life of the AIP-1 mRNA. Although we have not yet fully elucidated the mechanism behind QKI-6 destabilization of AIP-1 mRNA, we suspect that this effect may be due to competition for mRNA binding between QKI-6 and mRNA-stabilizing factors. To this end, we have observed several sequences neighboring QKI-6 QRE on AIP-1 mRNA that are reminiscent of HuR recognition sites (Brennan and Steitz, 2001). We thus suggest that QKI-6 binds to the AIP-1 QRE and prevents stabilizing factors such as HuR (Brennan and Steitz, 2001) from binding, leading to the observed reduction in AIP-1 half-life.
The highly regulated process of actin polymerization and depolymerization is important in many physiological processes, such as cytokinesis, motility, cell polarity, endocytosis, and differentiation (Pak et al., 2008). AIP-1, also called WDR1 (tryptophan-aspartate repeat protein 1), interacts with ADF and cofilin–actin complexes to facilitate actin filament severing and accelerate depolymerization (Ono, 2003). AIP-1 was identified to be regulated by acoustic overstimulation in the chick basilar papilla and for its association with actin (Adler et al., 1999). The AIP-1 homologue in Caenorhabditis elegans (Unc078) is a regulator of actin filament organization during myofibril development (Ono, 2001; Mohri et al., 2006). AIP-1-null mice are embryonic lethal; however, hypomorphic alleles have been generated and these mice develop macrothrombocytopenia and autoinflammatory disease (Kile et al., 2007). This study also identified AIP-1 in the actin dynamics of megakaryocytes and neutrophils (Kile et al., 2007). Although a role for AIP-1 in neuronal function was not reported, a role for cofilin 1 was demonstrated in neuronal growth and neural crest cell migration (Bellenchi et al., 2007).
Considerable information is known about the cytoskeleton in OL differentiation and myelination (Bauer et al., 2009) and on the function of RNA-binding proteins in regulation of the cytoskeleton (Carson et al., 2008). For example, OL process outgrowth has been shown to be impaired in cells with reduced Mayven and its related protein (MRP2) expression (Williams et al., 2005; Jiang et al., 2007). As a modulator of actin disassembly, AIP-1 is expected to be required for only a short period or in bursts (Kueh et al., 2008). This short stage of rapid actin disassembly would allow the cell to be primed for new actin filament formation and eventually new cell structure and morphology. The observed decrease in AIP-1 expression levels paralleled the increase in QKI expression during OL differentiation and CNS myelination. Global analysis of gene expression in differentiating OLs revealed that AIP-1 mRNA levels decrease during differentiation (Cahoy et al., 2008), consistent with our present observations. In addition, our observations in rat brains further provide evidence of AIP-1 importance during development as well as the role QKI plays in AIP-1 regulation.
AIP-1 has been shown to be required for cell shape morphology changes, mainly cell flattening (Ono, 2003). This phenomenon is a response probably needed during OL differentiation to extend the myelinating processes. To this end, AIP-1 and cofilin show a very similar localization throughout OLs, with the staining being enriched at branch points. This evidence together with our findings of reduction in AIP-1 during late stages of differentiation, suggests that AIP-1 may be required for restructuring of OL cytoskeleton for process outgrowth.
RNA-binding proteins are known to regulate a large array of mRNA targets within a conserved network to provide a rapid response to cellular needs (Keene and Lager, 2005). Previously, QKI has been reported to bind and regulate mRNA levels of several targets involved in OL differentiation, such as p27KIP1 (Larocque et al., 2005), MBP (Li et al., 2000; Larocque et al., 2002; Ryder and Williamson, 2004), myelin-associated glycoprotein (Wu et al., 2002), and MAP1B (Zhao et al., 2006a). Our analysis by 2D-DIGE has led to identification of several cytoskeletal proteins, including AIP-1, TPM1, ACTG1, and lamin B1 that are regulated by QKI-6 (Table 1). This interaction network is strengthened by our evidence that all of the proposed cytoskeletal targets contain a QRE site in their coding region or 3′-UTR (Table 2). These observations are consistent with cell adhesion as being a major annotation category of QKI mRNA targets (Galarneau and Richard, 2005). Although only AIP-1 mRNA has been confirmed as a target of QKI, we believe that we have uncovered the involvement of the QKI isoforms within a larger subset of cytoskeletal proteins, allowing regulation of morphological changes in OLs. Our findings provide a new regulatory network in OLs and further demonstrate a role for the QKI isoforms in OL function.
Table 2.
QREs identified in the putative QKI-6 targets
| Gene | Accession no. | QRE sequence |
|---|---|---|
| ACO2 | NM_001098 | (1030) ACUAAUUGAAAUUAAC (1045)–CDSa |
| ACTB | NM_173979 | (1622) ACAAAUUUUGUUUUUAAU (1640)–3′-UTR |
| ACTG1 | NM_001614 | None |
| AIP-1 | NM_017491 | (2903) UAAU-N26-ACUAAU (2940)–3′-UTR |
| ANXA2 | NM_001002858 | (533) ACCAAC-N17-UAAC (560)–CDS |
| CAPZB | NM_004930 | (1315) UAAA-N17-ACUAAC (1341)–3′-UTR |
| CCT6A | NM_001762 | (636) ACUAAA-N25-UAAC (671)–CDS |
| Eef2 | NM_001961 | (2972) ACUAAUUUUAAC (2983)–3′-UTR |
| GANAB | NM_198334 | (1570) ACUAAU-N23-UAAC (1603)–CDS |
| GSN | NM_001127662 | (1410) ACUACC-N24-UAAU (1444)–CDS |
| HNRNPH3 | NM_012207 | (865) UAAU-N11-ACUAAA (886)–CDS |
| HSPA2/8 | NM_021979 | (2674) ACUAAA-N13-UAAU (2696)–3′-UTR |
| HSPD1 | NM_002156 | (1867) ACUAGU-N13-UAAU (1890)–3′-UTR |
| Lamin B1 | NM_005573 | (2476) UAAU-N20-ACUAGC (2506)–3′-UTR |
| NPM1 | NM_002520 | (311) ACUAAA-N30-UAAU (350)–CDS |
| PKM2 | NM_182470 | (2133) ACUCACUCUGGGCUGUAAC (2152)–3′-UTR |
| PYGB | NM_002862 | (3413) UAAC-N16-ACUAGC (3439)–3′-UTR |
| TPI | NM_000365 | (990) UAAUGGUUGGAACUAAA (1006)–3′-UTR |
| TPM3 | NM_152263 | (1607) UAAGAUUGACUAAU (1621)–3′-UTR |
| VDAC2 | NM_003375 | (736) ACUAAUGUCAAC (748)–CDS |
| XRCC2 | NM_005431 | (2167) UAAUUAAACUAAA (2199)–3′-UTR |
Supplementary Material
ACKNOWLEDGMENTS
E. D., C. G., and J.D.H. are supported by studentships from the Multiple Sclerosis Society of Canada (MSSOC). This work was supported by grants from the MSSOC to G. A and S. R. S. R. is a chercheur-national of the Fonds de la Recherche en Santé au Québec.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0305) on July 14, 2010.
REFERENCES
- Adler H. J., Winnicki R. S., Gong T. W., Lomax M. I. A gene upregulated in the acoustically damaged chick basilar papilla encodes a novel WD40 repeat protein. Genomics. 1999;56:59–69. doi: 10.1006/geno.1998.5672. [DOI] [PubMed] [Google Scholar]
- Almazan G., Afar D.E.H., Bell J. C. Phosphorylation and disruption of intermediate filament protein in oligodendrocyte precursor cultures treated with calyculin A. J. Neurosci. Res. 1993;36:163–172. doi: 10.1002/jnr.490360206. [DOI] [PubMed] [Google Scholar]
- Bauer N. G., Richter-Landsberg C., Ffrench-Constant C. Role of the oligodendroglial cytoskeleton in differentiation and myelination. Glia. 2009;57:1691–1705. doi: 10.1002/glia.20885. [DOI] [PubMed] [Google Scholar]
- Bellenchi G. C., Gurniak C. B., Perlas E., Middei S., Ammassari-Teule M., Witke W. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev. 2007;21:2347–2357. doi: 10.1101/gad.434307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M., Steitz J. A. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy J. D., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson J. H., Gao Y., Tatavarty V., Levin M. K., Korza G., Francone V. P., Kosturko L. D., Maggipinto M. J., Barbarese E. Multiplexed RNA trafficking in oligodendrocytes and neurons. Biochim. Biophys. Acta. 2008;1779:453–458. doi: 10.1016/j.bbagrm.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenard C. A., Richard S. New implications for the QUAKING RNA binding protein in human disease. J. Neurosci. Res. 2008;86:233–242. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- Ebersole T. A., Chen Q., Justice M. J., Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 1996a;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Ebersole T. A., Chen Q., Justice M. J., Artzt K. The quaking gene unites signal transduction and RNA binding in the developing nervous system. Nat. Genetics. 1996b;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Galarneau A., Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- Hardy R. J., Lazzarini R. A., Colman D. R., Friedrich V. L., Jr. Cytoplasmic and nuclear localization of myelin basic proteins reveals heterogeneity among oligodendrocytes. J. Neurosci. Res. 1996a;46:246–257. doi: 10.1002/(SICI)1097-4547(19961015)46:2<246::AID-JNR13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Hardy R. J., Loushin C. L., Friedrich V. L., Jr., Chen Q., Ebersole T. A., Lazzarini R. A., Artzt K. Neural cell type-specific expression of QKI proteins is altered in the quaking viable mutant mice. J. Neurosci. 1996b;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan E. L., Greenfield S. Animal models of genetic disorders of myelin. Myelin. 1984:489–534. [Google Scholar]
- Jiang S., Seng S., Avraham H. K., Fu Y., Avraham S. Process elongation of oligodendrocytes is promoted by the Kelch-related protein MRP2/KLHL1. J. Biol. Chem. 2007;282:12319–12329. doi: 10.1074/jbc.M701019200. [DOI] [PubMed] [Google Scholar]
- Keene J. D., Lager P. J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Kile B. T., et al. Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia. Blood. 2007;110:2371–2380. doi: 10.1182/blood-2006-10-055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh H. Y., Charras G. T., Mitchison T. J., Brieher W. M. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque D., Galarneau A., Liu H. N., Scott M., Almazan G., Richard S. Protection of the p27KIP1 mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Larocque D., Pilotte J., Chen T., Cloutier F., Massie B., Pedraza L., Couture R., Lasko P., Almazan G., Richard S. Nuclear retention of MBP mRNAs in the Quaking viable mice. Neuron. 2002;36:815–829. doi: 10.1016/s0896-6273(02)01055-3. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Schedl T. RNA-binding proteins. WormBook. 2006;18:1–13. doi: 10.1895/wormbook.1.79.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Y., Li D., Feng Y. Destabilization and mislocalization of the myelin basic protein mRNAs in quaking dysmyelination lacking the Qk1 RNA-binding proteins. J. Neurosci. 2000;20:4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong K. E., Chang K. W., Khandjian E. W., Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Mohri K., Ono K., Yu R., Yamashiro S., Ono S. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol. Biol. Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland P. J., et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–791. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H., Volohonsky G., Reuveny A., Zaidel-Bar R., Volk T. Two isoforms of the Drosophila RNA binding protein, How, act in opposing directions to regulate tendon cell differentiation. Dev. Cell. 2002;2:183–193. doi: 10.1016/s1534-5807(01)00118-6. [DOI] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1, new blades for twisted filaments. Biochemistry. 2003;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Pak C. W., Flynn K. C., Bamburg J. R. Actin-binding proteins take the reins in growth cones. Nat. Rev. Neurosci. 2008;9:136–147. doi: 10.1038/nrn2236. [DOI] [PubMed] [Google Scholar]
- Pilotte J., Larocque D., Richard S. Nuclear translocation controlled by alternatively spliced isoforms inactivates the QUAKING apoptotic inducer. Genes Dev. 2001;15:845–858. doi: 10.1101/gad.860301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder S. P., Williamson J. R. Specificity of the STAR/GSG domain protein Qk1: implications for the regulation of myelination. RNA. 2004;10:1449–1458. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiere P. D., Bamburg J. R. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J. Neurobiol. 2004;58:103–117. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- Volk T., Israeli D., Nir R., Toledano-Katchalski H. Tissue development and RNA control: “HOW” is it coordinated? Trends Genet. 2008;24:94–101. doi: 10.1016/j.tig.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Webb A., Clark P., Skepper J., Compston A., Wood A. Guidance of oligodendrocytes and their progenitors by substratum topography. J. Cell Sci. 1995;108:2747–2760. doi: 10.1242/jcs.108.8.2747. [DOI] [PubMed] [Google Scholar]
- Williams S. K., Spence H. J., Rodgers R. R., Ozanne B. W., Fitzgerald U., Barnett S. Role of Mayven, a kelch-related protein in oligodendrocyte process formation. J. Neurosci. Res. 2005;81:622–631. doi: 10.1002/jnr.20588. [DOI] [PubMed] [Google Scholar]
- Wu J. I., Reed R. B., Grabowski P. J., Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc. Natl. Acad. Sci. USA. 2002;99:4233–4238. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian W., Schwertfeger K. L., Vargo-Gogola T., Rosen J. M. Pleitropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J. Cell Biol. 2005;171:663–673. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ku L., Chen Y., Xia M., LoPresti P., Feng Y. QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol. Biol. Cell. 2006a;17:179–186. doi: 10.1091/mbc.E06-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Tian D., Xia M., Macklin W. B., Feng Y. Rescuing qkV dysmyelination by a single isoform of the selective RNA-binding protein QKI. J. Neurosci. 2006b;26:11278–11286. doi: 10.1523/JNEUROSCI.2677-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.