Abstract
The cJun NH2-terminal kinase (JNK) signal transduction pathway has been implicated in mammary carcinogenesis. To test the role of JNK, we examined the effect of ablation of the Jnk1 and Jnk2 genes in a Trp53-dependent model of breast cancer using BALB/c mice. We detected no defects in mammary gland development in virgin mice or during lactation and involution in control studies of Jnk1−/− and Jnk2−/− mice. In a Trp53−/+ genetic background, mammary carcinomas were detected in 43% of control mice, 70% of Jnk1−/− mice, and 53% of Jnk2−/− mice. These data indicate that JNK1 and JNK2 are not essential for mammary carcinoma development in the Trp53−/+ BALB/c model of breast cancer. In contrast, this analysis suggests that JNK may partially contribute to tumor suppression. This conclusion is consistent with the finding that tumor-free survival of JNK-deficient Trp53−/+ mice was significantly reduced compared with control Trp53−/+ mice. We conclude that JNK1 and JNK2 can act as suppressors of mammary tumor development.
Introduction
The cJun NH2-terminal kinase (JNK) group of signaling enzymes are activated by cytokines/growth factors and also by exposure to environmental stress [1]. Targets of the JNK pathway include members of the activator protein 1 (AP1) group of transcription factors (e.g. cJun, JunB, and JunD). JNK is therefore a major regulatory mechanism of AP-1 dependent gene expression [1]. In addition, JNK can regulate many cytoplasmic and nuclear processes [2]. These studies have implicated the JNK signaling pathway in the regulation of cell growth and cell death [1]. Dysregulation of the JNK pathway may therefore contribute to the development of cancer [3].
The role of JNK in cancer has been studied using mouse models that are JNK-deficient. Two genes (Jnk1 and Jnk2) encode isoforms of JNK that are ubiquitously expressed [1]. Jnk1−/− mice and Jnk2−/− mice are viable, but compound mutant Jnk1−/− Jnk2−/− mice exhibit an early embryonic lethal phenotype [1]. Studies using Jnk1−/− mice and Jnk2−/− mice indicate that JNK may have isoform-dependent effects on cancer. Thus, Bcr-Abl-induced lymphoma [4] and carcinogen-induced hepatocellular carcinoma [5] are suppressed in Jnk1−/− mice. Moreover, carcinogen-induced skin cancer is suppressed in Jnk2−/− mice [6]. Similarly, important roles for JNK2 have been identified in studies of human glioblastoma, prostate cancer, and lung carcinoma cell lines [7]–[10]. Together, these data confirm that both JNK1 and JNK2 can play roles in tumor development.
The purpose of this study was to test the requirement of JNK1 and JNK2 in a mouse model of mammary carcinoma. Somatic mutation of the human p53 gene (TP53) is common in sporadic breast cancer [11]. Furthermore, mammary carcinoma is the most common form of cancer in women with heritable mutations in TP53 (Li-Fraumeni syndrome) [12]. Initial studies using mouse models demonstrated that Trp53−/− animals develop lymphoma with high frequency and that Trp53−/+ animals display a moderately broader tumor spectrum with slower onset of disease [13], [14]. Subsequent studies using Trp53−/+ mice on a BALB/c strain background demonstrated that, like humans with Li-Fraumeni syndrome, mammary carcinomas were frequently observed, together with some lymphomas and sarcomas [15]. The BALB/c mouse model can therefore be used to examine Trp53-dependent formation of mammary carcinoma.
We report that JNK1 and JNK2 are not required for the development of mammary carcinoma in the Trp53−/+ BALB/c mouse model. In contrast, the tumor-free survival of JNK-deficient Trp53−/+ mice was reduced compared with control Trp53−/+ mice. These data suggest that JNK may partially contribute to tumor suppression.
Materials and Methods
Mice
We have described Jnk1− /− mice [16] and Jnk2− /− mice [17] on a C57BL/6J strain background [18], and mice with Trp53 gene ablation [13] on a BALB/cMed strain background [19]. The mice used in this study were backcrossed (ten generations) to the BALB/cJ strain (Jackson Laboratories) and were housed in a facility accredited by the American Association for Laboratory Animal Care (AALAC). The Institutional Animal Care and Use Committee (IACUC) of the University of Massachusetts Medical School approved all studies using animals (Docket A-1032).
Genotype analysis
Genotype analysis was performed by PCR using genomic DNA as the template. The wild-type Jnk1 (460 bp) and knockout Jnk1 (390 bp) alleles were identified using the amplimers 5′-CGCCAGTCCAAAATCAAGAATC-3′, 5′-GCCATTCTGGTAGAGGAAGTTTCTC-3′, and 5′-CCAGCTCATTCCTCCACTCATG-3′. The wild-type Jnk2 (400 bp) and knockout Jnk2 (270 bp) alleles were identified using the amplimers 5′- GGAGCCCGATAGTATCGAGTTACC-3′, 5′-GTTAGACAATCCCAGAGGTTGTGTG-3′, and 5′-CCAGCTCATTCCTCCACTCATG-3′. The wild-type Trp53 (470 bp) and knockout Trp53 (700 bp) alleles were identified using the amplimers 5′-TATACTCAGAGCCGGCCT-3′, 5′-ACAGCGTGGTGGTACCTTAT-3′ and 5′-CTATCAGGACATAGCGTTGG-3′.
Analysis of tissue morphology
Mammary gland development was examined in virgin female mice (8 to 10 weeks of age), lactating mice (1 week post partum), and mice with mammary gland involution (pups removed at 1 week post-partum). The fourth inguinal mammary gland pair was dissected from each mouse; one gland was analyzed by whole mount and the other was formalin-fixed and paraffin-embedded.
Whole mounts were performed by spreading the gland on a glass slide and incubation (2–4 hrs.) with Carnoy's fixative (60% ethanol, 30% chloroform, 10% glacial acetic acid). The glands were then incubated with a graded series of 70%, 50% and 25% ethanol (15 mins each), followed by 5 minutes in water and stained with carmine alum overnight. The glands were washed in 70%, 90% and 100% ethanol (15 mins each), two changes of xylene (30 mins), and then mounted with Permount (Fisher Scientific).
Analysis of tissue sections was performed using tissue fixed in 10% formalin for 24 h, dehydrated, and embedded in paraffin. Sections (7 µm) were cut and stained using hematoxylin and eosin (Biocare Medical). Immunofluorescence analysis was performed using de-parafinized sections treated with the endogenous Biotin-Blocking kit (Invitrogen), staining (4°C, 12 h) with biotin-conjugated anti-PCNA (Invitrogen), and the incubation (25°C, 1 hr) with AlexaFluor633-conjugated Streptavidin (Invitrogen). The sections on coverslips were washed and mounted on slides using VectaShield medium containing DAPI (Vector Labs.). Images were examined using a Leica TCS SP2 confocal microscope.
Results
Effect of JNK-deficiency on mammary gland development
We backcrossed Jnk1− /− mice [16] and Jnk2− /− mice [17] to the BALB/cJ strain background. To test whether JNK-deficiency altered mammary gland development, we examined Jnk1−/− and Jnk2−/− BALB/c mice. No defects were detected in whole mount preparations of fourth inguinal mammary glands of JNK-deficient virgin female mice compared with control mice (Figure 1A). Sections prepared from these mammary glands confirmed that JNK-deficiency did not cause major defects in virgin mammary gland development (Figure 1B).
Figure 1. Effect of JNK-deficiency in virgin mice on breast development.
A) Whole mount preparations of the fourth inguinal mammary gland of 10 week-old female virgin mice were stained with carmine red. Representative images are presented. Scale bar: 5 mm (upper panel); 800 µm (lower panel). B) Sections of the breast tissue were stained with hematoxylin and eosin. Representative images are presented. Scale bar: 100 µm.
Pregnancy causes major changes in mammary gland development, including the formation of alveoli. Sections prepared from the fourth inguinal mammary glands of JNK-deficient lactating mice and control lactating mice were similar (Figure 2). Indeed, sections stained for proliferating cell nuclear antigen (PCNA) indicated that JNK-deficiency did not alter epithelial cell proliferation in the lactating mammary gland (Figure 2).
Figure 2. Effect of JNK-deficiency on breast development during lactation.
Sections of the fourth inguinal mammary gland of female mice at day 7 post-partum were examined by staining with hematoxylin and eosin (upper panels). Sections were also stained with an antibody to the proliferation marker PCNA (red) and the DNA stain DAPI (blue) (lower panels). Scale bar: 200 µm (upper panel); 50 µm (lower panel).
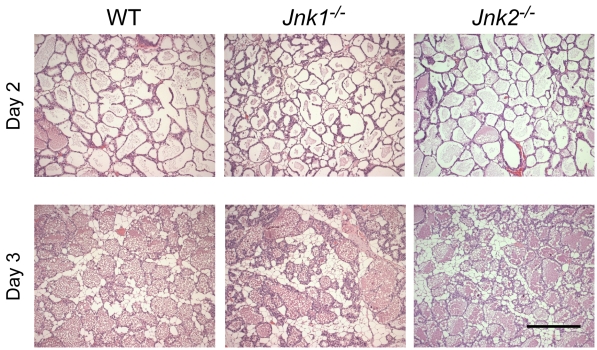
Involution of the lactating mammary gland occurs after weaning pups. We compared sections of the fourth inguinal mammary glands prepared on day 2 and day 3 following weaning. No defects in involution were detected in JNK-deficient mice compared with control mice (Figure 3).
Figure 3. Effect of JNK-deficiency on mammary gland involution.
The pups were removed from female mice at day 7 post-partum to induce mammary gland involution. Sections of the fourth inguinal mammary gland were examined at two days or three days post-weaning by staining with hematoxylin and eosin. Scale bar: 200 µm.
Together, these data demonstrate that JNK1-deficiency and JNK2-deficiency did not cause detected changes in mammary gland development. Similarly, no developmental defects caused by JNK1-deficiency or JNK2-deficiency were detected in Trp53−/+ mice.
Effect of JNK-deficiency on tumor development in Trp53−/− BALB/c mice
We examined the tumor-free survival of Trp53−/− mice, Jnk1−/− Trp53−/− mice, and Jnk2−/− Trp53−/− mice on a BALB/c strain background. The mice rapidly developed cancer and died (Figure 4A). No significant differences in tumor-free survival between control and JNK-deficient mice were detected. Pathological examination of the mice demonstrated, as expected, a high incidence of lymphoma (Figure 4B). The second most frequent type of tumor detected in Trp53−/− mice and Trp53−/− Jnk2−/− mice was hemangiosarcoma (Figure 4B). In contrast, Jnk1−/− Trp53−/− mice displayed fewer hemangiosarcomas and a higher incidence of lymphoma compared with Trp53−/− mice (Figure 4B). These data suggest that JNK1 may influence the tumor spectrum of Trp53−/− mice. Indeed, a low incidence of mammary carcinoma was observed in both male and female Jnk1−/− Trp53−/− mice, but not in Trp53−/− mice or Jnk2−/− Trp53−/− mice (Figure 4B). The presence of mammary carcinoma in Jnk1−/− Trp53−/− mice indicates that JNK may be relevant to breast cancer.
Figure 4. Effect of JNK-deficiency on Trp53−/− mouse survival.
A) Kaplan-Meier analysis of the tumor-free survival of wild-type (WT), Jnk1 −/−, Jnk2 −/−, Trp53 −/−, Jnk1 −/− Trp53 −/−, and Jnk2 −/− Trp53 −/− mice is presented. No statistically significant differences between Trp53 −/−, Jnk1 −/− Trp53 −/−, and Jnk2 −/− Trp53 −/− mice were detected (Log-rank test, p>0.05). The data represent groups of 44–62 mice. These groups include equal numbers of male and female mice. B) The spectrum of tumors detected in Trp53 −/−, Jnk1 −/− Trp53 −/−, and Jnk2 −/− Trp53 −/− mice following euthanasia is presented. No statistically significant differences in the tumor profiles between genotypes were detected using Fisher's exact test.
Effect of JNK-deficiency on tumor development in Trp53− /+ BALB/c mice
We performed studies of tumor-free survival of Trp53−/+ mice, Jnk1−/− Trp53−/+ mice, and Jnk2−/− Trp53−/+ mice on a BALB/c strain background. Tumor development in the Trp53−/+ mice was delayed compared with Trp53−/− mice (Figures 4 & 5). Interestingly, JNK1-deficiency (p = 0.026) and JNK2-deficiency (p = 0.012) caused significantly shortened tumor-free survival compared with control Trp53−/+ BALB/c mice (Figure 5A). Pathological analysis demonstrated that mammary carcinoma was the most common type of tumor detected. Mammary carcinomas were detected in 43% of control mice, 70% of Jnk1−/− mice, and 53% of Jnk2−/− mice (Figure 5B). Analysis of mammary carcinoma-free survival of Trp53−/+ mice, Jnk1−/− Trp53−/+ mice, and Jnk2−/− Trp53−/+ mice demonstrated that JNK1-deficiency (p = 0.018) and JNK2-deficiency (p = 0.039) significantly decreased survival compared with control Trp53−/+ mice (Figure 5C). No significant difference in mammary carcinoma-free survival between JNK1-deficient mice and JNK2-deficient mice was detected (Figure 5C).
Figure 5. Effect of JNK-deficiency on Trp53−/+ mouse survival.
A) Kaplan-Meier analysis of the tumor-free survival of wild-type (WT), Jnk1 −/−, Jnk2 −/−, Trp53 −/+, Jnk1 −/− Trp53 −/+, and Jnk2 −/− Trp53 −/+ mice is presented. The survival of Jnk1 −/− Trp53 −/+ mice and Jnk2 −/− Trp53 −/+ mice was reduced compared with Trp53 −/+ mice (Log-rank test, p = 0.026 and 0.012, respectively). The data represent groups of 14 - 20 female mice. B) The spectrum of tumors detected in Trp53 −/+, Jnk1 −/− Trp53 −/+, and Jnk2 −/− Trp53 −/+ female mice following euthanasia is presented. No statistically significant differences in the tumor profiles between genotypes were detected using Fisher's exact test. C) Kaplan-Meier analysis of the mammary carcinoma-free survival of Trp53 −/+, Jnk1 −/− Trp53 −/+, and Jnk2 −/− Trp53 −/+ mice is presented. Cohorts of Trp53 −/+ mice (n = 6), Jnk1 −/− Trp53 −/+ mice (n = 14), and Jnk2 −/− Trp53 −/+ mice (n = 8) with mammary carcinoma were examined. JNK1-deficiency and JNK2-deficiency caused reduced mammary carcinoma-free survival compared with Trp53 −/+ mice (Log-rank test, p = 0.018 and 0.039, respectively).
The increased mammary carcinoma detected in JNK1-deficient Trp53−/+ mice was associated with a decreased incidence of hemangiosarcoma (Figure 5B). No hemangiosarcomas were detected in JNK2-deficient Trp53−/+ mice (Figure 5B). These changes in tumor spectrum may reflect the shortened tumor-free survival of Trp53−/+ mice (Figure 5A,C).
Together, these data indicate that JNK1 and JNK2 are not required for mammary carcinoma development in the Trp53−/+ BALB/c mouse model of breast cancer. However, both JNK1 and JNK2 can influence breast cancer development. It appears that JNK can contribute to tumor suppression.
Discussion
JNK1 and JNK2 are not required for the development of mammary carcinoma in the Trp53 BALB/c mouse model
JNK plays a critical role in the development of some forms of cancer [1]. Thus, carcinogen-induced hepatocellular carcinoma [5] and BcrAbl-induced lymphoma [4] are strongly suppressed in Jnk1−/− mice and carcinogen-induced skin cancer is suppressed in Jnk2−/− mice [6]. Moreover, studies of glioblastoma, prostate cancer, and lung carcinoma cell lines have identified important roles for JNK2 [7]–[10]. Together, these data confirm that JNK plays an important role in cancer development.
The results of this study suggest that JNK may play a different role in mammary carcinogenesis because neither JNK1-deficiency nor JNK2-deficiency in the Trp53 BALB/c mouse model caused a reduction in the incidence of mammary carcinoma. This observation strongly contrasts with the finding that JNK-deficiency can markedly suppress hepatocellular carcinoma, lymphoma, and skin cancer [4]–[6].
Although JNK1-deficiency and JNK2-deficiency did not suppress mammary carcinogenesis in the Trp53 BALB/c mouse model, we cannot exclude the possibility that deficiency of both JNK1 plus JNK2 might reduce the formation of mammary carcinoma. Indeed, the Jnk1 and Jnk2 genes may have partially redundant functions [18], [20]–[22]. Studies of compound mutants with disruption of Jnk1 plus Jnk2 are required. The early embryonic lethal phenotype of Jnk1−/− Jnk2−/− mice [23] makes such studies difficult. Nevertheless, the effect of compound JNK-deficiency on mammary carcinoma development needs to be tested in future studies.
JNK and tumor suppression
The analysis of JNK1-deficiency and JNK2-deficiency in the Trp53 BALB/c mouse model of mammary carcinoma development demonstrates that neither JNK1 nor JNK2 is required for breast tumorigenesis (Figures 5). In contrast, the mammary carcinoma-free survival of both Jnk1−/− mice and Jnk2−/− mice was significantly reduced compared with control mice (Figure 5C). These data suggest that JNK may have a tumor suppressor role in breast cancer. This conclusion is consistent with the observation that JNK2-deficiency increases breast cancer in a transgenic mouse model with expression of polyoma virus T antigen [24]. Moreover, human genetic analysis has identified mutations in the JNK signaling pathway in breast cancer that correlate with tumor suppression and metastasis [3]. Specifically, loss-of-function mutations in MKK4, a human gene that encodes an activator of JNK, is mutated at low frequency in human breast cancer [25], [26], [27], [28]. It is likely that JNK1-deficiency and JNK2-deficiency in the mouse may phenocopy the effects of MKK4 gene mutation on breast cancer in humans. The molecular mechanism of tumor suppression by the JNK signaling pathway is unclear, but may be related to a requirement of JNK for genetic stability [24]. Indeed, it has been reported that genes that encode DNA repair enzymes are over-represented as targets of JNK pathway signaling [29]. A role for JNK in the maintenance of genetic stability is also consistent with the finding that a dominant genetic trait in the Trp53−/+ BALB/c mouse model of mammary carcinogenesis is loss of heterozygosity at the Trp53 locus [30]. This observation indicates that the formation of mammary carcinoma in JNK-deficient mice may be caused by accelerated loss of heterozygosity of tumor suppressor genes.
Conclusions
We tested the hypothesis that JNK1 or JNK2 plays a critical role during breast cancer development. We found that neither JNK1 nor JNK2 is required for mammary carcinoma in the Trp53 BALB/c mouse model. Breast tumor-free survival was significantly reduced by JNK1-deficiency or JNK2-deficiency. These data suggest that that JNK1 and JNK2 may play a role in mammary carcinoma suppression.
Acknowledgments
We thank Tammy Barrett for expert technical assistance, and Kathy Gemme for administrative assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Institutes of Health, National Cancer Institute (NCI) (www.nci.nih.gov) grant number R01CA65861 to RJD. RJD is an Investigator of the Howard Hughes Medical Institute. RJD and HKS are members of the Diabetes and Endocrinology Research Center of the University of Massachusetts Medical School that is funded by the National Institutes of Health, National Institute of Diabetes Digestive disorders and Kidney (NIDDK) (www.niddk.nih.gov) grant number P30DK52530. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 2.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Whitmarsh AJ, Davis RJ. Role of mitogen-activated protein kinase kinase 4 in cancer. Oncogene. 2007;26:3172–3184. doi: 10.1038/sj.onc.1210410. [DOI] [PubMed] [Google Scholar]
- 4.Hess P, Pihan G, Sawyers CL, Flavell RA, Davis RJ. Survival signaling mediated by c-Jun NH(2)-terminal kinase in transformed B lymphoblasts. Nat Genet. 2002;32:201–205. doi: 10.1038/ng946. [DOI] [PubMed] [Google Scholar]
- 5.Hui L, Zatloukal K, Scheuch H, Stepniak E, Wagner EF. Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation. J Clin Invest. 2008;118:3943–3953. doi: 10.1172/JCI37156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N, Nomura M, She QB, Ma WY, Bode AM, et al. Suppression of skin tumorigenesis in c-Jun NH(2)-terminal kinase-2-deficient mice. Cancer Res. 2001;61:3908–3912. [PubMed] [Google Scholar]
- 7.Bost F, McKay R, Bost M, Potapova O, Dean NM, et al. The Jun kinase 2 isoform is preferentially required for epidermal growth factor-induced transformation of human A549 lung carcinoma cells. Mol Cell Biol. 1999;19:1938–1949. doi: 10.1128/mcb.19.3.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui J, Han SY, Wang C, Su W, Harshyne L, et al. c-Jun NH(2)-terminal kinase 2alpha2 promotes the tumorigenicity of human glioblastoma cells. Cancer Res. 2006;66:10024–10031. doi: 10.1158/0008-5472.CAN-06-0136. [DOI] [PubMed] [Google Scholar]
- 9.Potapova O, Gorospe M, Bost F, Dean NM, Gaarde WA, et al. c-Jun N-terminal kinase is essential for growth of human T98G glioblastoma cells. J Biol Chem. 2000;275:24767–24775. doi: 10.1074/jbc.M904591199. [DOI] [PubMed] [Google Scholar]
- 10.Yang YM, Bost F, Charbono W, Dean N, McKay R, et al. C-Jun NH(2)-terminal kinase mediates proliferation and tumor growth of human prostate carcinoma. Clin Cancer Res. 2003;9:391–401. [PubMed] [Google Scholar]
- 11.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 12.Birch JM, Alston RD, McNally RJ, Evans DG, Kelsey AM, et al. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20:4621–4628. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 13.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 14.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 15.Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, et al. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–2159. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong C, Yang DD, Wysk M, Whitmarsh AJ, Davis RJ, et al. Defective T cell differentiation in the absence of Jnk1. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 17.Yang DD, Conze D, Whitmarsh AJ, Barrett T, Davis RJ, et al. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity. 1998;9:575–585. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]
- 18.Das M, Sabio G, Jiang F, Rincon M, Flavell RA, et al. Induction of hepatitis by JNK-mediated expression of TNF-alpha. Cell. 2009;136:249–260. doi: 10.1016/j.cell.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerry DJ, Kuperwasser C, Downing SR, Pinkas J, He C, et al. Delayed involution of the mammary epithelium in BALB/c-p53null mice. Oncogene. 1998;17:2305–2312. doi: 10.1038/sj.onc.1202157. [DOI] [PubMed] [Google Scholar]
- 20.Tournier C, Hess P, Yang DD, Xu J, Turner TK, et al. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–874. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- 21.Das M, Jiang F, Sluss HK, Zhang C, Shokat KM, et al. Suppression of p53-dependent senescence by the JNK signal transduction pathway. Proc Natl Acad Sci U S A. 2007;104:15759–15764. doi: 10.1073/pnas.0707782104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeschke A, Karasarides M, Ventura JJ, Ehrhardt A, Zhang C, et al. JNK2 is a positive regulator of the cJun transcription factor. Mol Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 23.Kuan CY, Yang DD, Samanta Roy DR, Davis RJ, Rakic P, et al. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 24.Chen P, O'Neal JF, Ebelt ND, Cantrell MA, Mitra S, et al. Jnk2 effects on tumor development, genetic instability and replicative stress in an oncogene-driven mouse mammary tumor model. PLoS One. 2010;5:e10443. doi: 10.1371/journal.pone.0010443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debies MT, Welch DR. Genetic basis of human breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2001;6:441–451. doi: 10.1023/a:1014739131690. [DOI] [PubMed] [Google Scholar]
- 26.Su GH, Song JJ, Repasky EA, Schutte M, Kern SE. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat. 2002;19:81. doi: 10.1002/humu.9002. [DOI] [PubMed] [Google Scholar]
- 27.Teng DH, Perry WL, 3rd, Hogan JK, Baumgard M, Bell R, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- 28.Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, et al. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339–2342. [PubMed] [Google Scholar]
- 29.Hayakawa J, Mittal S, Wang Y, Korkmaz KS, Adamson E, et al. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol Cell. 2004;16:521–535. doi: 10.1016/j.molcel.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Blackburn AC, McLary SC, Naeem R, Luszcz J, Stockton DW, et al. Loss of heterozygosity occurs via mitotic recombination in Trp53+/− mice and associates with mammary tumor susceptibility of the BALB/c strain. Cancer Res. 2004;64:5140–5147. doi: 10.1158/0008-5472.CAN-03-3435. [DOI] [PubMed] [Google Scholar]