Abstract
Macroautophagy (hereafter autophagy) is a cellular degradation process, which in yeast is induced in response to nutrient deprivation. In this process, a double-membrane vesicle, an autophagosome, surrounds part of the cytoplasm and fuses with the vacuole to allow the breakdown and subsequent recycling of the cargo. In yeast, many autophagy-related (ATG) genes have been identified that are required for selective and/or nonselective autophagy. In all autophagy-related pathways, core Atg proteins are required for the formation of the autophagosome, which is one of the most unique aspects of autophagy and is unlike other vesicle transport events. In contrast to nonselective autophagy, the selective processes are induced in response to various specific physiological conditions such as alterations in the carbon source. In this review, we provide an overview of the common aspects concerning the mechanism of autophagy-related pathways, and highlight recent advances in our understanding of the machinery that controls autophagy induction in response to nutrient starvation conditions.
1. Introduction
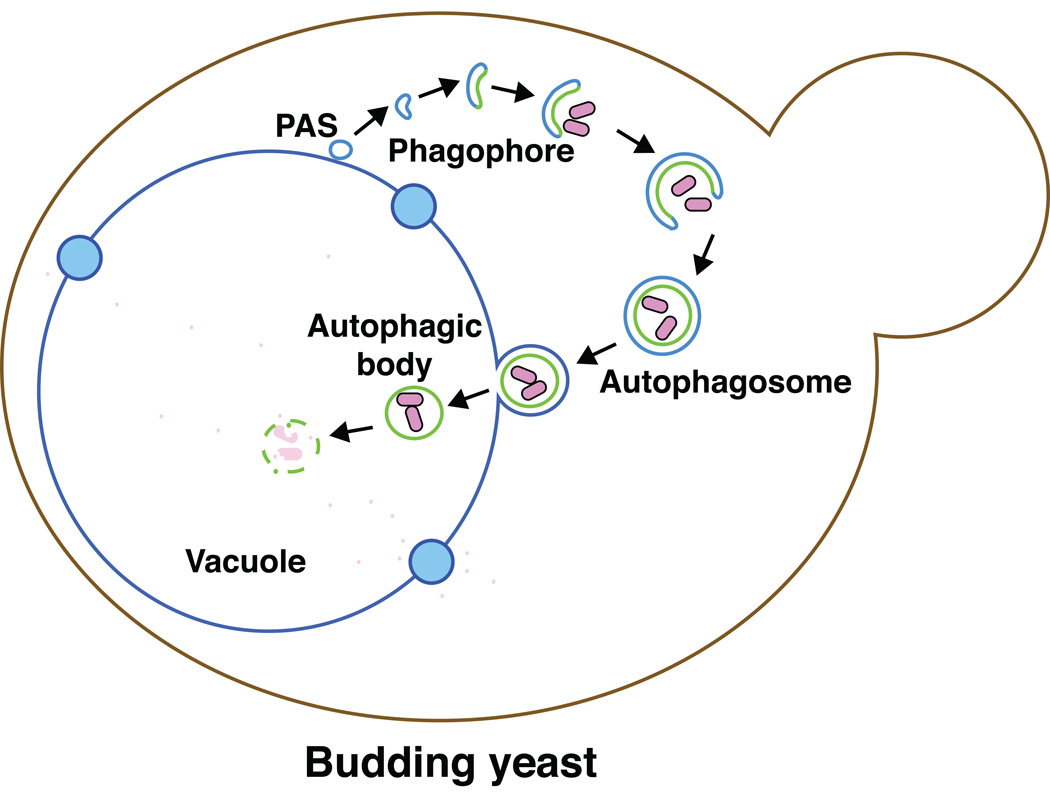
In many eukaryotic cells, the response to starvation conditions is one of the most important processes for their survival. To maintain life in unfavorable conditions such as nutrient deprivation, these cells degrade their own cellular components by a self-eating process via the lysosome/vacuole [1, 2, 3]. The sequestration and delivery of cellular components to the lytic compartment is performed by autophagy. Autophagy is a highly conserved mechanism in eukaryotic cells. In this process, the phagophore generates a double-membrane vesicle in the cytosol, which encloses cellular components. This double-membrane compartment is termed an autophagosome [4, 5]. Upon completion, the outer membrane of the autophagosome fuses with the lysosome/vacuole membrane. As a result, the inner membrane of the autophagosome and its cargo are exposed to a range of hydrolytic enzymes that lyse the membrane and degrade the macromolecules contained within the vesicle; the breakdown products are released back into the cytosol through various permeases, and are reused by the cell (Figure 1).
Figure 1. Autophagy in the yeast Saccharomyces cerevisiae.
In this process, a double-membrane phagophore is generated in the cytosol, which involves nucleation at the phagophore assembly site (PAS). The phagophore enwraps cellular components and expands to form a double-membrane vesicle termed an autophagosome. The outer membrane of the autophagosome ultimately fuses with vacuole membrane. As a result, the inner membrane, which encloses the cargo, is released into the vacuole lumen. The intravacuolar single-membrane vesicle, termed an autophagic body, is lysed, exposing the cargo to a range of hydrolytic enzymes. The resulting degradation products are released back into the cytosol for reuse.
In addition to bulk, nonselective degradation that occurs under starvation conditions, selective cargos, such as organelles, are also targeted and transported to the vacuole via autophagy. In this case, particular physiological conditions induce the processes through which organelles are sequestered into autophagosomes selectively. These specific targeting mechanisms play important roles in maintaining cellular homeostasis. Included among these auotphagy-related pathways are mitophagy [6], pexophagy [7, 8], and ribophagy [9], the selective degradation of mitochondria, peroxisomes and ribosomes, respectively. In yeast cells, α-mannosidase (Ams1) and the precursor form of aminopeptidase I (prApe1) are specific cargos that are delivered to the vacuole via an autophagy-related pathway in vegetative conditions. These hydrolases are synthesized in the cytosol and transported to the vacuole by the cytoplasm-to-vacuole targeting (Cvt) pathway, which is the only known biosynthetic use of autophagic delivery [10–12].
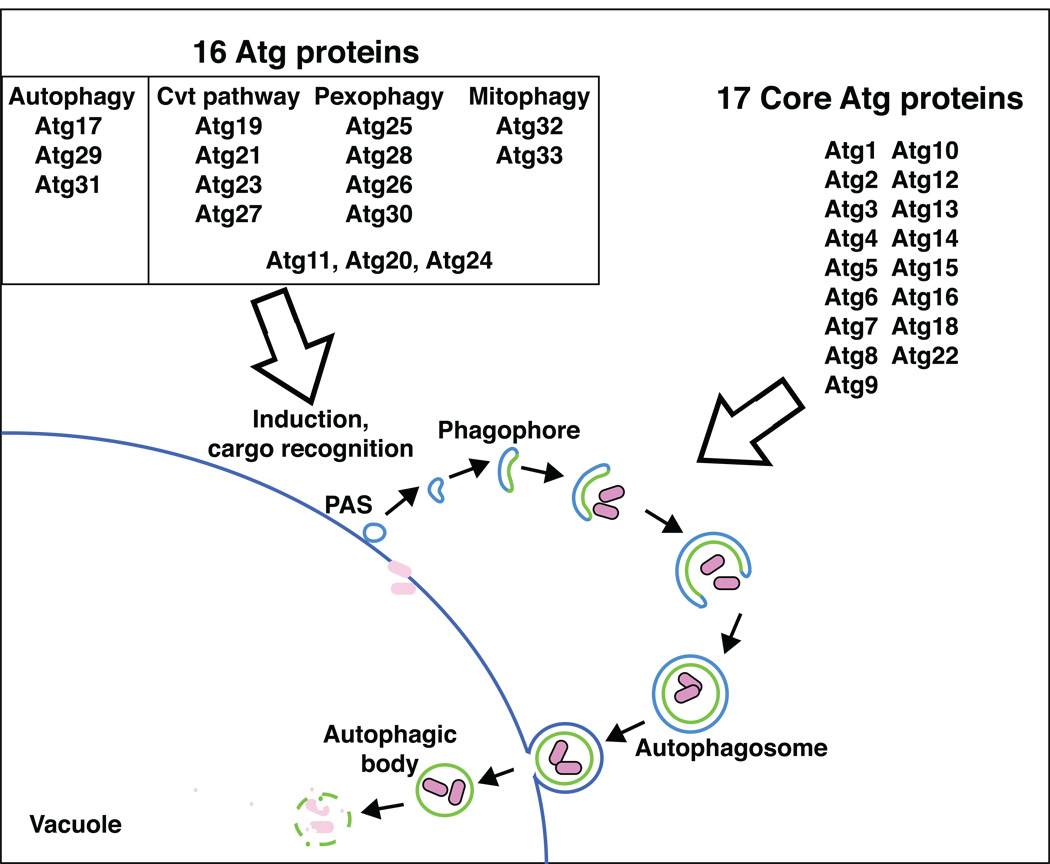
Our understanding of autophagy-related pathways has seen remarkable progress within the last ten to twelve years, coinciding with the molecular era of autophagy. A major breakthrough resulted from genetic screens in the budding yeast Saccharomyces cerevisiae, and the methylotrophic yeasts Pichia pastoris and Hansenula polymorpha [13–20]. From these studies, 33 autophagy-related genes have been identified [21]. The corresponding gene products function at several steps in the nonselective and selective autophagy-related pathways. Seventeen of the Atg proteins are commonly required for all of the autophagy-related pathways and are referred to as the core components, whereas another sixteen proteins have more specific roles.
During autophagosome formation, most of the Atg proteins can be detected at a single perivacuolar punctum by microscopy, when tagged with a fluorophore such as the green fluorescent protein. This site is referred to as the phagophore assembly site (PAS), and is thought to play a critical role in autophagosome formation, although neither the detailed image nor the function of the PAS has been fully realized [22, 23]. The PAS may be a nucleating site for autophagosome formation, or it may literally represent the phagophore, the initial compartment that matures into an autophagosome. Although the machinery of autophagosome formation is essentially the same for all autophagy-related pathways, the induction mechanisms vary depending on the specific physiological conditions. This review provides an overview of our understanding of the process of autophagosome formation, and the induction mechanisms that operate during nonselective and selective autophagy.
2. Roles of core autophagy-related proteins in autophagosome formation
Autophagy is unique in that formation of the double-membrane sequestering vesicle occurs through a process that is unlike any other intracellular membrane trafficking pathways. Thus, the mechanism of autophagosome formation is one of the most important aspects of autophagy with regard to understanding this dynamic process, and the significance of this step is reflected in the fact that the majority of the Atg proteins are involved in autophagosome biogenesis (Figure 2). Although the precise role of each Atg protein during autophagosome formation is not fully understood, several proteins have been relatively well characterized. One critical feature of the autophagosome is that its size can be modulated to accommodate various cargos. Thus, the phagophore is thought to expand rather than being generated in a single step. In this process, the PAS may play a role for assembly of the incoming membrane, and elongation of the phagophore. Various organelles, including the endoplasmic reticulum, mitochondria, and the Golgi complex, might act as a membrane source for autophagy [24–26]. In fact, it seems that the cell must mobilize membrane from multiple locations to meet the huge demand for autophagosome formation that occurs during starvation.
Figure 2. Identification of 33 Atg proteins.
Currently, there are 33 autophagy-related (Atg) proteins in fungi that have been identified by various screens. Seventeen core Atg proteins are required for all autophagy-related pathways, and most function at the stage of autophagosome formation. Another sixteen proteins have more specific roles. These latter components are typically involved in the induction of particular autophagy-related pathways in response to different physiological conditions and/or the recognition of selective cargos.
2.1. Atg9 functions in delivering membrane to the expanding phagophore
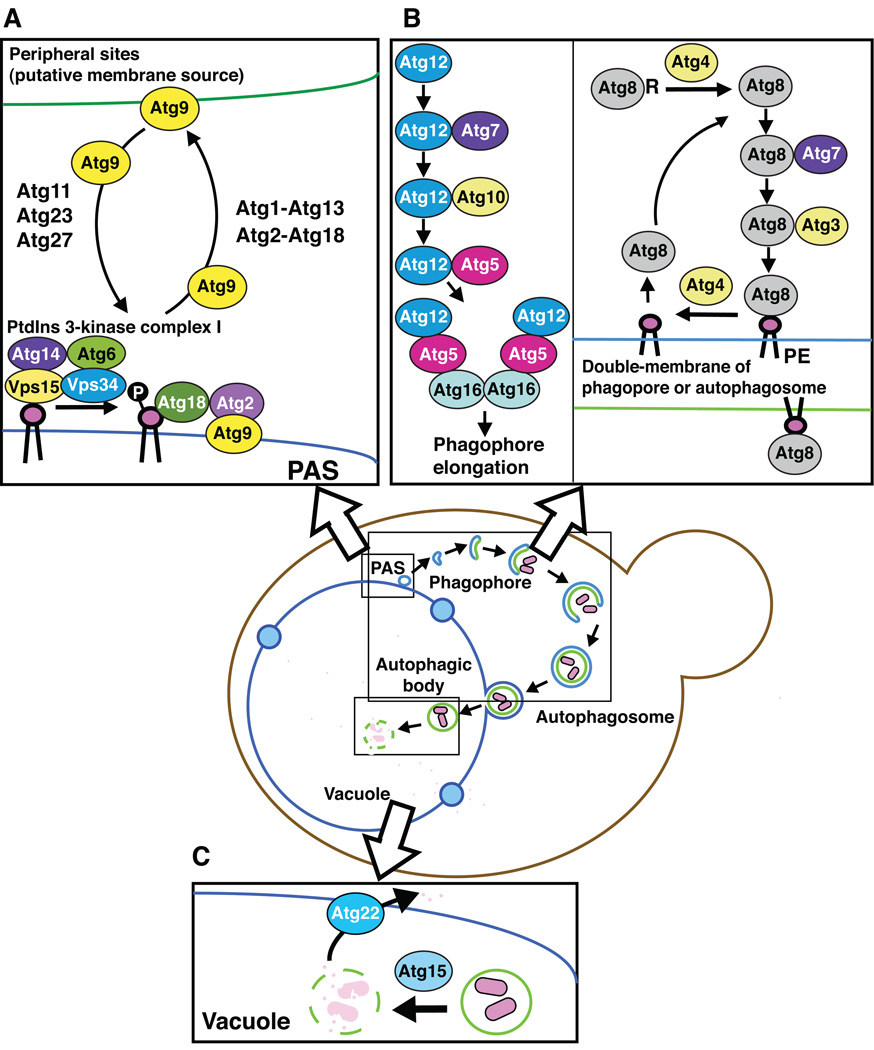
The autophagy-related protein that appears to be the most relevant with regard to the membrane source for phagophore expansion is Atg9, which is the only integral membrane protein of the core Atg proteins that are required for autophagosome biogenesis (Figure 3A) [27]. Atg9 localizes to the PAS similar to the other Atg proteins. However, it is also detected at multiple, peripheral (i.e., non-PAS) sites within the cell. At least some of these sites correspond to the mitochondria, or regions in close proximity to this organelle [28]. Atg9 shuttles between these sites and the PAS; thus, one hypothesis is that Atg9 acts as a carrier that transfers membrane from the donor sites to the expanding phagophore. Although the actual mechanism involved in this type of membrane shuttling is not known, several Atg proteins participate in regulating Atg9 movement. Atg11, Atg23 and Atg27 are involved in anterograde movement of Atg9 to the PAS, whereas Atg1-Atg13, Atg2-Atg18 and Atg14 are all required for retrograde movement back to the peripheral sites [29, 30].
Figure 3. Molecular mechanism of autophagosome formation.
Most of the core Atg proteins function at the stage of autophagosome formation.
A. Recycling of Atg9. Atg9 is an integral membrane protein, and it localizes to the PAS as well as multiple peripheral sites that are putative membrane sources for the forming autophagosome. Atg9 shuttles between these peripheral sites and the PAS. Atg11, Atg23 and Atg27 are involved in anterograde movement of Atg9 to the PAS, whereas Atg1-Atg13 and Atg2-Atg18 are required for retrograde movement back to the peripheral sites.
B. Two ubiquitin-like conjugation systems. Both Atg8 and Atg12 are ubiquitin-like proteins that are involved in two separate conjugation systems. Atg12 is activated by Atg7, and transferred to Atg10, which conjugates it to an internal lysine of Atg5. The Atg12–Atg5 conjugate binds Atg16, which self-oligomerizes to generate a dimer. The Atg12–Atg5-Atg16 complex localizes to the PAS and to the surface of the phagophore. After, or upon, the completion of the autophagosome, these proteins are released from the surface of the vesicle and are reused. Atg8 is initially processed by Atg4 to expose a C-terminal glycine residue. The processed Atg8 is then activated by Atg7, and conjugated by Atg3 to form an amide bond with phosphatidylethanolamine (PE). Atg4 also plays a role as a deconjugation enzyme; a second cleavage event removes Atg8 from PE. Thus, the Atg8 is released from the surface of the autophagosome, whereas the Atg8–PE that remains bound to the inner surface of the completed autophagosome is delivered to the vacuole and degraded.
C. Degradation of the autophagic body in the vacuole. Atg15 is a putative lipase, and is needed for lysis of the single-membrane vesicle, the autophagic body, in the vacuole lumen. After breakdown of the cargo by vacuolar hydrolases, the products are released back into the cytosol through permeases such as Atg22.
Atg9 colocalizes with the Atg2-Atg18 complex at the PAS, and the deletion of either ATG2 or ATG18 causes the accumulation of Atg9 at this site, similar to the phenotype of strains lacking Atg1, Atg13 or Atg14. These observations are the basis for suggesting that these proteins are needed for retrieval of Atg9 from the PAS. Atg18 is recruited to the PAS through its binding to phosphatidylinositol 3-phosphate (PtdIns(3)P) [30]. There is only one PtdIns 3-kinase, Vps34, identified in yeast. Vps34 forms two distinct PtdIns 3-kinase complexes; complex I contains Vps34, Vps15, Atg6/Vps30 and Atg14, whereas PtdIns 3-kinase complex II also has Vps34, Vps15 and Atg6, but Atg14 is replaced with Vps38 [31]. In the absence of Atg14, but not Vps38, the Atg2-Atg18 complex is not able to localize to the PAS. Because of this requirement for binding PtdIns(3)P, PtdIns 3-kinase complex I activity is required for the membrane assembly involved in the formation of autophagosomes through the movement of Atg9.
2.2. Two ubiquitin-like conjugation systems function in autophagy
Assembly of Atg proteins at the PAS involves a hierarchy; certain proteins such as Atg11 in vegetative conditions [32], and Atg1-Atg13-Atg17-Atg29-Atg31 in starvation conditions, assemble at the PAS first and allow the recruitment of subsequent Atg proteins [33]. Along these lines, other proteins are dependent upon Atg9 for their own assembly at the PAS. For example, Atg8 and Atg12 do not target efficiently to this site in the absence of Atg9 [22]. Atg8 and Atg12 are ubiquitin-like proteins that are involved in two separate conjugation systems (Figure 3B) [34, 35]. Atg12 is activated by Atg7 (an E1 ubiquitin activating enzyme homologue), and conjugated through the action of Atg10 (an E2 ubiquitin conjugating enzyme analogue) to an internal lysine of Atg5 [36]. The Atg12–Atg5 conjugate binds Atg16, which self-oligomerizes to generate a dimer [37]. The Atg12–Atg5-Atg16 complex localizes to the PAS and to the surface of the phagophore. After, or upon, the completion of the autophagosome, these proteins are released from the surface of the vesicle and are reused. This complex may act in part as an E3 ligase for the second ubiquitin-like protein, Atg8 [38].
Atg8 is synthesized as a precursor form, which has an additional sequence at the C terminus; in S. cerevisiae the penultimate glycine is followed by a single arginine residue. Cleavage by Atg4 (a cysteine protease) removes the arginine to expose the glycine [39]. The processed Atg8 is then activated by Atg7, and conjugated by Atg3 (a second E2 ubiquitin conjugating enzyme analogue) to form an amide bond with phosphatidylethanolamine (PE), one of the major phospholipids of cellular membranes [36, 40]. Atg8–PE localizes to both the phagophore and autophagosomes. Atg4 also acts as a deconjugation enzyme; a second cleavage event removes Atg8 from PE [41]. Thus, the Atg8 on the surface of the autophagosome is released and can be reused, whereas the Atg8–PE that remains bound to the inner surface of the completed autophagosome is delivered to the vacuole and degraded. The regulation of the second Atg4-dependent cleavage event is not understood. It is proposed that Atg4 activity might be regulated through a redox mechanism [42], although it is also suggested that some of the Atg proteins control access of Atg4 to its substrate [43].
2.3. Degradation and recycling are critical steps in starvation-induced autophagy
All of the core Atg proteins described above are necessary for the formation of the autophagosome, which is the morphological hallmark of autophagy. The final set of core Atg proteins are those that act after the fusion of the autophagosome with the vacuole membrane (Figure 3C). Under starvation conditions, degradation and recycling of the autophagosome cargo is essential to maintain cell viability [44]. Lysis of the single-membrane vesicle, the autophagic body, in the vacuole lumen depends on Atg15, a putative lipase [45]. After breakdown of the cargo by the vacuolar hydrolases, the products are released back into the cytosol through permeases such as Atg22. It is clear that there are hydrolases for the degradation of proteins and carbohydrates in the vacuole, but the presence of nucleases or general lipases has not been verified. Similarly, there are no identified channels for the release of nucleotides, and it is not known how lipids might be released from the vacuole or subsequently reused.
3. Molecular machinery for the induction of autophagy-related pathways
3.1. Induction of nonselective autophagy in response to starvation conditions
3.1.1. Regulation of core autophagy-related proteins
As described in the preceding section, all autophagy-related pathways appear to generate double-membrane vesicles via mechanisms that utilize the same Atg proteins. Although recent studies have identified possible pathways that are independent of at least some of the core proteins, these pathways may reflect bypass mechanisms that do not occur under normal physiological conditions. The primary differences between autophagosomes and selective sequestering vesicles (e.g., Cvt vesicles that are used in the Cvt pathway, pexophagosomes, etc.) are their sizes, the exclusion of nonspecific cargo and the frequency of generation. As an example, Cvt vesicles are approximately 140 to 160-nm in diameter, compared to 300 to 900-nm for autophagosomes [4, 22, 23, 46]. In addition, the sequestering vesicles formed for selective types of autophagy appear to closely appose the cargo, excluding bulk cytoplasm. Thus, in the case of the Cvt pathway, the cargo is comprised of the Cvt complex (prApe1, with its receptor Atg19) and Ams1 [47, 48]. Finally, these selective vesicles are generated constitutively at a low level. In contrast, when autophagy is induced there is a substantial increase in the frequency of autophagosome formation (autophagy also occurs at a low basal level in vegetative conditions) [49]. The Cvt complex can be incorporated into autophagosomes, but this still happens selectively through the action of Atg19, which binds to Atg8 after delivery of the cargo to the PAS [50]. Therefore, it is likely that the induction machinery for nonselective autophagy may work as a switch to regulate autophagosome formation at an early step, modulating both their size and the frequency of formation.
When bulk autophagy is induced in starvation conditions, the expression level of some of the core ATG genes increases. The greatest change is seen with the expression of ATG8 and ATG14 [51–53]. The Atg8 protein increases 10 to 20-fold following induction, but the transcription factor(s) that regulate the expression level of the ATG8 gene are still unknown. The Atg8 level affects the size of the autophagosome, but not the frequency of its generation [54]. The transcription of ATG14 is also increased more than 20-fold in nitrogen starvation conditions, although the relationship between the Atg14 level and autophagy induction is not known [53]. Gln3, a nitrogen catabolite repression-sensitive GATA-type transcription factor, controls the expression of ATG14.
In addition to ATG8 and ATG14, there are other ATG genes that are upregulated in response to starvation conditions, although the magnitude of the increase is less than is seen with these two genes. Microarray analyses indicate that ATG1, ATG3, ATG4, ATG5, ATG7, ATG12 and ATG13 are upregulated [55], but as with ATG14, the significance of increased expression is not known. Additional studies of transcriptional regulation of the ATG genes will likely clarify the relationship between increasing Atg protein levels and autophagy induction. While alterations in the expression levels of ATG genes whose products are involved in autophagosome formation might be regulated in starvation conditions, this type of change does not fully account for the mechanism of autophagy induction in response to starvation.
3.1.2. Atg1 kinase complex
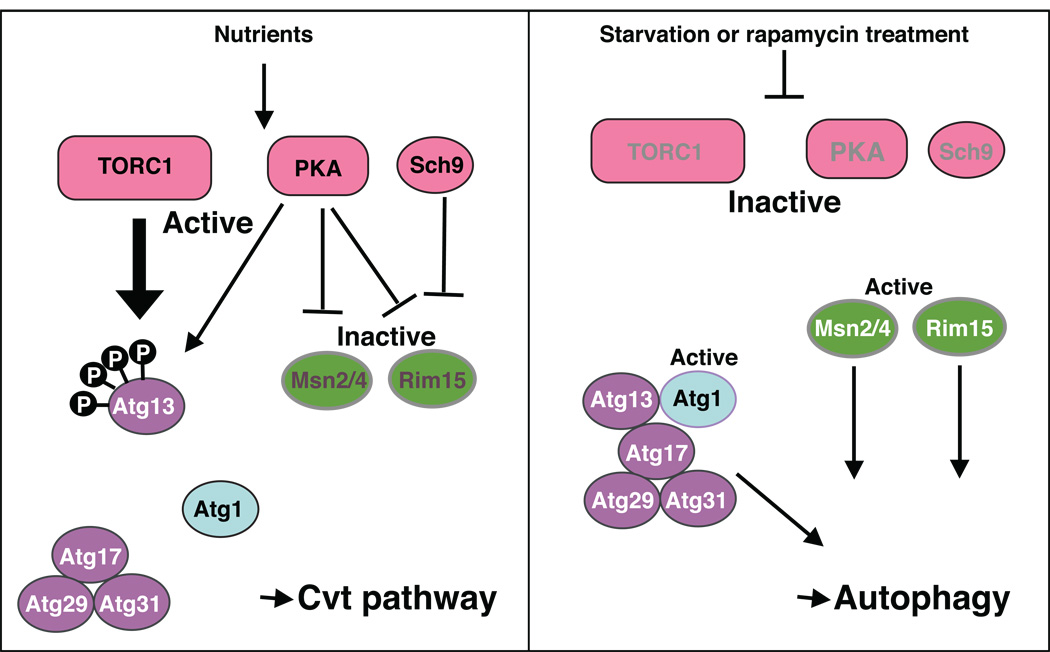
Atg13, one of the core Atg proteins, plays a role in directly receiving and transducing the signal that allows the cell to respond to changing nutrient conditions. In nutrient rich conditions, Atg13 is highly phosphorylated. When cells shift to starvation conditions, there is a rapid at least partial dephosphorylation of Atg13 (Figure 4) [56]. The dephosphorylated Atg13 is proposed to interact with Atg1 with a higher affinity and to regulate its kinase activity. Atg1 is another of the core Atg proteins, and the central member of a kinase complex. Atg1 is a Ser/Thr kinase, and this kinase activity is essential for autophagy [57]. Atg1 kinase activity is enhanced in nutrient starvation conditions or following rapamycin treatment [56]. Neither the function of the Atg1 kinase complex nor the physiologically-relevant target of Atg1 kinase are known, although the current model is that modulation of Atg1 kinase activity and the Atg1 interacting proteins plays a role in switching from the Cvt pathway to autophagy in starvation conditions.
Figure 4. Induction of autophagy in response to starvation conditions.
In nutrient rich conditions, TORC1 is active, and it phosphorylates Atg13. Phosphorylated Atg13 is not able to bind efficiently to Atg1. In starvation conditions, TORC1 is inactivated, and Atg13 is partially dephosphorylated, increasing its affinity for Atg1. Atg1-Atg13 interacts with Atg17-Atg29-Atg31 to form the Atg1 kinase complex; Atg1 kinase activity is enhanced in nutrient starvation conditions by the formation of this complex. PKA is also a negative regulator of autophagy, and its downstream targets include Rim15 kinase and the Msn2/Msn4 transcription factors. PKA also phosphorylates Atg1 and Atg13. It is likely that phosphorylation of a single substrate such as Atg1 is not the sole mechanism of PKA inhibition. In addition to PKA, Sch9 is another negative regulator involved in nutrient sensing. Thus, autophagy is suppressed by at least three parallel pathways, involving TORC1, PKA and Sch9, in nutrient-rich conditions.
It is likely that other members of the Atg1 kinase complex also play a role in the regulation of Atg1 kinase activity. In addition to Atg1 and Atg13, the Atg1 kinase complex includes Atg17, Atg29 and Atg31, which are required for autophagy, but not the Cvt pathway, and Atg11, Atg20 and Atg24, which are needed for the Cvt pathway, but not for nonselective autophagy; however, it is not known if all of these proteins are present in the complex at the same time, or if there are subcomplexes that vary in their composition depending on the nutrient conditions. The presence of the Atg1, Atg13, Atg17, Atg29 and Atg31 proteins are all necessary for their localization at the PAS [32]. This finding suggests that these five proteins are recruited to the PAS shortly after switching to starvation conditions, that they then form an Atg1 kinase complex, and that they subsequently act as a scaffold for the recruitment of additional Atg proteins to this site. Atg17, Atg29 and Atg31 always form a complex, even in nutrient-rich conditions [58]. According to a recent study, Atg17 interacts with Atg9, but the significance of this interaction is not known [59]. It will be important to understand the functions of the autophagy-specific proteins, how they receive the signal for starvation, and how they modulate Atg1 kinase activity to regulate the magnitude of the autophagosome formation process. One conclusion from these various studies, however, is that the kinases and phosphatases that regulate the phosphorylation of Atg13, and possibly other members of the Atg1 kinase complex, are some of the key factors controlling the induction mechanism of autophagy.
3.1.3. Signaling pathways regulating autophagy
TOR (target of rapamycin) is one of the major nutrient-signaling regulators in eukaryotes. The Tor proteins function as nutrient sensors, and in particular as sensors of nitrogen. The TOR signaling pathway plays a major role in controlling autophagy induction, although as with other aspects of regulation the molecular machinery of the induction in response to starvation conditions is not fully understood. S. cerevisiae has two TOR homologous genes, TOR1 and TOR2 [60]. Both TOR proteins are kinases, and they form two functionally distinct protein complexes, TORC1 (that includes Tor1 and/or Tor2) and TORC2 (containing only Tor2) [61]. Both of these complexes have various functions for cell growth and respond to the availability of nutrients as well as other types of stress. TORC1 has a much greater sensitivity to rapamycin, and inactivation of TORC1 by rapamycin treatment stimulates autophagy induction even in nutrient-rich conditions [62]. Therefore, TORC1 functions as a negative regulator of autophagy.
It is not clearly understood how nutrient metabolites act upstream of TORC1 to change its kinase activity, and in particular how TORC1 senses extracellular nutrient conditions, such as the levels of nitrogen. TORC1 apparently senses the intracellular level of glutamine, which is a major intermediate in yeast nitrogen metabolism, and in response regulates a range of transcription factors [63]. Although some downstream effectors of TORC1 are regulated by the concentration of glutamine, not all such factors are affected. Thus, the activity of TORC1 is likely controlled by various nutrient factors acting in parallel, not only by glutamine.
Protein phosphatase type-2A (PP2A) is a major downstream effector of TORC1, and the Tap42-PP2A pathway is involved in the regulation of autophagy [64]. In this pathway, TORC1 inhibits autophagy via the phosphorylation of Tap42, which in turn activates the catalytic subunit of PP2A. The target of PP2A that is involved in the negative regulation of autophagy, however, is not known.
In addition to indirect regulation mechanisms (i.e., those managed through transcriptional regulation), TORC1 directly phosphorylates Atg13 [56, 65]. The expression of an unphosphorylated Atg13 mutant induces autophagy via activation of Atg1 even in nutrient-rich conditions. This result suggests that Atg13 is an initial target of TORC1, and that it acts as a nutrient sensor of autophagy.
In addition to TORC1, the Ras/cAMP-dependent protein kinase A (PKA) signaling pathway also plays a regulatory role. The Ras/PKA pathway controls many aspects of cell growth in response to extracellular nutrients and stress conditions [66–68]. PKA consists of both catalytic and regulatory subunits [69]. The isoforms of the catalytic subunits are encoded by TPK1, TPK2 and TPK3, which are redundant genes, whereas the regulatory subunit is encoded by BCY1 [70]. In the presence of cAMP, Bcy1 binds to cAMP and releases the catalytic subunits as individual active monomers. Thus, the regulatory subunits inhibit the kinase activity of the Tpk proteins in a cAMP-dependent manner, and the cAMP concentration is regulated by the Ras signaling pathway. In S. cerevisiae, there are two RAS genes (RAS1 and RAS2), which are highly homologous to mammalian RAS genes [71]. The Ras proteins are small GTP binding proteins that localize to the plasma membrane. Ras protein activity depends on the form of nucleotide that they bind; the GTP-bound form is active, whereas the GDP-bound form is inactive, and this is regulated by the guanine nucleotide exchange factor Cdc25 in response to the glucose concentration [72]. Thus, when there is a higher concentration of glucose, Ras becomes a GTP-bound active form that upregulates the cAMP concentration [73]. The higher concentration of cAMP in turn upregulates the kinase activity of PKA via the suppression of Bcy1. PKA is another negative regulator of autophagy. Although the relevant targets of PKA are not fully known, they appear to include Rim15 kinase and the Msn2/Msn4 transcription factors [74]. Atg1 and Atg13 are also substrates of PKA, whereas Atg18 and Atg21 are putative substrates [75, 76]. It is likely that phosphorylation of a single substrate such as Atg1 is not the sole mechanism of PKA inhibition; the expression of a hyperactive Ras mutant still inhibits autophagy even in the presence of an Atg1 mutant that lacks the PKA phosphorylation sites [75]. In addition to PKA, Sch9 is also involved in nutrient sensing. The inactivation of both PKA and Sch9 induces autophagy [74], whereas inactivation of Sch9 alone does not appear to be sufficient. These results suggest that nonselective autophagy is suppressed by at least three parallel pathways, TORC1, PKA and Sch9, in nutrient-rich conditions (Figure 4).
3.2. Induction of selective autophagy
As discussed above, in addition to nonselective autophagy, specific cargos are transported to the vacuole via selective types of autophagy. The Cvt pathway is a biosynthetic process, wherein prApe1 and Ams1 are transported to the vacuole where they carry out their function as hydrolases. Precursor Ape1 is synthesized in the cytosol and subsequently assembles into a homo-dodecamer, which then form a larger oligomer (termed the Ape1 complex) [48]. The Atg19 receptor binds to the propeptide of prApe1 to form the Cvt complex; an Ams1 oligomer is also recruited to this complex via its binding to Atg19, but is a relatively minor component in terms of quantity. The Cvt complex interacts with Atg11 via its binding to Atg19, and localizes the complex to the PAS. Neither Atg19 nor Atg11 is necessary for the nonselective autophagy that occurs in response to starvation conditions, but Atg11 normally acts in assembly of the PAS in nutrient-rich conditions and facilitates the formation of the starvation-specific PAS involving Atg1-Atg13-Atg17-Atg29-Atg31.
Selective autophagy is also seen with the specific degradation of mitochondria, termed mitophagy [6]. Damaged or excessive mitochondria are a potential source of reactive oxygen species that can cause oxidative damage. Therefore, the degradation of damaged-mitochondria is critical for proper cellular homeostasis. Recent studies identified Atg32, a mitochondrial protein that is essential for mitophagy [77, 78]. Atg32 is a mitophagy-specific protein that appears to function as an organelle tag. Atg32 localizes at the outer membrane of mitochondria, and binds to Atg11 under mitophagy-inducing conditions, allowing the mitochondria to localize to the PAS; Atg11 functions as an adaptor in several types of selective autophagy. Atg32 contains a WXXL motif that is necessary for its interaction with Atg8, an interaction that is important for mitophagy [79]. It is still unclear whether mitophagy occurs through a macroautophagy- or microautophagy-like process, or both.
The mechanism of selective peroxisome turnover under nutrient-rich conditions by an autophagy-related pathway is termed pexophagy. In Pichia pastoris, methanol is one of the carbon sources that is metabolized in peroxisomes. Therefore, when cells are switched from growth on methanol to another carbon source, such as glucose or ethanol, the accumulated peroxisomes are largely superfluous, are another potential source of reactive oxygen species, and are rapidly eliminated by pexophagy. PpAtg30 is required for pexophagy in this organism, but a homologue has not been identified in S. cerevisiae [80]. This protein functions as an adaptor, linking PpAtg14, a peroxisomal tag, with the autophagy machinery in conjunction with Atg11.
4. Conclusion
The start of the molecular era of autophagy research in yeast allowed researchers to begin to address certain questions concerning the specific protein components of this pathway. Two aspects of autophagy that have been of particular interest are the mechanism of autophagosome formation, and the regulation of autophagy induction. Indeed, the double-membrane autophagosome is the morphological hallmark of macroautophagy. Nonetheless, the biogenesis of this vesicle is still not understood. For example, what is the source of the autophagosome membrane? How is curvature imposed during nonspecific autophagy? What determines the size of the autophagosome? Autophagy involves dynamic membrane rearrangement, and the majority of the autophagy-related proteins function at this stage of the process. Most of the core ATG genes are conserved from yeast to plants and animals. This fact suggests that although autophagy and autophagy-related pathways perform various functions in response to diverse physiological conditions in eukaryotic cells, the sequestering membrane may be formed by a common mechanism. Further studies on the core Atg proteins should provide additional information on this topic.
The issue of regulation is also one of great interest both in terms of our understanding of basic cell biology, and also because of its relationship to potential therapeutic modulation of autophagy. Regulation can be considered from various aspects including the action of upstream components such as the TOR kinase, direct control of the Atg proteins including phosphorylation of Atg13, and the temporal regulation of protein-protein interactions as occurs with the recognition of cargo by specific receptors as with the binding of Atg11 and Atg32. There are large gaps in our knowledge of the regulatory pathways and how the overall network of components interacts in a coordinated manner. Continued studies on the mechanisms that transduce extracellular signals via the actions of various kinases, phosphatases and transcription factors, and by direct modifications of the autophagy-related proteins, will provide us with a better understanding of how autophagy is precisely regulated to maintain optimal cellular physiology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Deter RL, Baudhuin P, de Duve C. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 4.Baba M, Takeshige K, Baba N, Ohsumi Y. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba M, Osumi M, Ohsumi Y. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- 6.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai Y, Oku M, van der Klei IJ, Kiel JAKW. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Dunn WA, Jr, Cregg JM, Kiel JAKW, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 9.Kraft C, Deplazes A, Sohrmann M, Peter M. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 10.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 11.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Proc Natl Acad Sci U S A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klionsky DJ, Emr SD. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukada M, Ohsumi Y. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 14.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 15.Harding TM, Morano KA, Scott SV, Klionsky DJ. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 17.Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA., Jr J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]
- 18.Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukaiyama H, Oku M, Baba M, Samizo T, Hammond AT, Glick BS, Kato N, Sakai Y. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- 20.Titorenko VI, Keizer I, Harder W, Veenhuis M. J Bacteriol. 1995;177:357–363. doi: 10.1128/jb.177.2.357-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. J Biol Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reggiori F, Shintani T, Nair U, Klionsky DJ. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Duve C, Wattiaux R. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 26.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juhasz G, Neufeld TP. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn WA, Jr, Klionsky DJ. J Biol Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 31.Kihara A, Noda T, Ishihara N, Ohsumi Y. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yorimitsu T, Klionsky DJ. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohsumi Y. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 35.Yorimitsu T, Klionsky DJ. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 37.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 38.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 39.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 41.Nakatogawa H, Ichimura Y, Ohsumi Y. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 42.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nair U, Cao Y, Xie Z, Klionsky DJ. J Biol Chem. 2010 doi: 10.1074/jbc.M109.080374. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Epple UD, Suriapranata I, Eskelinen E-L, Thumm M. J Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 51.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang W-P, Scott SV, Kim J, Klionsky DJ. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 53.Chan TF, Bertram PG, Ai W, Zheng XF. J Biol Chem. 2001;276:6463–6467. doi: 10.1074/jbc.M008162200. [DOI] [PubMed] [Google Scholar]
- 54.Xie Z, Nair U, Klionsky DJ. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Proc Natl Acad Sci U S A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 58.Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 59.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 60.Weisman R, Choder M. J Biol Chem. 2001;276:7027–7032. doi: 10.1074/jbc.M010446200. [DOI] [PubMed] [Google Scholar]
- 61.Reinke A, Anderson S, McCaffery JM, Yates J, III, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 62.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 63.Crespo JL, Daicho K, Ushimaru T, Hall MN. J Biol Chem. 2001;276:34441–34444. doi: 10.1074/jbc.M103601200. [DOI] [PubMed] [Google Scholar]
- 64.Yorimitsu T, He C, Wang K, Klionsky DJ. Autophagy. 2009;5:616–624. doi: 10.4161/auto.5.5.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santangelo GM. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Estruch F. FEMS Microbiol Rev. 2000;24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 68.Norbeck J, Blomberg A. Yeast. 2000;16:121–137. doi: 10.1002/(SICI)1097-0061(20000130)16:2<121::AID-YEA511>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 69.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 70.Kuret J, Johnson KE, Nicolette C, Zoller MJ. J Biol Chem. 1988;263:9149–9154. [PubMed] [Google Scholar]
- 71.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 72.Lai CC, Boguski M, Broek D, Powers S. Mol Cell Biol. 1993;13:1345–1352. doi: 10.1128/mcb.13.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Broek D, Samiy N, Fasano O, Fujiyama A, Tamanoi F, Northup J, Wigler M. Cell. 1985;41:763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- 74.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. Proc Natl Acad Sci U S A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. Proc Natl Acad Sci U S A. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 78.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noda NN, Ohsumi Y, Inagaki F. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 80.Farré JC, Manjithaya R, Mathewson RD, Subramani S. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]