Abstract
Membrane fusion plays an essential role in the entry of enveloped viruses into target cells. The merging of viral and target cell membranes is catalyzed by viral fusion proteins, which involves sequential multiple steps in the fusion process. However, the fusion mechanisms mediated by different fusion proteins involve multiple transient intermediates that have not been well characterized. Here we report a synthetic virus platform that allows us to better understand the different fusion mechanisms driven by the diverse types fusion proteins. The platform consists of lentiviral particles co-enveloped with a surface antibody, which serves as the binding protein, along with a fusion protein derived from either influenza virus (HAmu) or Sindbis virus (SINmu). By using a single virus tracking technique, we demonstrated that both HAmu- and SINmu-bearing viruses enter cells through clathrin-dependent endocytosis, but they required different endosomal trafficking routes to initiate viral fusion. Direct observation of single viral fusion events clearly showed that hemifusion mediated by SINmu upon exposure to low pH occurs faster than that mediated by HAmu. Monitoring sequential fusion processes by dual labeling the outer and inner leaflets of viral membranes also revealed that the SINmu-mediated hemifusion intermediate is relatively long-lived as compared with that mediated by HAmu. Taken together, we have demonstrated that the combination of this versatile viral platform with the techniques of single virus tracking can be a powerful tool for revealing molecular details of fusion mediated by various fusion proteins.
Keywords: membrane fusion, virus fusion proteins, single virus tracking, targeted gene delivery, lentiviral vectors
INTRODUCTION
An efficient transport of genetic materials to the target cells is a key requirement for viral infection. Many viruses enter cells through endocytosis and utilize different cellular environments to optimize their infection process. During the entry process of membrane-enveloped viruses, fusion of virus envelope with the endosomal membrane is essential for the release of the viral genome into cytosol prior to nuclear transport. Although merging the two membranes is thermodynamically favorable, the fusion reaction is hampered by a huge energy barrier (Chernomordik and Kozlov, 2003; Chernomordik and others, 2006; Harrison, 2008). Viral fusion proteins can lower this barrier through the activation energy provided by their conformational change to reach an energetically stable state (Carr and others, 1997; Dimitrov, 2004; Hogle, 2002; Mothes and others, 2000). It is generally believed that virus membrane fusion proceeds through sequential and multiple steps (Chernomordik and Kozlov, 2003; Dimitrov, 2004; Harrison, 2008; Jahn and others, 2003; Kielian and Rey, 2006; Kuhn and others, 2002). Initially, viral fusion proteins undergo a conformational change induced by receptor-binding and/or a pH change, which yields a point of contact between the viral and the target membranes. This induces the process of hemifusion (or lipid mixing), in which the proximal leaflets of two distinct lipid membranes are merged while the distal leaflets remain unchanged. The subsequent merging of the distal leaflets causes fusion pore formation (or content mixing), which establishes the first aqueous connection between the two apposing membranes (Jackson and Chapman, 2008; Lentz and others, 2000; Zimmerberg and others, 1994). This general fusion reaction scheme is relatively well documented, but the detailed mechanisms and pathways underlying each fusion step, which may vary significantly among the diverse types of viral fusion proteins, have been less well-studied.
The influenza and Sindbis viruses have been commonly used as model viruses to study the underlying mechanism of viral membrane fusion; cell-cell (Blumenthal and others, 1996; Chernomordik and others, 1998; Mittal and others, 2003; Zaitseva and others, 2005) and virus-liposome fusion assays (Shangguan and others, 1996; Smit and others, 1999; 2001; Smit and others, 2002; White and others, 1982) have been utilized for the study of the viral membrane fusion process. These conventional fusion assays have been instrumental for analyzing averaged results of a large number of fusion events, but they cannot be employed to study the kinetics of single viral fusion process or monitor the series of transient fusion intermediates formed during single virus fusion with target cell membranes. Direct visualization of the viral fusion process can expand our ability to reveal molecular details of virus fusion. The observation of the actual fusion event and its kinetic characterization at the single virus level can provide crucial understanding of the underlying mechanisms of virus infection. One complication of comparing the fusion reactions of different viruses in living cells is the differences in their receptor bindings, which may subsequently result in different intracellular viral trafficking mechanisms being involved in the fusion process. For example, the influenza virus HA1 protein has receptor-binding specificity to sialic acid (Skehel and Wiley, 2000), while the E2 protein of Sindbis virus has binding recognition to heparan sulfate structures on the cell surface (Byrnes and Griffin, 1998).
In this study, we test a virus platform for the study of molecular fusion mediated by the different types of fusogens. This platform is based on a lentivirus designed to incorporate a cell binding and a cell fusion protein as two separate molecules on the same viral particle (Fig. 1A). We have recently reported such a virus system displaying a CD20-specific surface antibody (αCD20) as the binding molecule and a binding-deficient fusion-competent glycoprotein as the fusogen (Yang and others, 2006). This recombinant virus could achieve targeted transduction of CD20-expressing cells in vitro and in vivo. We have also visualized the late stages of intracellular tracking and fusion of this virus (Joo and Wang, 2008). Beyond the application for targeted gene delivery, we hypothesize that this virus system can be used for the comparative study of different fusogen-mediated viral fusion. We envision that such a system would offer an opportunity to directly compare the fusion processes of various fusogens by allowing the production of viruses with the same binding proteins but different fusogens. Such designer viruses would undergo the same pathway of initial internalization induced by the interaction between the binding protein and the target receptor. The present study is to test this hypothesis by investigating the fusion properties of two fusion proteins: one is the class I fusogen derived from influenza virus hemagglutinin (designated as HAmu) and the other is the class II fusogen derived from Sindbis virus glycoprotein (designated as SINmu). The single virus tracking study of the early internalization process indicates that both HAmu- and SINmu-lentiviruses enter cells through clathrin-dependent endocytosis. This study further identifies the different requirements of endosomal trafficking for the membrane fusion of these two lentiviruses. The planar fusion assay utilizing dual labeling of outer and inner leaflets of viral membranes allows us to reveal the different kinetics of hemifusion and fusion pore formation triggered by these two fusogens in living cells.
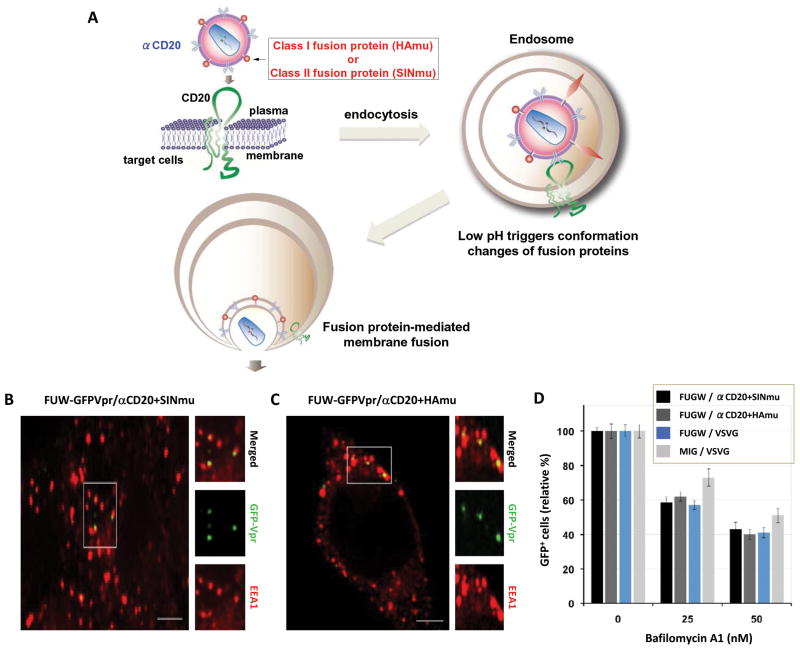
Fig. 1.
Engineered lentiviruses can enter target cells via endocytosis. (A) Schematic representation of a proposed entry mechanism for engineered lentiviruses enveloped with a CD20-specific surface antibody (αCD20) and a fusion protein (HAmu or SINmu). (B and C) GFP-Vpr-labeled viruses (green) enveloped by αCD20 and either SINmu (FUW-GFPVpr/αCD20+SINmu, B) or HAmu (FUW-GFPVpr/αCD20+HAmu, C) were added to 293T/CD20 cells at 4°C for 30 min to synchronize the binding. The cells were then warmed to 37°C for 30 min, fixed, permeabilized, and immunostained against the early endosome antigen 1 (EEA1; red). The boxed regions are enlarged in the right panels. Scale bar represents 5 μm. (D) The effect of the vacuolar proton ATPases inhibitor bafilomycin A1 on viral infection. 293T/CD20 cells (2 × 105) were pretreated for 30 min with 0, 25, and 50 nM of bafilomycin A1. The cells were then spin-infected with supernatants of lentiviruses bearing αCD20 and fusogenic protein (SINmu: FUGW/α CD20+SINmu, or HAmu: FUGW/αCd20+HAmu), VSVG-pseudotyped lentivirus (FUGW/VSVG), or VSVG-pseudotyped gamma-retrovirus (MIG/VSVG). After 1.5 h of spin-infection and the additional 3 h of incubation in the presence of the drug, the cells were washed with PBS and resupplied with fresh media. The percentage of GFP+ cells was analyzed by FACS.
MATERIALS AND METHODS
Cell lines, Antibodies and Other Reagents
The 293T/CD20 cell line was generated previously (Yang and others, 2006). Cells were maintained in a 5% CO2 environment in Dulbecco’s modified Eagle’s medium (Mediatech, Inc., Manassas, VA, USA) with 10% FBS (Sigma, St Louis, MO, USA) and 2 mM L-glutamine (Hyclone, Logan, UT, USA). Mouse monoclonal antibodies against early endosomal antigen 1 (EEA1), clathrin, caveolin-1, and lysosome-associated membrane protein 1 (Lamp-1) were purchased from Abcam (Cambridge, MA, USA). Texas red-conjugated goat anti-mouse immunoglobulin G (IgG) antibody was obtained from Molecular Probes (Carlsbad, CA, USA). Bafilomycin A1, chlorpromazine, and filipin were purchased from Sigma.
Plasmids
Assembly PCR was employed to fuse GFP to the N-terminus of Vpr. The PCR product was then inserted into the expression plasmid pcDNA3 (Invitrogen, Carlsbad, CA, USA). The cDNAs for Rab5 and Rab7 were PCR-amplified and cloned into pcDsRed-monomer-C1 (Clontech, Mountain View, CA, USA) as described (Joo and others, 2008). The plasmid encoding the dominant-negative mutant of DsRed-Rab7 (Rab7T22N) was generated by site-directed mutagenesis using the forward primer (5′-GTCGGGAAGAACTCACTCATGAACC-3′) and the backward primer (5′-GGTTCATGAGTGAGTTCTTCCCGAC-3′). The integrity of the DNA sequence for this mutant was confirmed by DNA sequencing. The constructs for GFP-Rab7 and the dominant-negative mutant of DsRed-Rab5 were obtained from Addgene (Cambridge, MA, USA).
Virus Production
GFP-Vpr-labeled lentivectors enveloped with αCD20 and fusogenic protein (SINmu or HAmu) were produced by transfecting 293T cells by a calcium phosphate precipitation method. 293T cells at 80% confluence in 6 cm culture dishes were transfected with 5 μg of the lentiviral vector FUW, together with 2.5 μg each of pcDNA3-GFPVpr, pαCD20 which encodes a mouse/human chimeric anti-CD20 antibody, pIgαβ which encodes human Igα and Igβ, two immunoglobulin associated proteins that are required for the surface expression of antibodies, pSINmu or pHAmu, and the packaging vector plasmids (pMDLg/pRRE and pRSV-REV) (Yang and others, 2006). For the production of VSVG-pseudotyped lentiviruses or retroviruses, 293T cells were transfected with 5 μg of the lentiviral vector FUGW or the retroviral vector MIG, along with 2.5 μg each of the packaging plasmids (pMDLg/pRRE, pRSV-REV, and gag-pol for lentiviruses; pClEco for retroviruses), and the envelope plasmid (VSVG). The cells were washed at 4 h post-transfection, and the medium was replaced. The viral supernatant was collected after 48 h post-transfection and filtered through a 0.45-μm pore size filter. The viral supernatant could be concentrated by ultracentrifugation (Optima L-90 K ultracentrifuge, Beckman Coulter) for 90 min at 82,700 × g and resuspended in an appropriate volume of PBS containing 5 mM MgCl2.
Viral Transduction
For the viral transduction of drug-treated cells, 293T/CD20 cells were preincubated with drugs (bafilomycin A1: 25 and 50 nM; chlorpromazine: 10 and 25 μg/ml; filipin: 1 and 5 μg/ml) for 30 min at 37°C and the cells (0.2 × 106 per well) were then spin-infected with 2 ml of viral supernatants in a 24-well culture dish at 2,500 rpm and 30°C for 90 min with a Sorval Legend centrifuge. The drug concentration was maintained during the spin-infection. The cells were further incubated for 3 h at 37°C. The drugs were then removed and replaced with fresh D10 media. For the viral transduction with Rab protein-treated cells, 293T/CD20 cells were transfected with DsRed-Rab5 or -Rab7 (either wild type or dominant-negative mutant), then seeded (0.2 × 106 per well) and spin-infected with 2 ml of viral supernatants in a 24-well culture dish. The percentage of GFP+ cells was analyzed by FACS at day 3 post-infection. All transduction assays were performed in triplicate and the results are presented as mean values with ± standard deviations.
Confocal Imaging
Fluorescent images were acquired on a Zeiss LSM 510 META laser scanning confocal microscope equipped with Argon, red HeNe, and green HeNe lasers using a Plan-apochromat 63×/1.4 oil immersion objective. A Coherent Chameleon Ti-Sapphire laser was attached as well for multiphoton imaging. Some images were collected by a Yokogawa spinning-disk confocal scanner system (Solamere Technology Group, Salt Lake City, UT) using a Nikon eclipse Ti-E microscope equipped with a 60×/1.49 Apo TIRF oil objective and a Cascade II: 512 EMCCD camera (Photometrics, Tucson, AZ, USA). An AOTF (acousto-optical tunable filter) controlled laser-merge system (Solamere Technology Group Inc.) was used to provide illumination power at each of the following laser lines: 491 nm, 561 nm, and 640 nm solid state lasers (50mW for each laser).
For the viral trafficking studies with different endocytic markers, GFP-Vpr-labeled viruses (FUW-GFPVpr/αCD20+SINmu or FUW-GFPVpr/αCD20+HAmu) at a multiplicity of infection (MOI) ~ 20 were added to 293T/CD20 cells at 4°C for 30 min. The cells were then kept at 37°C for various incubation periods, fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, and immunostained with antibodies against early endosomes, clathrin, or caveolin-1. Texas red-conjugated goat anti-mouse IgG antibody was used as the secondary antibody. To remove viral aggregates, virus-containing media were filtered by 0.45-μm pore size centrifuge tube filters (Costar, NY, USA) before the experiments were conducted.
To image virus-endosome fusion, the concentrated GFP-Vpr-labeled viruses were incubated with 50 μM of 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD) (Molecular Probes) for 1 h at room temperature. Unbound dye was removed by Microcon filter units with a 50kDa cutoff (Millipore, Billerica, MA). Double-labeled viruses were incubated with 293T/CD20 cells at 4°C for 30 min to synchronize infection, after which the cells were moved to 37°C for various time periods and fixed with 4% formaldehyde. All samples were scanned under the same conditions for magnification, laser intensity, brightness, gain, and pinhole size. To observe viral fusion in different endosomal compartments, 293T/CD20 cells were transfected with DsRed-Rab5 and GFP-Rab7 plasmids for early endosome and late endosome markers, respectively. At 48 h post-transfection, cells were seeded onto glass-bottom culture dishes (MatTek Corporation, Ashland, MA, USA) and grown at 37°C overnight. DiD-labeled viruses (FUW/αCD20+SINmu or FUW/αCD20+HAmu) at MOI ~ 20 were then incubated with the cells at 4°C for 30 min to synchronize infection. The cells were then kept at 37°C for 30 or 60 min and fixed. Images were analyzed with the use of the Zeiss LSM 510 software version 3.2 SP2 or the Nikon NIS-Elements software.
Live Cell Imaging of Virus-endosome Fusion
For the real-time detection of viral fusion in different endosomes, 293T/CD20 cells were transfected with DsRed-Rab5 and GFP-Rab7 plasmids. DiD-labeled viruses (FUW/αCD20+SINmu or FUW/αCD20+HAmu) at MOI ~ 20 were then incubated with the cells at 4°C for 30 min to allow for virus binding. The cells were then warmed to 37°C for 20 or 50 min to induce the viral internalization of SINmu- or HAmu-displaying lentiviruses. Confocal time-lapse images were recorded by spinning-disk confocal microscopy.
Imaging of Virus-cell Fusion
For the real-time detection of virus-cell fusion, 293T cells were transfected with plasmids for virus production as described above. To label the inner leaflets of the virus membrane, cells were cotransfected with a plasmid encoding DsRed-monomer-F (Clontech, Mountain View, CA, USA). The transfected cells were washed at 4 h post-transfection and incubated with 2.5 μM of DiO (3,3′-dioctadecyloxacarbocyanine perchlorate) or DiD in serum-free medium for 3 h at 37°C to label the virus at levels below self-quenching concentrations. The cells were washed again and regular D10 medium was added. To label the virus with pH-sensitive CypHer5E mono NHS ester (GE Healthcare), the concentrated viruses were incubated with the dye in 0.1 M sodium bicarbonate buffer (pH 9.3) for 1 h at room temperature. Unbound dye was removed via buffer exchange into PBS (pH 7.4) using a gel filtration column.
To detect lipid mixing upon exposure to low pH, the DiO-labeled and CypHer5-labeled viral particles (total ~ 106 infectious units) were mixed at a particle number ratio of 10 to 1 and adhered to poly-lysine-coated no. 1 coverslips for 1 h at 37°C. To monitor the sequential steps of lipid and content mixing, DiD-/DsRed-F-labeled viruses (~106 infectious units) were immobilized on poly-lysine-coated coveslips. After removing unbound viruses by PBS washing, 293T/CD20 cells were overlaid on the viruses and incubated for 30 min at room temperature. Virus-cell fusion was triggered by adding the appropriate volume of 0.2 M acetic acid, pretitrated to achieve the desired final pH. Time-lapse images were captured at ~ 3 s intervals. Fluorescent intensity versus time within the regions of interest was analyzed using the Nikon NIS-Elements software.
RESULTS
Engineered Lentiviruses Require Low pH to Trigger Endosomal Fusion
To image the intracellular trafficking of individual engineered lentiviruses displaying different fusion proteins (HAmu or SINmu) within target cells, we constructed a labeling protein GFP-Vpr by fusing green fluorescent protein (GFP) with the HIV accessory protein Vpr (viral protein R). GFP-Vpr-labeled lentiviruses enveloped with αCD20 along with HAmu (FUW-GFPVpr/αCD20+HAmu) or SINmu (FUW-GFPVpr/αCD20+SINmu) were produced as described (Joo and Wang, 2008; Yang and others, 2006). To examine whether both types of engineered lentiviruses entered target cells via endocytosis, GFP-Vpr-labeled lentiviruses were incubated with 293T/CD20 cells on ice for 30 min to synchronize infection. Cells were then warmed to 37°C for 30 min and immunostained against early endosome antigen 1 (EEA1), an early endosomal marker (Lakadamyali and others, 2006; Sieczkarski and Whittaker, 2003; Vonderheit and Helenius, 2005). As shown in Figs. 1B and 1C, most of viral particles were colocalized with EEA1, as evidenced by the appearance of yellow color in the merged images, suggesting that both viruses were located in the EEA1+ endosomes. To further demonstrate that a low pH compartment is required to allow engineered lentiviruses to penetrate into cells, bafilomycin A1, a specific inhibitor of vacuolar proton ATPases, was used to block low pH endosomal fusion (Bowman and others, 1988). To assay the transduction, we replaced the lentiviral backbone plasmid FUW with FUGW, which carries a GFP reporter gene under the control of the human ubiquitin-C promoter (Lois and others, 2002), in our transfection protocol to generate viruses. 293T/CD20 cells were pretreated with bafilomycin A1 at different concentrations and transduced with lentiviruses bearing αCD20 and a fusion protein (FUGW/αCD20+SINmu or FUGW/αCD20+HAmu), VSVG-pseudotyped lentivirus (FUGW/VSVG), or VSVG-pseudotyped gamma-retrovirus (MIG/VSVG). Viruses pseudotyped with vesicular stomatitis virus glycoprotein (VSVG) are known to enter cells through pH-dependent endocytosis (Sieczkarski and Whittaker, 2003; Sun and others, 2005) and were therefore utilized as a positive control. The FACS analysis of GFP+ cells showed that the viral transduction was inhibited in a dose-dependent manner (Fig. 1D), indicating that both types of engineered lentiviruses require low-pH-triggered endosomal fusion to complete transduction.
Entry of Engineered Lentiviruses is Clathrin-dependent
Clathrin- and caveolin-mediated endocytosis have been observed in the internalization of many viruses into cells (Blanchard and others, 2006; DeTulleo and Kirchhausen, 1998; Doxsey and others, 1987; Kirchhausen, 2000; Nabi and Le, 2003; Nichols and Lippincott-Schwartz, 2001; Rust and others, 2004). To investigate the role of clathrin- or caveolin-mediated endocytosis in the entry of engineered lentiviruses bearing HAmu or SINmu fusion proteins to target cells, we visualized the individual lentiviral particles (FUW-GFPVpr/αCD20+HAmu or FUW-GFPVpr/αCD20+SINmu) and endocytic structures (clathrin or caveolin) in target cells after different incubation time periods (0, 10, 30 min). As shown in Figs. 2A and 2C, significant colocalization of SINmu-bearing viruses (67%, n=59) or HAmu-bearing viruses (61%, n=64) with the discrete clathrin structures was detected after 10 min of incubation. After incubation for 30 min, less colocalization of SINmu-viruses (20%, n=71) or HAmu-viruses (19%, n=82) with the clathrin structures was observed, suggesting that many of the viruses had already been dissociated from uncoated clathrin vesicles and had likely been transported to early endosomes. However, no significant colocalization of engineered lentiviruses and caveolin was observed during these time periods (Figs. 2B and 2D), suggesting that caveolin might not be involved in the entry of these engineered lentiviruses. Thus, these time-course images showed that both types of engineered lentiviruses enter target cells through clathrin-mediated endocytosis.
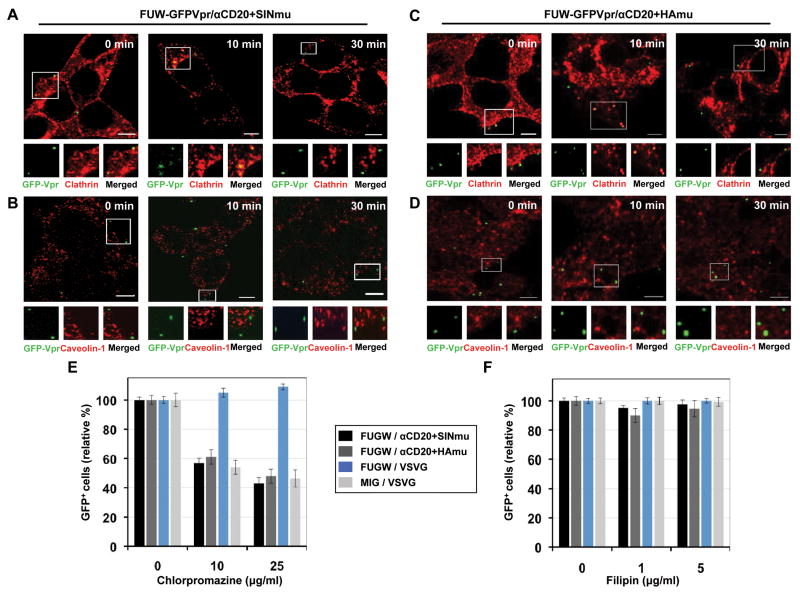
Fig. 2.
Clathrin/caveolin-dependent entry of engineered lentiviruses. GFP-Vpr-labeled lentiviruses (green) enveloped with αCD20 and SINmu (A and B) or HAmu (C and D) were incubated with 293T/CD20 cells at 4°C for 30 min. The cells were warmed to 37°C for various time periods (0, 10, and 30 min), fixed, permeabilized, and immunostained with anti-clathrin (A and C; red) or anti-caveolin-1 (B and D; red) antibodies. The boxed regions are magnified and shown below in individual panels. Scale bar represents 5 μm. Inhibition of clathrin-dependent internalization by chlorpromazine (E) or caveolin-dependent internalization by filipin (F). 293T/CD20 cells were preincubated with chlorpromazine or filipin for 30 min at 37°C. The cells (2 × 105) were then spin-infected with supernatants of lentiviruses (FUGW/αCD20+SINmu, FUGW/αCd20+HAmu, FUGW/VSVG, or MIG/VSVG). Both drug concentrations were maintained during the spin-infection as well as for the additional 3 h incubation, after which the drug was removed and replaced with fresh media. The percentage of GFP+ cells was analyzed by FACS.
To confirm the results observed from confocal imaging of the individual viral particles with clathrin or caveolin, we examined the effect of inhibitory drugs on viral entry (Figs. 2E and 2F). Chlorpromazine is a drug known to prevent clathrin polymerization and inhibit internalization mediated by clathrin-coated vesicles (CCV) (Wang and others, 1993), while filipin is a cholesterol-binding reagent that blocks caveolin-dependent internalization (Orlandi and Fishman, 1998). The FACS analysis showed that the entry of both types of lentiviruses (FUGW/αCD20+HAmu or FUGW/αCD20+SINmu) was markedly inhibited by the chlorpromazine treatment (Fig. 2E), whereas no inhibitory effect of filipin was observed (Fig. 2F), indicating that the entry of both types of viruses is dependent on clathrin. We found that VSVG-pseudotyped gamma-retrovirus (MIG/VSVG) enters cells via the clathrin-dependent route (Fig. 2E), which is in agreement with previous reports (Blanchard and others, 2006; Lee and others, 1999) and is consistent with the endocytic nature of the vesicular stomatitis virus (VSV) (Sun and others, 2005). However, no inhibitory effect by chlorpromazine was observed for the entry of the VSVG-pseudotyped lentivirus (FUGW/VSVG) (Fig. 2E), which is surprising but consistent with previous biochemical studies by us and others (Daecke and others, 2005; Joo and others, 2008). Overall, this set of drug inhibition studies confirmed the results of the imaging study that the entry of both types of engineered lentiviruses relies on a clathrin-dependent route.
Different Fusion Proteins Modulate Different Kinetics of Virus-endosome Fusion
To compare the endosomal fusion kinetics of engineered lentiviruses bearing different fusion proteins (SINmu or HAmu), we visualized the actual fusion event of internalized viruses within endosomes by labeling viruses with a lipophilic dye, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD). The spontaneous incorporation of DiD into the viral membranes of GFP-Vpr-tagged lentiviral particles offers a double-labeling scheme (Joo and Wang, 2008). Viral fusion with endosomal membranes can be detected by fluorescent dequenching of DiD (Joo and Wang, 2008; Lakadamyali and others, 2003; Sakai and others, 2006). It has been reported that Vpr remains largely associated with the preintegration complex after viral fusion (McDonald and others, 2002). This property enables us to track the viral cores that have been released to the cytosol from the fused endosomes. GFP-Vpr/DiD-labeled viruses were initially added to 293T/CD20 cells in the cold to synchronize binding. Cells were then incubated at 37°C for different time periods (0, 30, 60, and 90 min), and then fixed. The acquired images with both green and red fluorescent signals are shown in Fig. 3A. At 30 min of incubation, SINmu-bearing viruses (FUW-GFPVpr/αCD20+SINmu) were fused with endosomes, as indicated by the appearance of the red signal (Fig. 3A, upper, 30 min). However, although HAmu-bearing viruses (FUW-GFPVpr/αCD20+HAmu) also appeared to be endocytosed, the majority of the viral particles were detected solely by the green signal, suggesting that fusion had not yet occurred (Fig. 3A, lower, 30 min). The images obtained after 60 min of incubation showed that many HAmu-bearing viral particles were fused with endosomes (Fig. 3A, lower, 60 min). After 90 min of incubation, pure green signals were seen for some of the GFP-Vpr-carrying viral particles bearing SINmu, suggesting that certain viral particles were presumably released into the cytosol after endosomal fusion (Fig. 3A, upper, 90 min), whereas significant colocalization of HAmu-displaying viruses and fusion signals remained (Fig. 3A, lower, 90 min).
Fig. 3.
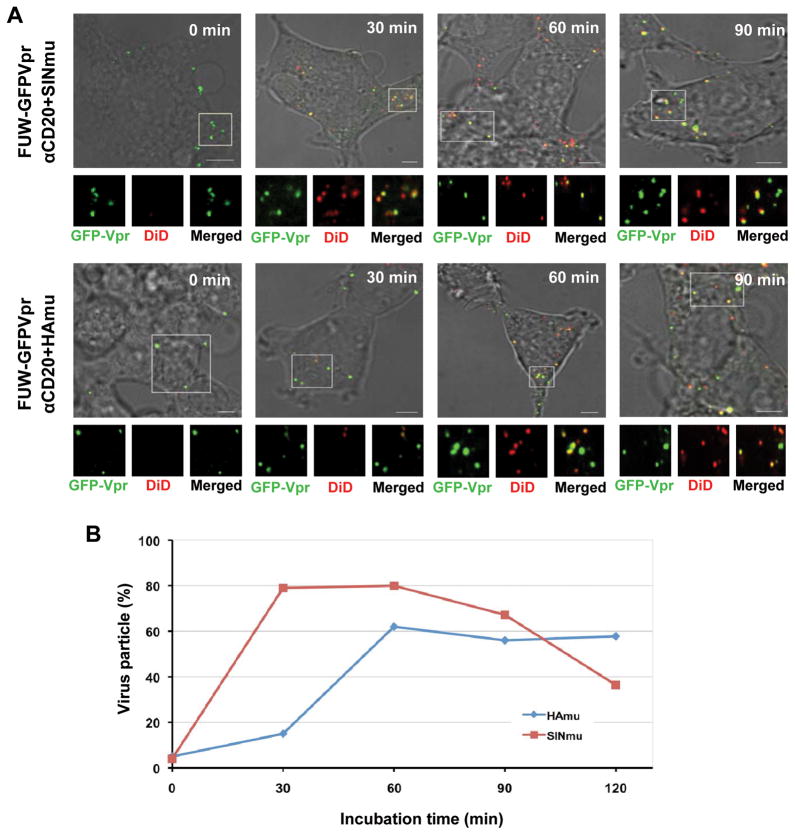
Visualization of virus-endosome fusion at different time points for engineered lentiviruses displaying SINmu or HAmu. (A) GFP-Vpr-labeled engineered lentiviruses (FUW-GFPVpr/αCD20+SINmu or FUW-GFPVpr/αCD20+HAmu; green) were labeled with DiD (red) for 1 h at room temperature. Double-labeled viruses were added to 293T/CD20 cells at 4°C for 30 min to synchronize the binding. The cells were then warmed to 37°C for 0, 30, 60, or 90 min, then fixed and imaged. The boxed regions are magnified and shown below in individual panels. Yellow signals indicate viral particles fused with endosomes. Scale bar represents 5 μm. (B) Quantification of the fused viral particles at different incubation time periods. GFP-Vpr+ viral particles with the fusion signal were quantified by viewing more than 100 viral particles at each time point. The viral particles positive for both GFP-Vpr and DiD were considered to be fused with endosomes, whereas particles that were only GFP-Vpr+ were considered to be unfused viruses. The results were collected from three independent experiments.
We quantified the endosomal fusion of the engineered viruses by viewing more than 100 viral particles for each time point and counting the GFP-Vpr-labeled viruses colocalized with or without the DiD signal (Fig. 3B). This quantification method showed that the virus-endosome fusion of SINmu-bearing viruses peaked at 30 min after incubation. This fusion event was maintained for up to 60 min of incubation, but decreased significantly between 90 and 120 min of incubation. The lower percentage of colocalization might be attributed to the release of viral particles from the endosomes. On the other hand, the fusion of HAmu-bearing viruses peaked at 60 min after incubation, and the colocalization of GFP-Vpr with the fusion signal (DiD) remained at the same level for up to 120 min of incubation. The results of this direct visualization of viral fusion indicated that fusion of SINmu-bearing viruses occurs faster than that of HAmu-bearing viruses. This quantification study also showed that the fusion yield at peak time points for SINmu-viruses (~ 80%) is higher than that of HAmu-viruses (~ 60%), and that the release of SINmu viral cores from fused endosomes into the cytosol seems to occur earlier than that of HAmu viral cores.
Expression of Dominant-negative Rab7 Inhibits the Transduction of HAmu-bearing Viruses
Viruses that enter cells in a pH-dependent manner are believed to fuse in either early or late endosomes for a successful infection. To determine which of these compartments are necessary for the productive transduction of engineered lentiviruses, the dominant-negative mutants of Rab proteins were used to disable either the early, Rab5, (Stenmark and others, 1994) or the late, Rab7, endosome function (Press and others, 1998). 293T/CD20 cells transfected with either the wild-type or dominant-negative form of Rab5 or Rab7 were incubated with FUGW/αCD20+SINmu, FUGW/αCD20+HAmu, FUGW/VSVG, or MIG/VSVG. Expression of the Rab5 dominant-negative mutant reduced the transduction rate of SINmu- or HAmu-bearing viruses by 60~70% as compared to the transduction of wild-type Rab5-expressing cells (Fig. 4A), suggesting that the engineered lentiviruses must be trafficked through early endosomes for a successful transduction. The transduction by VSVG-pseudotyped gamma-retrovirus (MIG/VSVG) was inhibited by the over-expression of dominant-negative Rab5 (Fig. 4A), consistent to the notion that VSVG-pseudotyped viruses are internalized into low-pH endosomes to infect target cells. However, over-expression of the Rab5 mutant did not result in significant inhibition to the transduction of 293T/CD20 cells by VSVG-pseudotyped lentivirus (Fig. 4A). This result is somewhat unexpected, although a similar observation was reported previously (Vidricaire and Tremblay, 2005).
Fig. 4.
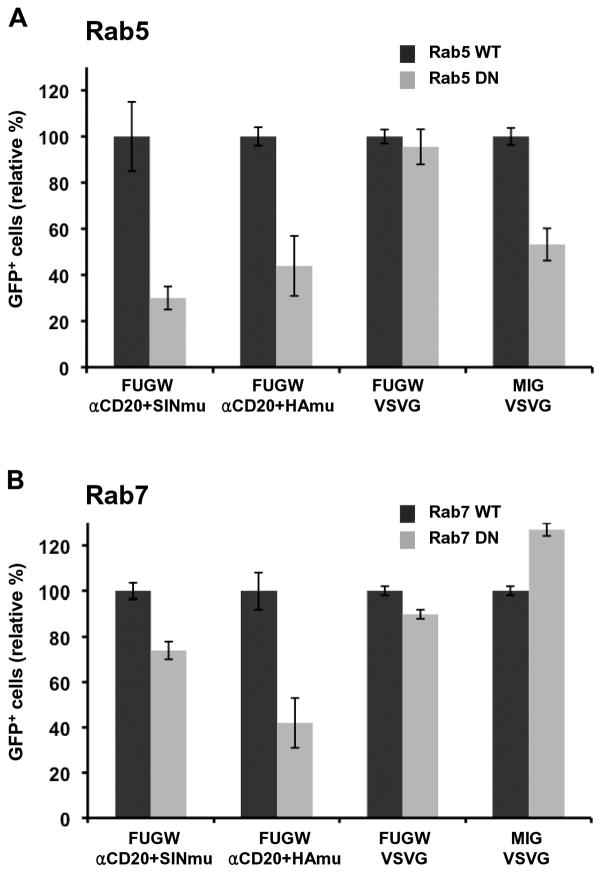
Inhibition of virus entry by Rab5 and Rab7 dominant-negative mutants. (A) 293T/CD20 cells transiently transfected with wild-type or dominant-negative mutant Rab5 were spin-infected with various lentiviruses (FUGW/αCD20+SINmu, FUGW/αCd20+HAmu, FUGW/VSVG, or MIG/VSVG). (B) 293T/CD20 cells transiently transfected with wild-type or dominant-negative mutant Rab7 were spin-infected with different viruses as indicated above. The percentage of GFP+ cells was analyzed by FACS.
As shown in Fig. 4B, expression of the dominant-negative Rab7 mutant resulted in significant transduction inhibition of HAmu-bearing viruses, indicating that the HAmu virus requires a functional late endosome for infection. Expression of mutant Rab7 also blocked transduction by SINmu-bearing viruses, although to a lesser degree than HAmu-bearing viruses, suggesting that infection by the SINmu virus is also associated with late endosomal trafficking. We found that the entry of VSVG-pseudotyped viruses (FUGW/VSVG or MIG/VSVG) was not affected by the expression of the dominant-negative Rab7 mutant, which is not too surprising because VSV is known to fuse in early endosomes (Sieczkarski and Whittaker, 2003).
Lentivirus Displaying the Fusion Protein HAmu, but not SINmu, Requires Functional Late Endosome Trafficking for Viral Fusion
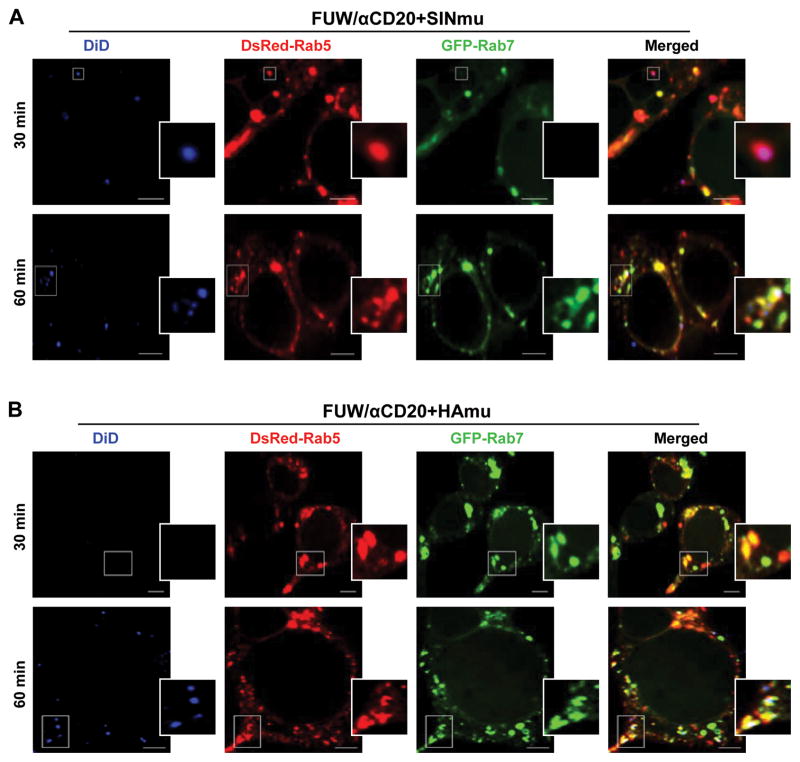
To further characterize the endosomal compartment where fusion of the engineered lentiviruses occurs, the individual fusion events were imaged with fluorescent protein-tagged endocytic structures (DsRed-Rab5 and GFP-Rab7) in target cells. We generated engineered lentiviruses lacking GFP-Vpr (FUW/αCD20+SINmu or FUW/αCD20+HAmu) for this study. DiD-labeled lentiviruses were incubated with 293T/CD20 cells transfected to express DsRed-Rab5 and GFP-Rab7 for 30 min at 4°C. The cells were then moved to 37°C for different time periods and analyzed by confocal microscopy (Fig. 5). The acquired images showed that at 30 min, 62% of the viral fusion signals (n=98) for FUW/αCD20+SINmu were detected in DsRed-Rab5+ organelles (Fig. 5A, 30 min), and after 60 min, 78% of the fusion signals (n=93) were observed in both DsRed-Rab5+ and GFP-Rab7+ organelles (Fig. 5A, 60 min). On the other hand, viral fusion of FUW/αCD20+HAmu was rarely detected at 30 min of incubation (Fig. 5B, 30 min) but after 60 min, 73% of the fusion signals (n=86) were observed in endosomes positive for both DsRed-Rab5 and GFP-Rab7 (Fig. 5B, 60 min). Based on these observations, we concluded that fusion of SINmu-bearing viruses takes place in early endosomes and the virus is further transported to intermediate endosomes containing both early and late endosome markers, whereas fusion of HAmu-bearing viruses does not occur until the viruses reach the intermediate endosomes.
Fig. 5.
Detection of viral fusion in DsRed-Rab5- and GFP-Rab7-expressing cells. DiD-labeled lentiviruses displaying SINmu (A) or HAmu (B) were incubated with 293T/CD20 cells transiently transfected with DsRed-Rab5 (red) and GFP-Rab7 (green) at 4°C for 30 min to synchronize the binding. The cells were then moved to 37°C for 30 or 60 min, fixed, and analyzed for the colocalization of the viral fusion signal (blue) with the different endosomal markers. The boxed regions are enlarged in the right panels. Scale bar represents 5 μm.
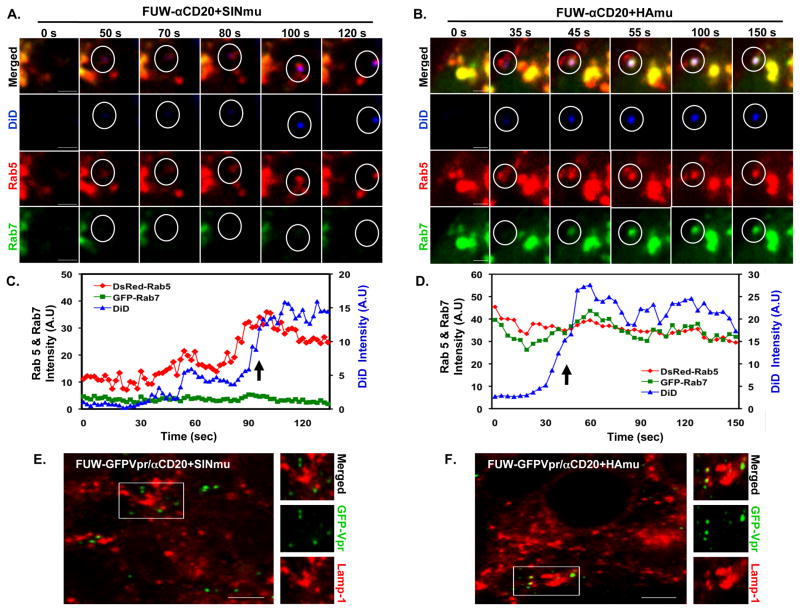
To further confirm our observations, viral fusion with Rab5- and Rab7-endosomes was monitored in real-time. DiD-labeled lentiviruses bearing SINmu were incubated with 293T/CD20 cells transfected with DsRed-Rab5 and GFP-Rab7 for 20 min at 37°C to allow for the initial internalization of the viruses, then live cell imaging was conducted using spinning-disk confocal microscopy. Selected images obtained from a time-series are shown in Fig. 6A. At 100 s, viral fusion with endosomes was identified by the dramatic increase in signals as the result of DiD dequenching. The images clearly show that the fusion signals (DiD) were colocalized with DsRed-Rab5+ endosomes but not with GFP-Rab7+ endosomes. The quantification of the fluorescent signals associated with fusion sites indicated that viral fusion was detected in endosomes positive for DsRed-Rab5, but not GFP-Rab7 (Fig. 6C), confirming that viral fusion of the SINmu-bearing virus occurs in early endosomes. In contrast, analysis of the HAmu-bearing virus imaged after 50 min of incubation at 37°C showed that viral fusion was detected in endosomes where both DsRed-Rab5 and GFP-Rab7 were present (Figs. 6B and 6D), supporting the conclusion that functional late endosomes are required for HAmu-bearing viral fusion. In addition, to determine the accumulation of viral particles in the lysosomal compartments, we monitored the colocalization of GFP-Vpr-labeled lentiviruses with lysosome associated membrane protein 1 (Lamp-1), a marker for lysosomes (Sieczkarski and Whittaker, 2003). After incubating the GFP-Vpr-labeled lentiviruses (FUW-GFPVpr/αCD20+SINmu or FUW-GFPVpr/αCD20+HAmu) with 293T/CD20 cells at 37°C for 4 h, the cells were immunostained against Lamp-1 (Figs. 6E and 6F). The images showed that no significant colocalization (< 5%, n=97) of SINmu-bearing viruses with lysosomes was observed (Figure 6E), whereas some viruses bearing HAmu (16.8%, n=103) were detected in lysosomes. This seemingly low colocalization (16.8%) could be significant if one considers that only portions of the degrading viruses in the lysosomes could be seen at a particular time point. The imaging results suggest that HAmu-bearing lentiviruses are more prone to degradation in the lysosomal compartments than SINmu-displaying lentiviruses.
Fig. 6.
Selected frames from the monitoring of viral fusion in DsRed-Rab5- and GFP-Rab7-expressing cells. DiD-labeled lentiviruses displaying SINmu (A) or HAmu (B) were added to 293T/CD20 cells transiently transfected with DsRed-Rab5 (red) and GFP-Rab7 (green) in the cold to synchronize the infection. Time series imaging was acquired 20 or 50 min after incubation at 37°C to initiate the internalization of SINmu- or HAmu-bearing lentiviruses, respectively. Scale bar represents 2 μm. (C, D) Fluorescent time trajectories of DiD (blue), DsRed-Rab5 (red), and GFP-Rab7 (green) signals associated with viral fusion (arrow) of SINmu-displaying lentivirus (FUW/αCD20+SINmu, C) or HAmu-displaying lentivirus (FUW/αCD20+HAmu, D) shown in A or B, respectively. (E, F) Viral degradation in the lysosomal compartments. GFP-Vpr-labeled viruses (green) enveloped by αCD20 and SINmu (E), or HAmu (F) were added to 293T/CD20 cells at 4°C for 30 min to synchronize the binding. The cells were then warmed to 37°C for 4 h, fixed, permeabilized, and immunostained with lysosome associated membrane protein 1 (Lamp-1; red). The boxed regions are enlarged in the right panels. Scale bar represents 5 μm.
Hemifusion Mediated by the SINmu Fusogen Occurs Faster than that by the HAmu Fusogen at Low pH
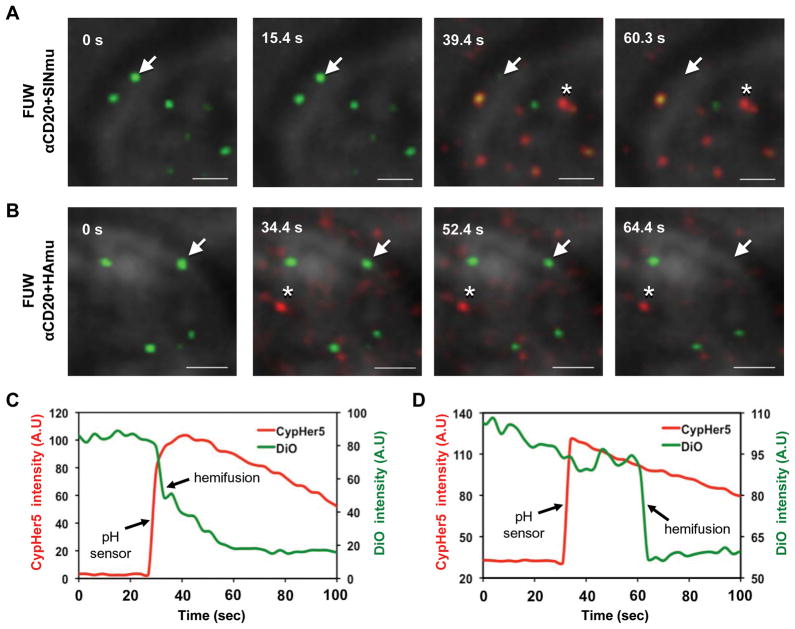
Membrane fusion proceeds through at least two steps: lipid mixing (i.e. hemifusion) and content mixing (i.e. fusion pore formation and/or enlargement) (Dimitrov, 2004; Harrison, 2008; Melikyan and others, 2005). Analysis of both lipid and content mixings between a single virus and its target membrane is useful for elucidating the molecular details of the membrane fusion process, but direct observation of such processes within endosomes is difficult due to the constant movement of the virus-endosome complex. To overcome this difficulty, we adhered viral particles to a planar substrate and placed the target cells above the viruses, then triggered virus-cell membrane fusion by lowering the pH environment. This setup allowed us to monitor both lipid- and content-mixing processes within the same focal plane (Markosyan and others, 2005; Melikyan and others, 2005). With this experimental setup, we compared the kinetics of hemifusion mediated by HAmu and SINmu fusion proteins after exposure to the optimal low pH at which influenza (pH 5.0) and Sindbis viruses (pH 5.5) are known to be able to fuse with cells (Chernomordik and others, 1998; Düzgüneş and others, 1992; Smit and others, 1999; White and others, 1982; Zaitseva and others, 2005). The kinetics of hemifusion (lipid mixing) was determined by observing the decrease of fluorescence signals of the lipophilic carbocyanine dye (DiO) incorporated into the viral membrane at a low concentration (below the self-quenching level). For the precise measurement of the time at which the environmental pH drop occurred, we included virions labeled with a pH sensitive dye, CypHer5, as a pH sensor. The mixture of DiO-labeled viruses and CypHer5-labeled viruses (at the particle number ratio of 10:1) was adhered to the poly-lysine coated coverslip and 293T/CD20 cells were overlaid on the viruses for 30 min. Virus-cell fusion was triggered by adding the appropriate volume of 0.2 M acetic acid, pre-titrated to provide the desired pH, and monitored by pH drop (CypHer5, red) and lipid transfer (DiO, green) (Figs. 7A and 7B). There was no observable lipid transfer at neutral pH (data not shown). Lipid mixing, indicated by the disappearance of the green signal, for viruses bearing SINmu occurred quickly upon exposure to low pH, as seen by the sudden increase in CypHer5 fluorescence (Figs. 7A and 7C), whereas HAmu-bearing viruses exhibited a longer lag time (~30 sec) between pH drop and lipid mixing (Figs. 7B and 7D). Thus, these results indicate that the hemifusion of lentiviruses with SINmu is faster than that of HAmu-bearing virus.
Fig. 7.
Detection of hemifusion between viruses and target cells with a pH sensor. DiO-labeled (green) and CypHer5-labeled (red) viruses (mixing ratio 10:1) bearing SINmu (A) or HAmu (B) were prebound to 293T/CD20 cells for 30 min at room temperature. During live-cell imaging, virus-cell fusion was triggered by adding the appropriate volume of acidic buffer, pre-titrated to provide the desired pH (HAmu: pH 5.0, SINmu: pH 5.5), and monitored by pH drop (asterisk) and lipid transfer (arrow). Scale bar represents 2 μm. (C, D) The fluorescent intensity of the pH-sensitive CypHer5 (red) and the hemifusion (green) signals associated with the viral particles bearing SINmu (C) or HAmu (D), which are indicated by the asterisk and the arrow in A or B, respectively.
Fusion Pore Formation Mediated by SINmu, but not by HAmu, is Delayed after Lipid Mixing
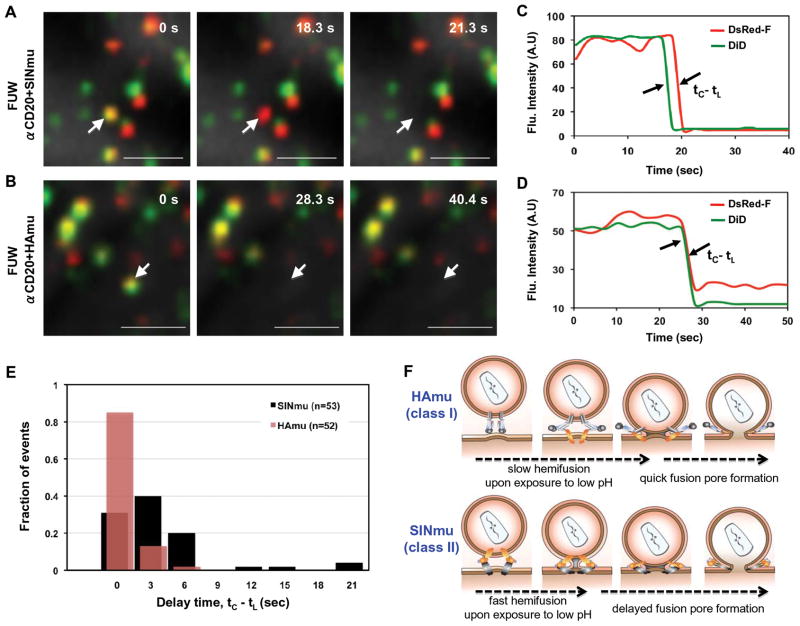
For a more detailed analysis of hemifusion and fusion pore formation catalyzed by Hamu and SINmu fusion proteins, viral particles were double-labeled with lipophilic dye (DiD), for the detection of hemifusion, and DsRed-F (DeRed fused at the N-terminus with the farnesylation sequence of p21 (Ras) (Harvey and others, 2001)), which could be incorporated into the inner leaflet of the viral membrane, for the detection of fusion pore formation. This labeling scheme allowed for the analysis of both lipid- and content-mixing trajectories of single viruses within target cells (Campbell and others, 2007). The kinetics between hemifusion and fusion pore formation were measured by observing the loss in DiD and DsRed-F signals as the result of lipid- and content-mixing, respectively. DiD/DsRed-F-labeled viruses were adhered to a poly-lysine-coated coverslip and were then prebound to 293T/CD20 cells for 30 min. Colocalization analysis of the viral particles showed that 40~60% of the DsRed-F-labeled particles contained DiD. Virus-cell fusion was then triggered by low pH (HAmu: pH 5.0, SINmu: pH 5.5) and monitored by lipid (DiD, green) and content (DsRed-F, red) mixing (Figs. 8A and 8B). Interestingly, it was observed that the decay of the DsRed-F signals on SINmu-bearing viruses was usually not instantaneous after the transfer of DiD (Figs. 8A and 8C), while the loss of the DiD and the DsRed-F signals on HAmu-bearing viruses usually occurred simultaneously (Figs. 8C and 8D). Quantitative analysis of the delay times between lipid mixing (tL) and content mixing (tC) for single viruses showed that ~60% of the DsRed-F transfer events mediated by SINmu (n= 53) were delayed around 3 to 6 sec after the DiD transfer (resolution, 3 sec/frame), whereas ~85% of the DsRed-F transfer events mediated by HAmu (n= 52) were detected simultaneously (in the same frame) with the DiD transfer (Fig. 8E). These results suggest that the hemifusion intermediate (i.e. lipid stalk, before fusion pore formation) mediated by SINmu is maintained relatively longer than that induced by HAmu (Fig. 8F).
Fig. 8.
Hemifusion and fusion pore formation between viruses and target cells. DiD/DsRed-F-labeled viruses displaying SINmu (A) or HAmu (B) were prebound to 293T/CD20 cells for 30 min at room temperature, and then virus-cell fusion was triggered by adding the appropriate volume of acidic buffer, pre-titrated to provide the desired pH (HAmu: pH 5.0, SINmu: pH 5.5). Lipid (DiD, green) and content (DsRed-F, red) mixing was monitored, as indicated by the disappearance of signals. Yellow signals indicate viral particles labeled by both DiD and DsRed-F. Viral particles that contain either DiD or DsRed-F are shown in green or red, respectively. The viral particles that transferred the DiD and DsRed-F signals are indicated by arrows in A and B. Scale bar represents 2 μm. (C, D) The fluorescent intensity of DiD (green) and DsRed-F (red) signals associated with the viral particles bearing SINmu (C) and HAmu (D), as indicated by the arrows in A or B, respectively. (E) Distribution of the delay times between lipid mixing (tL) and content mixing (tC) of individual SINmu (black bar, n=53) or HAmu (red bar, n=52) particles. The time intervals (tC-tL) between DiD and DsRed-F signal disappearance were measured as shown in C and D. (F) A proposed sequence of events in membrane fusion driven by HAmu and SINmu fusion proteins.
DISCUSSION
The goal of this study is to utilize a direct visualization technique via confocal microscopy to compare the intracellular trafficking of lentiviruses enveloped with fusogen derived from the influenza virus glycoprotein (HAmu) or the Sindbis virus glycoprotein (SINmu), along with αCD20. The entry mechanism of this engineered virus is hypothesized to begin with the binding of αCD20 to its cognate CD20 antigen on the target cell surface, and then the virus is internalized into the acidic endosomes where the surface-displayed fusogen triggers membrane fusion.
We investigated the early entry stage of these engineered lentiviruses by visualizing the interactions of single viral particles with endocytic structures in living cells. This study clearly showed that engineered lentiviruses containing either SINmu or HAmu enter cells through pH-dependent and clathrin-mediated endocytosis, which was also confirmed by the drug inhibition study. It was perhaps not surprising that we observed that both types of engineered lentiviruses exploited the same entry route because they contained the same binding molecule, αCD20, which likely determined the early stage of virus entry.
The actual fusion event of the engineered lentiviruses was detected in target cells by a fluorescence-dequenching assay. This experiment revealed that viral fusion of the SINmu-bearing virus occurred faster and exhibited relatively higher fusion efficiency than that of the HAmu-bearing virus. In addition, viral transduction to cells overexpressing the dominant-negative mutants of Rab proteins showed that the HAmu-displaying virus required functional late endosome trafficking for productive infection. This was further confirmed by real-time imaging of viral fusion, which showed that the HAmu-bearing virus had to travel at least to intermediate endosomes containing both early and late endosomal markers to initiate fusion. In contrast, viral fusion of the SINmu-displaying virus started at the early endosome stage, which is consistent with our previous observation (Joo and Wang, 2008). In addition, the involvement of the late endosomal compartments in trafficking the SINmu virus (Figs. 4B and 5B, 60min) is in line with our previous observation, in which the release of the core of the engineered SINmu lentivirus was associated with the maturing process of the early to the intermediate endosomes (Joo and Wang, 2008). Additionally, the colocalization assay with lysosomal markers suggests that HAmu-bearing viruses seem to be more prone to degradation in the lysosomes than SINmu-displaying viruses.
Many studies have proposed that influenza and Sindbis virus glycoproteins undergo a conformational change upon exposure to the low pH within endosomes, which exposes the fusion peptide or loop(s) and allows it to interact with the target membrane to cause hemfusion (Dimitrov, 2004; Harrison, 2008). By a real-time imaging method along with a pH sensor, we were able to demonstrate the precise kinetics of the hemifusion of single viruses upon exposure to the optimal low-pH environment at which influenza and Sindbis viruses are known to be able to fuse with cells (Chernomordik and others, 1998; Düzgüneş and others, 1992; Smit and others, 1999; White and others, 1982; Zaitseva and others, 2005). Our results indicated that hemifusion mediated by SINmu in response to low pH is faster than that induced by HAmu.
A method of dual labeling the outer and inner leaflets of single virus membranes enabled us to sequentially detect hemifusion (lipid mixing) and fusion pore formation (content mixing) between individual viruses bearing HAmu or SINmu fusion proteins and target cells. Although the kinetics of hemifusion measured with the immobilized viruses on the planar substrate may differ from virus fusion with the limited membrane of endosomes, the observation of sequential multiple steps in membrane fusion at the single virus level could provide useful information for understanding the underlying fusion behaviors mediated by different fusion proteins in a detailed manner. We observed that more than 60% of the fusion pore formation events mediated by SINmu were remarkably delayed after hemifusion, while ~85% of the HAmu-mediated fusion pores were formed almost simultaneously with hemifusion, indicating that SINmu-mediated hemifusion intermediate is relatively long-lived compared with that of HAmu (Fig. 8F). This imaging result might provide an explanation for the fusion process leakiness observed by an experimental model of liposome-based virus fusion, which suggested that the influenza virus-mediated fusion process was quite leaky (Shangguan and others, 1996), whereas the fusion process mediated by alphaviruses is non-leaky (Smit and others, 2002). The long-lived hemifusion intermediate caused by SINmu, in which the proximal (outer) leaflets of the interacting membranes have exchanged their lipid components while the distal (inner) leaflets remain unopened, can yield a non-leaky fusion process. On the other hand, the occurrence of the very transient hemifusion intermediate induced by HAmu suggests that the interactions between the outer and inner leaflets of the two distinct membranes are nearly simultaneous, resulting in unstable contact of the two lipid bilayers, thereby creating a leaky fusion process. One possible explanation for the marked delay between lipid mixing (hemifusion) and content mixing (fusion pore formation) observed in SINmu-virus fusion could be that there is a requirement for membrane components such as cholesterol and sphigolipids in the target membrane for alphavirus fusion (Phalen and Kielian, 1991; Smit and others, 1999), but not for influenza virus fusion (Sieczkarski and Whittaker, 2002). Some studies have suggested that cholesterol and sphingolipids are needed for the formation of fusion pores, which supports our explanation (Nieva and others, 1994; Smit and others, 1999; White and Helenius, 1980). We are currently conducting experiments to investigate the exact effects of these membrane lipid components on the formation of the hemifusion intermediate upon virus fusion.
The recent study by Floyd et al. has developed a two-color fluorescent assay to monitor kinetics of single influenza virus particles fusing with a target planar bilayer and suggested a delay between hemifusion and pore formation for influenza HA. The reason for this difference is likely due to a different labeling method employed to detect fusion pore formation (Floyd and others, 2008); the pore formation was monitored by the loss of sulforhodamine B signal (viral interior labeling), resulted from the dye-diffusion into the fluid support of the bilayer after the fusion pore open, that may be kinetically slower than the loss of the dye incorporated into the inner leaflet of the viral membrane. The different experimental set-up for the viral fusion assay (e.g. target membranes) could also be another cause for the difference in kinetics of hemifusion and/or fusion pore formation.
To summarize, our findings suggest that engineered lentiviruses enveloped with either HAmu or SINmu fusion proteins have different requirements of endosomal trafficking for viral fusion and exhibit distinct kinetics of multistep processes in membrane fusion, which is likely correlated with the efficiency of virus infection. In addition, we have demonstrated that this imaging approach can provide a useful technological platform to dissect the mechanisms of viral membrane fusion, which can also be beneficial to the design of more efficient gene delivery vectors.
Acknowledgments
We thank Lili Yang for critical reading of the manuscript and the USC Norris Center Cell and Tissue Imaging Core. This work was supported by a National Institute of Health Grant.
LITERATURE CITED
- Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80(14):6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R, Sarkar D, Durell S, Howard D, Morris S. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman E, Siebers A, Altendort K. Bafilomycins: a class of inhibitors fo membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes AP, Griffin DE. Binding of sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360:286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Zimmerberg J, Kozlov MM. Membrane of the world unite! J Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daecke J, Fackler OT, Dittmar MT, Krausslich HG. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol. 2005;79(3):1581–1594. doi: 10.1128/JVI.79.3.1581-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTulleo LT, Kirchhausen T. The clathrin endocytic pathway in viral infection. EMBO J. 1998;17:4585–4593. doi: 10.1093/emboj/17.16.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov DS. Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Brodsky FM, Blank GS, Helenius A. Inhibition of endocytosis by anti-clathrin antibodies. Cell. 1987;50:453–463. doi: 10.1016/0092-8674(87)90499-5. [DOI] [PubMed] [Google Scholar]
- Düzgüneş N, Lima MCPd, Stamatatos L, Flasher D, Alford D, Friend DS, Nir S. Fusion activity and inactivation of influenza virus: kinetics of low pH-induced fusion with cultured cells. J Gen Virol. 1992;73:27–37. doi: 10.1099/0022-1317-73-1-27. [DOI] [PubMed] [Google Scholar]
- Floyd DL, Ragains JR, Skehel JJ, Harrison SC, Oijen AMv. Single-particle kinetics of influenza virus membrane fusion. Proc Natl Acad Sci USA. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KJ, Lukovic D, Ucker DS. Membrane-targeted green fluorescent protein reliably and uniquely marks cells through apoptotic death. Cytometry. 2001;43:273–278. doi: 10.1002/1097-0320(20010401)43:4<273::aid-cyto1059>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB, Chapman ER. The fusion pores of Ca2+-triggered exocytosis. Nat Struct Mol Biol. 2008;15:684–689. doi: 10.1038/nsmb.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Joo KI, Lei Y, Lee C-L, Lo J, Xie J, Hamm-Alvarez SF, Wang P. Site-specific labeling of enveloped viruses with quantum dots for single virus tracking. ACS nano. 2008;2:1553–1562. doi: 10.1021/nn8002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KI, Wang P. Visualization of targeted transduction by engineered lentiviral vectors. Gene Ther. 2008;15:1384–1396. doi: 10.1038/gt.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Zhao Y, Anderson WF. Receptor-mediated Moloney murine leukemia virus entry can occur independently of the clathrin-coated-pit-mediated endocytic pathway. J Virol. 1999;73(7):5994–6005. doi: 10.1128/jvi.73.7.5994-6005.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz BR, Malinin V, Haque ME, Evans K. Protein machines and lipid assemblies: current views of cell membrane fusion. Curr Opin Struct Biol. 2000;10:607–615. doi: 10.1016/s0959-440x(00)00138-x. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159(3):441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan RM, Cohen FS, Melikyan GB. Time-resolved imaging of HIV-1 env-mediated lipid and content mixing between a single virion and cell membrane. Mol Biol Cell. 2005;16(5502–5513) doi: 10.1091/mbc.E05-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, Barnard RJO, Abrahamyan LG, Mothes W, Young JAT. Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc Natl Acad Sci USA. 2005;102:8728–8733. doi: 10.1073/pnas.0501864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A, Leikina E, Chernomordik LV, Bentz J. Kinetically differentiating influenza hemagglutinin fusion and hemifusion machines. Biophys J. 2003;85:1713–1724. doi: 10.1016/S0006-3495(03)74601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JAT. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- Nabi IR, Le PU. Caveolae/raft-dependent endocytosis. J Cell Biol. 2003;161:673–677. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Lippincott-Schwartz J. Endocytosis without clathrin coats. Trends Cell Biol. 2001;11:406–412. doi: 10.1016/s0962-8924(01)02107-9. [DOI] [PubMed] [Google Scholar]
- Nieva JL, Bron R, Corver J, Wilschut J. Membrane fusion of Semliki Forest virus requires sphingllipids in the target membrane. EMBO J. 1994;13:277–2804. doi: 10.1002/j.1460-2075.1994.tb06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi PA, Fishman PH. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J Cell Biol. 1998;141(4):905–915. doi: 10.1083/jcb.141.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11(6):567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Ohuchi M, Imai M, Mizuno T, Kawasaki K, Kuroda K, Yamashina S. Dual wavelength imaging allows analysis of membrane fusion of influenza virus inside cells. J Virol. 2006;80:2013–2018. doi: 10.1128/JVI.80.4.2013-2018.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan T, Alford D, Bentz J. Influenza virus liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry. 1996;35:4956–4965. doi: 10.1021/bi9526903. [DOI] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76:10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Differential requirements of rab5 and rab7 for endocytosis of influenza and other enveloped viruses. Traffic. 2003;4:333–343. doi: 10.1034/j.1600-0854.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Smit JM, Bittman R, Wilschut J. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol. 1999;73:8476–8484. doi: 10.1128/jvi.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JM, Bittman R, Wilschut J. Deacylation of the transmembrane domains of Sindbis virus envelope glycoproteins E1 and E2 does not affect low-pH-induced viral membrane fusion activity. FEBS Lett. 2001;2001:57–61. doi: 10.1016/s0014-5793(01)02495-4. [DOI] [PubMed] [Google Scholar]
- Smit JM, Li G, Schoen P, Corver J, Bittman R, Lin K-C, Wilschut J. Fusion of alphaviruses with liposomes is a non-leaky process. FEBS Lett. 2002;521:62–66. doi: 10.1016/s0014-5793(02)02823-5. [DOI] [PubMed] [Google Scholar]
- Stegmann TM, HWM, Scholma J, Wilschut J. Fusion of influenza virus in an intracellular acidic compartment measured by fluorescence dequenching. Biochem Biophys Acta. 1987;904:165–170. doi: 10.1016/0005-2736(87)90100-3. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Grunberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endosomes. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Yau VK, Briggs BJ, Whittaker GR. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338(1):53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Vidricaire G, Tremblay MJ. Rab5 and Rab7, but not ARF6, govern the early events of HIV-1 infection in polarized human placental cells. J Immunol. 2005;175:6517–6530. doi: 10.4049/jimmunol.175.10.6517. [DOI] [PubMed] [Google Scholar]
- Vonderheit A, Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of semliki forest virus to late endosomes. PLos Bio. 2005;3:1225–1238. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123(5):1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Helenius A. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci USA. 1980;77:3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Kartenbeck J, Helenius A. Membrane fusion activity of influenza virus. EMBO J. 1982;1:217–222. doi: 10.1002/j.1460-2075.1982.tb01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Baiely L, Baltimore D, Wang P. Targeting lentiviral vectors to specific cell types in vivo. Proc Natl Acad Sci USA. 2006;103:11479–11484. doi: 10.1073/pnas.0604993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Ohnishi S. Uncoating of influenza virus in endosomes. J Virol. 1984;51:497–504. doi: 10.1128/jvi.51.2.497-504.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva E, Mittal A, Grifın DE, Chernomordik LV. Class II fusion protein of alphaviruses drives membrane fusion through the same pathway as class I proteins. J Cell Biol. 2005;169:167–177. doi: 10.1083/jcb.200412059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Blumenthal R, Sarkar DP, Curran M, Morris SJ. Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J Cell Biol. 1994;127:1885–1894. doi: 10.1083/jcb.127.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]