Abstract
The human complement system plays an essential role in innate and adaptive immunity by marking and eliminating microbial intruders. Activation of complement on foreign surfaces results in proteolytic cleavage of complement component 3 (C3) into the potent opsonin C3b, which triggers a variety of immune responses and participates in a self-amplification loop mediated by a multi-protein assembly known as the C3 convertase. The human pathogen Staphylococcus aureus has evolved a sophisticated and potent complement evasion strategy, which is predicated upon an arsenal of potent inhibitory proteins. One of these, the Staphylococcal Complement INhibitor (SCIN), acts at the level of the C3 convertase (C3bBb) and impairs downstream complement function by trapping the convertase in a stable but inactive state. Previously, we have shown that SCIN binds C3b directly and competitively inhibits binding of human factor H, and to a lesser degree that of factor B to C3b. Here, we report the co-crystal structures of SCIN bound to C3b and C3c at 7.5 and 3.5 Å limiting resolution, respectively, and show that SCIN binds a critical functional area on C3b. Most significantly, the SCIN binding site sterically occludes the binding sites of both fH and fB. Our results give insight into SCIN binding to activated derivatives of C3, explain how SCIN can recognize C3b in the absence of other complement components, and provide a structural basis for the competitive C3b-binding properties of SCIN. In the future, this may suggest templates for the design of novel complement inhibitors based upon the SCIN structure.
Keywords: Complement, C3 convertase, Immune evasion, Staphylococcus aureus, complement inhibitor
The complement cascade is a centerpiece of the immune system and functions not only as the first line of response to foreign materials in the body, but also in the initiation of inflammation and stimulation of adaptive immunity. This versatility of complement is based on a highly regulated network of soluble or cell-surface-bound proteins that either serve as substrates, enzymes, or regulators of a hierarchical series of proteolytic and protein-protein recognition events. Initiation of the cascade may be triggered by three distinct pattern recognition mechanisms, which have been denoted the classical, lectin, and alternative pathways. Despite the precise nature of the initiating event, all three of these pathways lead to the formation of homologous multi-protein complexes (named C3 convertases) that enzymatically-cleave complement component C3 (184 kDa) into its active fragments C3a (10 kDa) and C3b (175 kDa) (Reviewed in 1). Release of the C3a anaphylatoxin fragment exposes an occluded, but reactive thioester moiety in C3b. It is the nucleophilic attack of this group which results in covalent deposition of the C3b on the surface of nearby biomaterial and ultimately triggers its phagocytosis via specific complement receptor-expressing cells 2; 3.
Structural studies have shown that the conversion from C3 to C3b is accompanied by substantial conformational changes, which themselves expose a number of critical ligand binding sites 4; 5; 6. A particularly prominent example of this is typified by the alternative pathway, where interaction of the pro-enzyme factor B with one of these sites enables its cleavage by factor D (fD) to generate the active alternative pathway C3 convertase complex (C3bBb). Subsequent binding and cleavage of C3 by the convertase produces additional C3b and thereby propagates a self-amplification loop of complement activation on foreign surfaces 7. Unhindered complement activation in this manner leads to the formation of C5 convertases, generation of additional anaphylatoxins (i.e. C3a and C5a), induction of signaling, chemotaxis, and phagocytosis, and assembly of a lytic membrane attack complex. However, the convertase assemblies by themselves are only transiently stabile and undergo irreversible dissociation with a half-life of approximately one minute8. Moreover, on host cells this dissociation is further catalyzed by a series of “regulators of complement activation” (RCA) that either destabilize the convertases or mediate the stepwise degradation of the essential C3b substituent (Reviewed in 1; 9). One pivotal regulator of this activation cycle is factor H (fH), which both accelerates the decay of the convertase complex and act as a cofactor for the factor I (fI)-mediated proteolysis of C3b that produces the inactive C3c fragment 2. Given this tenuous balance that leads to the “double-edged sword” nature of the immune system, many pathological conditions are now known to arise from insufficient regulation or aberrant activation of complement 10. Therefore, inhibition of complement initiation or amplification has been suggested as an attractive means of protection against these diseases 11.
Staphylococcus aureus is a widespread and verstaile human pathogen capable of colonizing a remarkable number of different tissues 12. Diseases caused by S. aureus span a broad landscape of relatively minor, localized skin infections to more life-threatening conditions such as disseminated bacteremia and endocarditis 12. S. aureus cells activate all three complement pathways and stimulate chemotaxis of leukocytes due to the presence of pathogen associated molecular patterns. However, it has become increasingly clear over the last decade that this pathogen has evolved an array of successful strategies to evade the surveillance and elimination mechanisms that are fundamental to innate immune system function 13; 14. In this regard, S. aureus secretes nearly one dozen factors which bind to and interfere with the various steps needed to activate C3 and to propagate various inflammatory responses within its host 14; 15. Subsequent structure/function analyses on several of these proteins have demonstrated that they are diverse in their mechanisms of action and, in some cases, has identified entirely novel modes of complement modulation 14; 15.
The Staphylococcal Complement Inhibitors (SCINs) are a family of small (~9.8 kDa), secreted proteins that adopt an anti-parallel three α-helical bundle fold 16, variants of which appear to be common among a larger group of Staphylococcal immune modulators 14; 15; 16. The prototypic member of this family, SCIN (also called SCIN-A), is a human-specific complement inhibitor that disrupts bacterial opsonization as well as associated downstream effects of complement activation by blocking function of the solid-phase C3 convertases derived from all three initiation pathways 17; 18. Previous studies originally suggested that SCIN binds only to fully-assembled C3 convertases (e.g. C3bBb) 17; however, this raised significant questions as to the nature of this interaction because all other characterized C3 convertase regulators or inhibitors bind to one or more of the complement components directly. Indeed, while SCIN does not bind native C3, we have recently shown that SCIN binds directly to activated C3b with an apparent nanomolar affinity in near physiological buffers (KD≈180 nM) 19. Functional dissection of the direct interaction between SCIN and C3b has revealed a multi-faceted nature to the SCIN inhibitory mechanism. First, while SCIN impairs the initial rate of C3 convertase assembly, it nevertheless stabilizes the assembled convertase against decay-acceleration by factor H (fH). Secondly, SCIN reduces the efficiency of factor I- and H-mediated degradation of C3b to iC3b. Finally, SCIN also prevents the proteolytic cleavage, but not the initial binding of the C3 substrate by the convertase 19.
In this manner, SCIN contributes to Staphylococcal complement inhibition by trapping the C3 convertase in a stabilized yet inactive state 19; 20. Taking advantage of these properties, Rooijakkers et al. recently described the structure of the SCIN-inhibited alternative pathway C3 convertase20. While this work gave important insights into the molecular architecture and function of the alternative pathway C3 convertase, it provided no conclusive structural information regarding the direct interaction between C3b and SCIN. Because of this, the precise nature of the SCIN binding site on C3b and C3c, its relationship to native C3, and the structural basis for several important effects of SCIN on C3 convertase assembly and dynamics remained incompletely described. Here, we report the crystal structures of SCIN bound to the C3 derivatives C3b and C3c. Analysis of these structures provides a molecular framework to understand SCIN inhibition of C3b binding to fH and factor B (fB), and demonstrates the structural basis for specific recognition of activated C3 fragments by SCIN.
Structures of SCIN bound to human complement fragments C3b and C3c
To gain further insight into molecular mechanisms of SCIN activity, we pursued crystal structures of SCIN bound to complement fragments C3b and C3c. Binary complexes of SCIN bound to C3b and C3c were reconstituted by incubation of purified components followed by concentration by ultrafiltration 21. The presence of each protein component in the resulting samples was verified by both native and SDS-polyacrylamide gel electrophoresis 21 prior to crystallization of both complexes by vapor-diffusion of hanging drops. Three distinct C3c-SCIN crystal forms were identified which diffracted X-rays to between 3.4 and 4.1 Å limiting resolution, while crystals of C3b-SCIN provided reasonably complete data to 7.5 Å resolution (Table 1). Initial phases were obtained by molecular replacement, and models for each crystal form were iteratively improved and refined to the values reported in Table 1.
Table 1.
Diffraction Data Collection and Refinement Statistics for the C3c-SCIN and C3b-SCIN Complexesa
| C3c-SCIN | C3b-SCINb | |||
|---|---|---|---|---|
| Data collection | ||||
| Beamline | 22-BM | 22-ID | 22-BM | 22-ID |
| Wavelength (Å) | 1.00 | 1.00 | 1.00 | 1.00 |
| Space group | P21212 | P21212 | P21 | P41212 |
| Cell dimensions | ||||
| a,b,c (Å) | 231.16, 231.50, 68.89 | 214.07, 68.43, 114.93 | 69.11, 217.03, 115.65 | 128.31, 128.31, 469.76 |
| β (°) | 89.99 | |||
| Resolution (Å) | 50-3.5 | 50-4.1 | 50-3.4 | 50-7.5 |
| Unique Reflections | 44,393 | 13,684 | 45,397 | 5,337 |
| Completeness (%) | 93.4 (85.1) | 98.4 (99.0) | 97.5 (93.1) | 97.2 (81.4) |
| Rmerge (%)c | 12.1 (46.6) | 18.1 (49.6) | 16.0 (50.0) | 11.6 (45.3) |
| Redundancy | 7.4 (3.7) | 5.4 (5.1) | 3.4 (2.8) | 10.4 (7.8) |
| I/σI | 10.5 (2.4) | 8.7 (4.1) | 5.0 (2.4) | 13.2 (3.0) |
| Refinement Statistics | ||||
| RCSB Accession Code | 3NSA | 3NMS | 3L3O | 3L5N |
| Complexes per A.U. | 2 | 1 | 2 | 1 |
| Rwork/Rfree (%)d | 26.6/28.8 | 27.1/28.9 | 22.3/27.1 | 26.5/26.8 |
| Mean B-factors (Å2) | 68.6 | 121.2 | 108.1 | 172.3 |
| RMSD | ||||
| bond lengths (Å) | 0.012 | 0.012 | 0.009 | 0.008 |
| bond angles (°) | 1.31 | 1.37 | 1.20 | 1.14 |
| Ramachandran plot (%) | ||||
| Favored | 85.9 | 86.7 | 88.9 | 85.7 |
| Allowed | 11.5 | 11.1 | 10.4 | 12.7 |
| Outlier | 2.6 | 2.2 | 0.8 | 1.6 |
Numbers in parentheses are for the highest-resolution shell.
Crystallization and data collection on the C3b-SCIN crystals has been published separately 21.
Rmerge = ΣhΣi|Ii(h)−<I(h)>|/ΣhΣiIi(h), where Ii(h) is the ith measurement of reflection h and <I(h)> is a weighted mean of all measurements of h.
R = Σh|Fobs(h)−Fcalc(h)|/Σh|Fobs|.
Rcryst and Rfree were calculated from the working and test reflection sets, respectively. The test set constituted 5% of the total reflections not used in refinement.
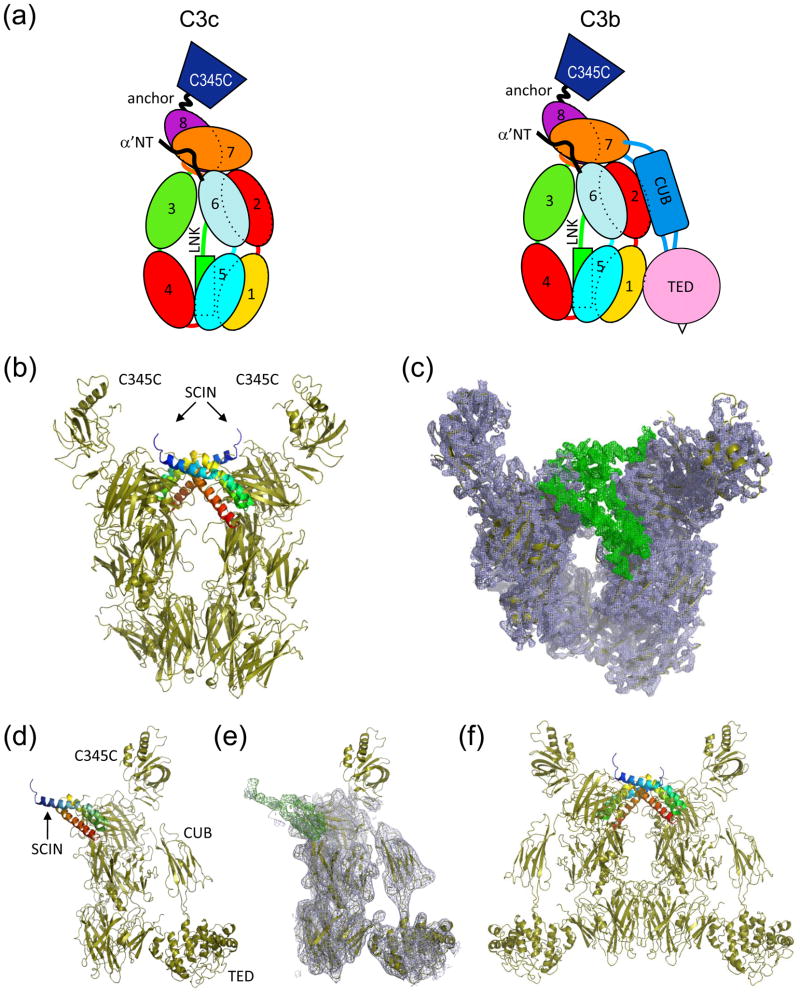
The C3c-SCIN structure is comprised of a heterotetramer, where two copies of the C3c-SCIN complex are aligned with, but rotated 180° about the long axis of the complement component (Figure 1a, b). This arrangement is observed in all three C3c-SCIN structures, even though the heterotetramer is divided between neighboring asymmetric units in the second orthorhobmic crystal form (Supplemental Figure 1, Table 1, and Supplemental Table 1). Similarly, though the asymmetric unit of the C3b-SCIN structure is defined by a single copy of each protein arranged as a heterodimer (Figure 1c, d), inspection of nearby symmetry-related complexes likewise indicates that a similar mode of tetramerization to that of C3c-SCIN is present within the C3b-SCIN crystal lattice (Figure 1e). In fact, superposition of the symmetry-related C3b-SCIN tetramer with that of C3c-SCIN indicates that these two structures share a high level of identity to one another.
Figure 1. Crystal structures of complement fragments C3c and C3b bound to SCIN.
(a) Schematic representation of the domains which comprise C3c and C3b. (b) Crystal structure of C3c-SCIN (drawn from RCSB code 3NSA), where two copies of SCIN (blue to red rainbow from amino to carboxy terminus) and two copies of C3c (sand) are found in the asymmetric unit. (c) Electron density map of the same structure generated by refinement of models not containing SCIN molecules. 2Fo−Fc density (blue cage; 1.5σ contour) fits two copies of C3c, while the Fo−Fc map (green cage; 2.0σ contour) shows clear positive density corresponding to the location of the SCIN polypeptide. Note that the viewing plane of this panel is inclined slightly relative to panel b for clarity. (d) Crystal structure of C3b-SCIN, where a single copy of both SCIN (blue to red rainbow from amino to carboxy terminus) and C3b (sand) are present in the asymmetric unit. (e) Electron density map of the same structure generated by refinement of the model without the SCIN polypeptide. Map parameters are identical to those in panel c. (f) Generation of the symmetry mate (x,y,−z) from the C3b-SCIN crystal indicates a mode of tetramerization similar to that seen in all C3c-SCIN structures (please see Supplemental Figure 1).
With the exception of the C345C domains, readily interpretable density is observed in 2Fo−Fc electron density maps (contoured at 1.5σ) for all functional domains of C3c and C3b present in the final models of all structures (Figure 1b, d). In all C3c-SCIN structures, the model versus map correlation is somewhat weaker for both copies of the C345C domain; however, these areas are clearest for orthorhombic form 1, and so this crystal is discussed in the remainder of the text unless otherwise indicated. In the case of C3b-SCIN, little contiguous density is observed in this region, therefore in the final model this domain has zero occupancy (residues 1497-1641). Conformational flexibility of C345C has been reported previously, as this domain adopts alternative conformations relative to the remainder of the molecule in various crystal structures of C3 and its activation and degradation fragments 20 (Supplemental Figure 1f). This is perhaps not surprising since this domain projects away from the “key-ring” like core of C3c, and it does not make stable contacts with SCIN in either of the structures reported here or in that of the SCIN-inhibited C3 convertase (RCSB code 2WIN) 20. Most significantly, however, SCIN occupies the same binding site and orientation relative to its complement target in both structures, and calculation of Fo−Fc difference maps where the SCIN chains were omitted from either model reveals contiguous regions of positive density (contoured at 2.0σ) that correspond to the location of the SCIN polypeptide in each complex (Figure 1b,d). Together, this strongly suggests that the C3c-SCIN and C3b-SCIN structures are directly comparable to one another.
A series of crystallographic studies have demonstrated that significant structural changes accompany activation of native C3 into C3b 5; 6; 22, and interactions of C3b with certain ligands, in particular the Staphylococcal complement inhibitor proteins Efb 23 and Ehp 24, have been shown to induce additional conformational changes in the complement component. We therefore used superimposition analysis to examine whether additional conformational changes accompany SCIN binding in the structures described above. Quantitative comparisons of each SCIN-bound C3 fragment with the structure of its unbound counterpart 4; 5 reveal no obvious changes to the complement components upon SCIN binding, however; without including the poorly ordered C345C domains in the calculations, the mainchain atoms’ root mean square deviations (RMSD) in the SCIN-bound to unbound structures are 1.80 and 2.60 Å for the C3c 5 and C3b 4 complexes, respectively. Separately, a recent crystal structure of a SCIN-inhibited form of the alternative pathway convertase (C3bBb-SCIN) has also been reported 20. Although SCIN is indeed bound to C3b in this structure, the additional presence of the large Bb fragment (~ 57 kDa) raised the possibility that structural rearrangements of the C3b-SCIN interaction may be necessary to form the resulting ternary complex. Because of this possibility, no direct conclusions could be drawn regarding the nature of C3b-SCIN binding from this previous work. We again used superimposition analysis to address these questions (Supplemental Figure 2). By and large, all three SCIN-bound structures superimpose well with one another; the mainchain atoms’ RMSD in the SCIN-C3c and SCIN-C3b binary complexes when compared to those of C3bBb-SCIN are 0.65 and 0.80 Å, respectively. Differences are visible between the structures in terms of the relative orientations of the C345C domain (which was not modeled in the C3b-SCIN structure), although this could be due to packing effects in the C3c-SCIN crystal lattices. Still, it is worth noting that this domain appears to serve as a swinging platform, which binds and orients Bb in a manner so that its proteolytic active site extends away from the remainder of the C3b molecule 20. Because the location of the SCIN protein is similar in both the binary and ternary complexes, it seems that SCIN binding is accommodated, even in an assembled and active convertase, through minor reorientation of this C345C domain; indeed, real time measurements using surface plasmon resonance (SPR) have shown that SCIN binds readily to C3bBb 19. When these data are considered along with recent functional analyses 19, the structures presented here provide firm evidence that SCIN is capable of binding C3c and C3b in the absence of any additional complement proteins or cofactors.
Nature of SCIN binding to activated forms of C3
The structures reported here and elsewhere reveal that each molecule of SCIN contacts both copies of the complement protein found in the rotationally symmetric tetramers presented in Figure 1. An examination of available data strongly suggests that each of these SCIN binding sites on C3b is relevant: both isothermal titration calorimetry and SPR analyses have shown that the C3b-SCIN interaction is most accurately described by a two-site binding model 19, and small-angle X-ray scattering studies on the C3b-SCIN complex similarly indicate a mixture between 1:1 and 2:2 species in solution 19. To better understand the physical basis for each SCIN binding site, we used the protein interfaces, surfaces, and assemblies (PISA) 25 server to analyze the intermolecular contacts found in the C3c-SCIN and C3b-SCIN structures.
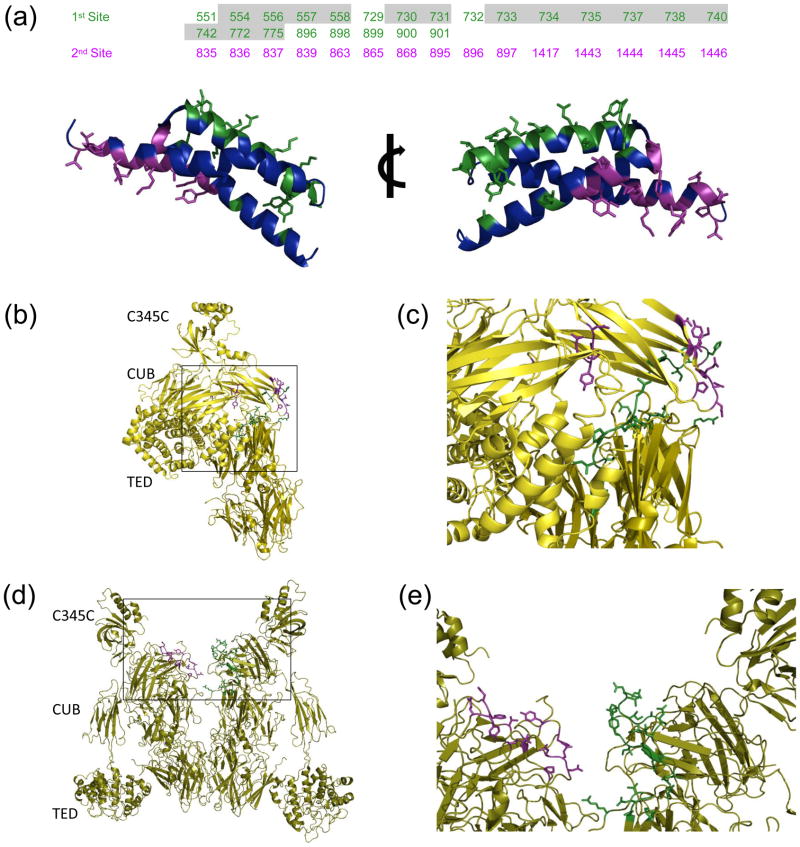
The first SCIN “footprint” on the C3b surface masks approximately 900 Å2, where 700 and 200 Å2 are derived from the α and β chains of the complement component, respectively (Figure 2a and Supplemental Figure 3a). Closer inspection of this interface (as judged by the C3c-SCIN structures which provide higher resolution) indicates that it arises from contacts made almost entirely by the second α-helix of SCIN. While 22 SCIN residues participate in complex formation in this manner, 11 of these form interactions that are polar in nature (e.g. hydrogen bonds and/or salt bridges). Conversely, the second SCIN “footprint” arises through interaction of the first and third α-helices of SCIN with the symmetry-related complement protein (Figure 2a and Supplemental Figure 3b). Interface analysis reveals that this additional site buries approximately 750 Å2, and that all 20 residues which comprise it are located on C3b α-chain. While the 25 total residues SCIN residues that form this interaction are derived from both the first and third helices of the bacterial protein, the physical nature of the contacts formed are very much similar to the first site described above (Supplemental Figure 3b). In particular, the 8 polar interactions found here are similarly distributed between both sidechain-to-sidechain and sidechain-to-mainchain contacts. In general, the proportion of polar contacts found in the SCIN interfaces are somewhat higher than that of the related Staphylococcal complement inhibitor protein, Efb, which binds to the C3d/TED domain of C3/C3b via 23 total residues that form a total of 8 polar interactions 23. Such a difference may partly explain the lower apparent affinity of SCIN for its C3b target relative to Efb (KD≈180 nM 19 versus 10 nM 23 in near physiological buffers, as judged by SPR).
Figure 2. Molecular nature of SCIN binding sites to C3c and C3b.
The EBI PISA server was used identify and analyze intermolecular contacts within the C3b-SCIN and C3c-SCIN structures. (a) In the top panel, residues from the complement components are classified as belonging to either the first (green) or the second (purple) SCIN binding site described in the text; the residues shown here are greater than 5% buried in the structure of C3b-SCIN relative to unbound C3b. Numbering is based upon the sequence of human pre-pro-C3, and those which are buried in the structure of native C3 are shaded grey. Two rotated views of the SCIN protein are provided in the bottom panel. The sidechains from residues comprising the first site (green) and the second site (purple) are represented in ball-and-stick convention. (b) Location of residues comprising the first (green) and second (purple) SCIN binding sites in native C3. (c) Magnified view of the boxed region from panel b. (d) Location of residues comprising the first (green) and second (purple) SCIN binding sites within a C3b dimer generated by crystal symmetry. (e) Magnified view of the boxed region from panel d.
Considerations of SCIN binding aside, activation of native C3 to C3b is accompanied by significant conformational changes that alter the large set of interactions which give rise to the nearly 5,000 Å2 interface between the α and β chains of C3 6. One consequence of such changes is that many residues previously buried in the C3 α/β interface become solvent accessible in C3b. This raised questions as to the nature of the SCIN binding sites in native C3 relative to its activated counterpart, C3b. Significantly, whereas 15 of the 23 C3b-derived residues which constitute the first C3b-SCIN interaction described above are buried in the large α/β interface of native C3, all of the C3b residues that comprise the second SCIN-binding site are solvent exposed in the structure of the native complement protein (Figure 2).
At first glance, this appears to suggest that the second site is actually the primary site of C3b-SCIN interaction. However, three independent lines of evidence reveal that this is not the case. First, unlike the related Staphylococcal complement inhibitors Efb, Ehp, and Sbi 14; 15, SCIN itself cannot bind to native C3 17; 19. The fact that the second binding pocket is exposed in native C3, but that the first is assembled only upon activation to C3b strongly suggests that this neoantigen is required for initial C3b-SCIN binding. Second, a previous study using chimeric proteins of SCIN and its non-functional homolog, ORF-D, has shown that residues 31-48 of SCIN are critical for complement inhibition 16. The structures presented here demonstrate that this region of SCIN, in particular the sidechain of R42, forms several contacts with the first binding pocket on C3b (Supplemental Figure 3). Finally, recent data strongly suggest that SCIN blocks C3 cleavage (but not substrate binding) by surface-bound C3 convertases in their monomeric form 19. Because complement inhibition requires the SCIN region that binds to the first site on C3b, it follows that this interaction is responsible for the convertase inhibitory activity of SCIN.
The observations above suggest that the secondary SCIN binding site on C3b is involved in tetramerization of this complex rather than convertase inhibition per se. However, aside from its potent anti-complement properties, SCIN has also been shown to function as an effective inhibitor of phagocytosis 16; 17; 18. A mechanism for this activity has recently been proposed, which requires the formation of inhibited convertase pseudo-dimers (C3bBb-SCIN)2 to block C3b binding by the complement receptors, CR1 and CRIg 26. Although no crystal structure is currently available for C3b-CR1, examination of the C3b-CRIg complex 27 reveals that the quaternary structure of C3b dimers apparently masks the CRIg-binding site 26. In this manner, C3b binding by complement receptors is disrupted, and phagocytosis of C3b-opsonized bacteria becomes less efficient. While this clearly suggests that the second C3b-binding site of SCIN plays an important role in blocking phagocytosis of C3b-opsonized bacteria, it is worth noting that these (C3b-SCIN)2 tetramers form in the absence of contributions by fB (Figure 1 and Supplemental Figure 1) 19. Although this hypothesis may form the basis for future work, whether or not these (C3b-SCIN)2 structures alone are sufficient to inhibit phagocytosis has not been established at this time.
Structural basis for SCIN effects on C3b binding by Factor H and Factor B
Precise regulation of complement activation is critical to maintaining the delicate balance between the ability to opsonize foreign cells without directing complement attack against healthy host cells and tissues. fH acts as an important host regulator of complement activity by both accelerating the decay of convertases and promoting the factor I mediated degradation of C3b to iC3b. The fact that SCIN binds to C3b substituent of the C3 convertase and renders this otherwise labile multi-component assembly insensitive to both the decay acceleration by fH and impairs its cofactor activity in the degradation of C3b 19 suggested that SCIN binding site might overlap, at least partly, with a region that is crucial for fH function. Indeed, SPR studies revealed that SCIN can efficiently compete with fH for binding to C3b 19.
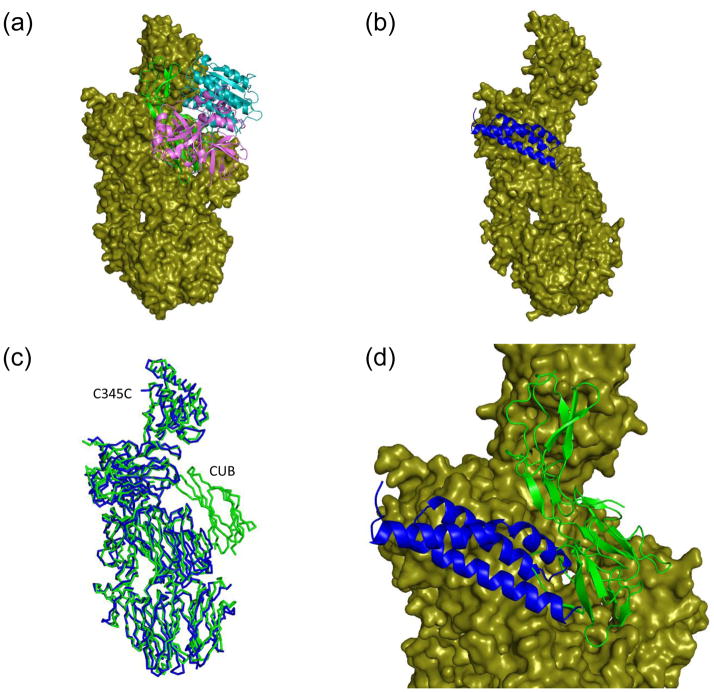
To examine the physical basis for the competition between of fH and SCIN for C3b binding, we superimposed the C3b-SCIN structure with those of C3b bound to an active fragment of fH (fH(1–4)) (Figure 3). Aside from the C345C domains (which were omitted from superposition due to poor map/model quality in this area), the remainder of the molecule (including the flexible CUB and TED domains) adopts a nearly identical conformation in each structure (Figure 3a–c). Interestingly, inspection of the superposed C3b-bound structures of SCIN and fH(1–4) reveals a clear steric clash between these two proteins (Figure 3d). In particular, binding of the second α-helix of SCIN prevents the extensive contacts made by the first domain of fH(1–4) with the MG7 domain of C3b, and to a lesser extent those made between second domain of fH(1–4) and the MG6β domain of C3b. Even though SCIN is not in position to impede the interactions between the third and fourth domains of fH(1–4) and C3b, the apparently large discrepancy in KD in near physiological buffers (180 nM for C3b-SCIN 19 versus 11 μM for C3b-fH(1–4) 28) between these two ligands strongly suggests that SCIN would bind preferentially over fH in situ. Thus, it appears that inhibition of fH activity could block both generation of additional C3b by preventing reassembly of functional convertases on that particular C3b molecule and prevent any entrapped C3b from participating in downstream inflammatory events 19.
Figure 3. SCIN masks the C3b binding site for factor H (fH).
Refined coordinates for the C3b-fH(1–4) (RCSB code 2WII) 28 and the C3b-SCIN structure were superimposed by least-squares methods. (a) Representation of the C3b-fH(1–4) structure, with C3b drawn as a molecular surface and fH(1–4) drawn as a red ribbon. The N-terminus of fH(1–4) lies toward the top of this panel as drawn. (b) Representation of the C3b-SCIN structure, with C3b drawn as a molecular surface and SCIN drawn as a blue ribbon. (c) Superposition of the Cα positions from the C3b component of panel a (red) and panel b (blue) demonstrates a high degree of structural identity. Note that the C345C domain has been omitted as this region is not readily interpretable in the C3b-SCIN structure. (d) Magnified view of a merged superposition from panels a and b illustrates the steric clash between SCIN and the first two domains of fH. The molecular surface shown here is drawn from the C3b-fH(1–4) structure.
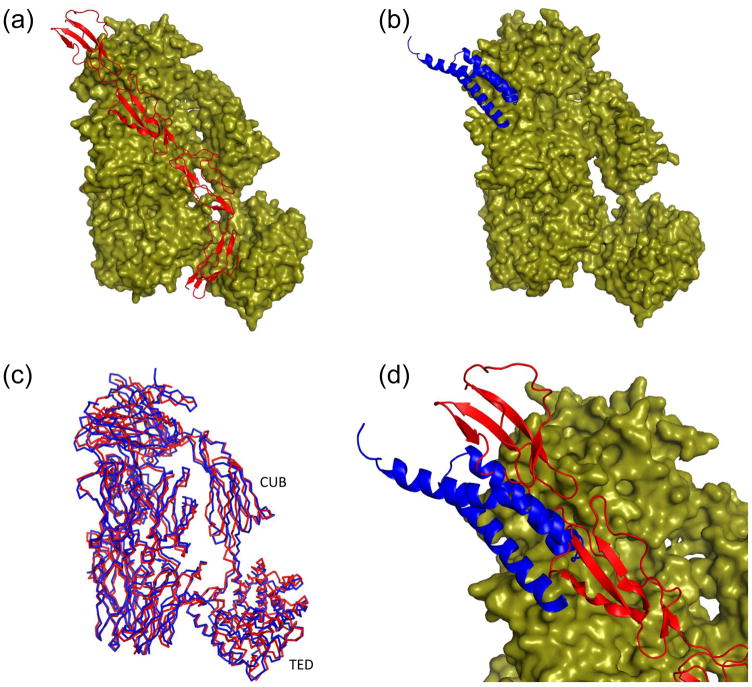
Though the major effect of SCIN may be attributed to the non-functional trapping of assembled convertases, recent data have also shown that this protein also impedes the rate of C3 convertase formation on surfaces 19. This feature most likely arises from the direct competition for C3b binding between SCIN and fB, which binds to C3b and is converted to C3bBb through the action of fD 19. In the absence of the stabilizing properties of SCIN 19; 20; 29, the C3bB pro-convertase is a kinetically labile complex that dissociates rather rapidly, which has long precluded high-resolution structural studies of this interaction. Cobra venom factor (CVF) is a strong complement activator found in certain cobra species and a structural homolog of C3c that forms highly stable complexes with fB, which are more amenable to structural analysis 30. To examine the physical basis for competition between SCIN and fB, we superimposed the CVF-fB structure 30 with that of C3c-SCIN (Figure 4a–c). This shows that while much of the fB interface is not perturbed, the loop which connects the first and second α-helices of SCIN would again create a steric clash with fB (Figure 4d). In particular, the Ba segment of fB appears to be affected to the greatest extent by SCIN binding. This is significant because it is the Ba segment that is responsible for the initial fast and metal-independent loading interaction of fB onto C3b 20. Thus, it seems that by steric impedance of the Ba segment, SCIN can slow the rate of fB loading onto the C3b, which in turns lowers the rate of C3 convertase formation.
Figure 4. SCIN partially occludes the C3b binding site for factor B (fB).
Refined coordinates for the CVF-fB (RCSB code 3HRZ) 30 and the C3c-SCIN structure (drawn from RCSB code 3NMS) were superimposed by least-squares methods. (a) Representation of the CVF-fB structure, with CVF drawn as a molecular surface and fB drawn as ribbon. Domains which comprise the Ba (green) and Bb segments (cyan and purple) are colored separately. (b) Representation of the C3c-SCIN structure, with C3c drawn as a molecular surface and SCIN drawn as a blue ribbon. (c) Superposition of the Cα positions from the C3c-like component of panel a (green) and panel b (blue) demonstrates a high degree of structural identity. Note that C3c lacks the CUB domain found in CVF. (d) Magnified view of a merged superposition from panels a and b illustrates the steric clash between SCIN and the Ba region of fB. The Bb domains have been removed from this panel for clarity. The molecular surface shown here is drawn from the CVF-fB structure.
Conclusions
Throughout their ages-long struggle against formidable host defenses, many pathogens have evolved elegant and powerful means to inhibit, disrupt, or evade the numerous activities required for proper immune function. As a central component of immunity, the complement system has been repeatedly targeted by diverse microbial evasion mechanisms 14. Detailed analyses on a growing number of these proteins have provided a unique perspective through which the structure/function relationships of central complement proteins can be better understood. In several cases this work has shown that quite unexpected features can play a critical role in the evasion proteins’ mechanism of action. For example, the secreted S. aureus proteins Efb and Ehp both bind to C3b and surprisingly induce a conformational change that impairs the activity of the alternative pathway C3 and C5 convertases 18; 23; 24. While SCIN also binds directly to C3b and blocks the activity of surface-assembled convertases 19, its mechanism of action seems to present a fascinating paradox. Indeed, kinetic stabilization of the ordinarily labile C3 convertase assembly 17; 19 would not at first appear to be advantageous to the pathogen. This strategy actually makes a great deal of sense, however: an assembled but non-functional convertase cannot effectively activate C3 into C3b, which both abrogates the self-amplifying nature of the complement response 19; 31 and prevents efficient generation of downstream pro-inflammatory mediators (i.e. C3a and C5a) 19. Furthermore, by competing with fH and inhibiting its decay acceleration activity 19, SCIN ensures that the assembled convertases stay locked in their non-functional state. Though many potent regulators of complement activity are known 2, none of them appear to involve a mechanism that is conceptually similar to SCIN. This further emphasizes the value of elucidating its functional properties, as well as understanding the precise nature of how SCIN recognizes various complement components at a structural level.
One peculiar feature of microbial immune modulator proteins is that they are often capable of targeting multiple aspects of host defense and repair simultaneously 32. On one hand, unique activities can be localized within discrete functional regions of the same protein. For example, S. aureus Efb binds to fibrinogen via its N-terminal region 33, whereas its C-terminal domain provides its anti-complement activity 23; 34. One the other hand, a single functional region of a protein can also disrupt independent processes within the host. In this manner, the helical bundle domains of the S. aureus complement inhibitors Efb and Ehp not only block the activity of C3 convertases 18; 23; 24, but also block C3d recognition by CR2, which interferes with downstream activation of the adaptive immune response 35. By targeting the activity of complement convertases and phagocytosis simulataneously, SCIN not only blocks complement amplification and inflammation, but also clearance of opsonized bacterial cells as well. The structures presented here reveal that these distinct activities localize to separate regions of SCIN, yet together occupy nearly 1/3 of available surface area of this bacterial protein. The fact that such potent, but uniquely valuable activities coexist in a protein of this size further highlights the exquisitely tuned nature of the S. aureus immune evasion arsenal.
Insufficiently controlled activation of complement is a major contributor in the pathophysiology of many inflammatory, immune, ischemia/reperfusion and age-related disorders 10. In all cases, these events culminate in directing the destructive effects of complement-mediated immune responses against host cells and structures, and thereby result in severe tissue damage. Complement inhibitors have long been recognized as a promising therapeutic approach to the prevention of these effects 11; 36. Surprisingly, bacteria have provided a considerable source of insight into the nature of complement regulation and inhibition. Through its secretion of a collection of small proteins, S. aureus deploys ingenious mechanisms to interfere with the central components of the complement cascade, in particular the pivotal conversion of C3 into C3b. Development of complement-directed therapeutics may therefore be greatly aided by the study of the structure, function, and mechanisms of pathogen-derived proteins. In this regard, analysis of the SCIN protein and the physical basis for its interactions with C3b and C3c presented here has provided clues into the recognition of the activated C3b relative its native C3 precursor. In the future, this may suggest templates for the design of novel complement inhibitors that mimic or expand upon SCIN activities toward therapeutic applications.
Methods
Recombinant SCIN was expressed in Escherichia coli and purified as previously described 21. Purified C3b was prepared by activated-thiol sepharose (GE Biosciences) separation of a limited trypsin digestion of isolated human C3 according to previously established protocols 37. Purified C3c was obtained from Complement Technology (Tyler, TX, USA). Solutions of each purified monomer were mixed to yield an equimolar complex of C3b-SCIN and C3c-SCIN prior to buffer exchange by ultrafiltration into 10 mM HEPES-NaOH (pH 7.4) to a final concentration of 5mg ml−1 complex. Crystallization was achieved using hanging drop vapor-diffusion. For each complex the precipitant was diluted 1:1 with ddH2O and 1 μl of this solution was mixed with 1μl of protein solution. C3b-SCIN co-crystals grew over the course of 21 days at 293 K using 0.1 M HEPES-NaOH (pH 7.0), 30% (v/v) Jeffamine ED-2001-HCl as a precipitant; this precipitant solution was used directly for cryopreservation of crystal samples. C3c-SCIN co-crystals were obtained in 2–5 days at 277K using 0.1 M HEPES-NaOH (pH 7.5), 10% (w/v) polyethylene glycol 6000, and 5% (v/v) 2-methyl-1,3-propanediol as a precipitant; these crystals were cryoprotected by a brief incubation in an analogous buffer that contained 20% (v/v) glycerol. Following flash-cooling to 93K, diffraction data were collected using either beamline 22-ID or 22-BM of the Advanced Photon Source (Argonne National Laboratory) and processed using HKL2000 38. Three different lattice types of C3c-SCIN co-crystals were identified from a single crystallization condition (one monoclinic and two orthorhombic; Table 1). Structures were solved by molecular replacement with PHASER 39 using the previously published structures of SCIN (RCSB code 2QFF)16, C3b (RCSB code 2IO7)4, and C3c (RCSB code 2A74)5 as search models. The final models were obtained after manual building in COOT 40 and refinement in PHENIX 41; 42; non-crystallographic symmetry restraints were employed where applicable during all steps of solving the C3c-SCIN structures. Models were analyzed and validated using MOLPROBITY 43. All structural representations were prepared using PyMOL [http://www.pymol.org/], while quantitative comparisons of superimpositions were determined using STAMP 44.
Supplementary Material
(a) Crystal structure of C3c-SCIN (monoclinic form, RCSB code 3L3O; Table 1), where two copies of SCIN (blue to red rainbow from amino to carboxy terminus) and C3c (sand) are found in the asymmetric unit. (b) Electron density map of the same structure generated by refinement of the model without the SCIN molecules. 2Fo−Fc density (blue cage; 1.5σ contour) both copies of C3c, while the Fo−Fc map (green cage; 2.0σ contour) shows clear positive density corresponding to the location of the SCIN polypeptides. Note that the viewing plane of this panel is inclined slightly relative to panel a for clarity. (c) Crystal structure of C3c-SCIN (orthorhombic form 2, RCSB code 3NMS; Table 1), where a single copy of SCIN (blue to red rainbow from amino to carboxy terminus) and C3c (sand) is present in the asymmetric unit. (d) Electron density map of the same structure generated by refinement of the model without the SCIN polypeptide. Map parameters are identical to those in panel b. (e) Generation of the symmetry mate (−x,−y+1, z; drawn at the left) for orthorhombic form 2 indicates a mode of tetramerization similar to that seen for orthorhombic form 1 (Figure 1b), and for the monoclinic form of C3c-SCIN (panel a above). (f) Least-squares superposition of the refined coordinates for orthorhombic form 1 of C3c-SCIN (red), the monoclinic form (silver), and the symmetry-generated tetramer of orthorhombic form 2 (blue). Subtle differences in the orientation of C345C domains in each structure are readily visible.
Refined coordinates of the C3c-SCIN and C3b-SCIN structures were superimposed with those of the SCIN-inhibited convertase (C3b-SCIN-Bb) (RCSB code 2WIN) by least-squares methods. (a) Cartoon representations analogous to those in Figure 1 depicting C3c-SCIN, C3b-SCIN, and C3b-SCIN-Bb pseudo-dimers are shown. The C345C domain in the C3b-SCIN depiction is not colored to reflect its occupancy of zero in this structure. (b) Ribbon diagram showing overlays of C3c-SCIN (silver; drawn from RCSB code 3NSA), C3b-SCIN (blue), and the SCIN-inhibited convertase (red). SCIN molecules are colored in green for all three structures. The C345C domain has been omitted as this region is not visible in the C3b-SCIN structure, while Bb has been removed from the convertase structure for clarity. (c) RMSD values for mainchain atoms relative to C3bBb-SCIN (2WIN). Due to the poorly ordered nature of the various C345C domains, residues corresponding to this region were removed from all structures prior to calculation of RMSD values.
The refined coordinates for the C3c-SCIN structure (drawn from RCSB code 3L3O) were analyzed using the protein interfaces, surfaces, and assemblies (PISA) 25 server to identify likely hydrogen bonds and salt bridges within each SCIN “footprint”. Polar interactions atoms derived from sidechains are shown as dashed red lines, while interactions involving mainchain atoms are drawn as dashed orange lines. (a) Magnified view of the first SCIN binding site on C3c. SCIN is shown as a blue cartoon, with its interacting residues in green according to ball-and-stick convention. C3c (sand) is also shown as a cartoon, however its interacting residues are colored in cyan. (b) Magnified view of the second SCIN binding site on C3c. The color scheme is identical to panel a, except that interacting SCIN residues are shown in purple. Note that the C3c-SCIN structure was analyzed instead of C3b-SCIN because it was determined and refined at significantly higher resolution.
Acknowledgments
We thank Samuel Bouyain, Zheng-Qing Fu, and Zhongmin Jin for helpful discussions during the course of these studies. This work was supported by grants from the National Institutes of Health (AI071028, AI30040, AI068730, and AI072106) and the Missouri Life Sciences Research Board (13238). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamlines at the Advanced Photon Source, Argonne National Laboratory. A list of supporting member institutions may be found at www.ser-cat.org/members.html.
Footnotes
Accession Codes
Refined coordinates and structure factors have been deposited in the RCSB under the accession codes 3NSA, 3NMS, and 3L3O for C3c-SCIN, and 3L5N for C3b-SCIN.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lambris JD, Sahu A, Wetsel R. The Chemistry and Biology of C3, C4, and C5. In: Volanakis JE, Frank M, editors. The Human Complement System in Health and Disease. Marcel Dekker, Inc; New York: 1998. [Google Scholar]
- 2.Morikis D, Lambris JD. In: Structural Biology of the Complement System. Morikis D, Lambris JD, editors. Taylor & Francis; Boca Raton, FL: 2005. [Google Scholar]
- 3.Wetsel RA, Kildsgaard J, Haviland DL. Complement Anaphylatoxins (C3a, C4a, C5a) and Their Receptors (C3aR, C5aR/CD88) as Therapeutic Targets in Inflammation. In: Lambris JD, Holers VM, editors. Therapeutic Interventions in the Complement System. Humana Press; Totowa: 2000. [Google Scholar]
- 4.Janssen BJC, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b Reveals Conformational Changes Underlying Complement Activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 5.Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Ekdahl-Nilsson K, Nilsson B, Gros P. Structures of Complement Component C3 Provide Insights Into the Function and Evolution of Immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 6.Gros P, Milder FJ, Janssen BJC. Complement Driven by Conformational Changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 7.Thurman JM, Holers VM. The Central Role of the Alternative Complement Pathway in Human Disease. J Immunol. 2006;176:1305–1310. doi: 10.4049/jimmunol.176.3.1305. [DOI] [PubMed] [Google Scholar]
- 8.Pangburn MK, Muller-Eberhard HJ. The C3 Convertase of the Alternative Pathway of Human Complement. Enzymic Properties of the Biomolecular Proteinase. Biochem J. 1986;235:723–730. doi: 10.1042/bj2350723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahu A, Lambris JD. Structure and Biology of Complement Protein 3, a Connecting Link Between Innate and Acquired Immunity. Immunol Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 10.Mollnes TE, Song WC, Lambris JD. Complement in inflammatory tissue damage and disease. Trends Immunol. 2002;23:61–64. doi: 10.1016/s1471-4906(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 11.Ricklin D, Lambris JD. Complement-targeted Therapeutics. Nat Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 13.Foster TJ. Immune Evasion by Staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 14.Lambris JD, Ricklin D, Geisbrecht BV. Complement Evasion by Human Pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisbrecht BV. Staphylococcal Complement Inhibitors: Biological Functions, Recognition of Complement Components, and Potential Therapeutic Implications. Adv Exp Med Biol. 2008;632:221–236. [PubMed] [Google Scholar]
- 16.Rooijakkers SHM, Milder FJ, Bardoel BW, Ruyken M, van Strijp JAG, Gros P. Staphylococcal Complement Inhibitor: Structure and Active Sites. J Immunol. 2007;179:2989–2998. doi: 10.4049/jimmunol.179.5.2989. [DOI] [PubMed] [Google Scholar]
- 17.Rooijakkers SH, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJ, van Kessel KP, van Strijp JA. Immune Evasion by a Staphylococcal Complement Inhibitor that Acts on C3 Convertases. Nat Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 18.Jongerius I, Köhl J, Pandey MK, Ruyken M, van Kessel KP, van Strijp JA, Rooijakkers SH. Staphylococcal Complement Evasion by Various Convertase-blocking Molecules. J Exp Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricklin D, Tzekou A, Garcia BL, Hammel M, McWhorter WJ, Sfyroera G, Wu YQ, Holers VM, Herbert AP, Barlow PN, Geisbrecht BV, Lambris JD. A Molecular Insight into Complement Evasion by the Staphylococcal Complement Inhibitor Protein Family. J Immunol. 2009;183:2565–2574. doi: 10.4049/jimmunol.0901443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P. Structural and Functional Implications of the Alternative Complement Pathway C3 Convertase Stabilized by a Staphylococcal Inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia BL, Tzekou A, Ramyar KX, McWhorter WJ, Ricklin D, Lambris JD, Geisbrecht BV. Crystallization of Human Complement Component C3b in the Presence of a Staphylococcal Complement-inhibitor Protein (SCIN) Acta Cryst F Struct Biol Cryst Commun. 2009;65:482–485. doi: 10.1107/S174430910901207X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen BJ, Halff EF, Lambris JD, Gros P. Structure of Compstatin in Complex with Complement Component C3c Reveals a New Mechanism of Complement Inhibition. J Biol Chem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 23.Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. A Structural Basis for Complement Inhibition by Staphylococcus aureus. Nat Immunol. 2007;8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 24.Hammel M, Sfyroera G, Pyrpassopoulos S, Ricklin D, Ramyar KX, Pop M, Jin Z, Lambris JD, Geisbrecht BV. Characterization of Ehp: a Secreted Complement Inhibitory Protein from Staphylococcus aureus. J Biol Chem. 2007;202:30051–30061. doi: 10.1074/jbc.M704247200. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Jongerius I, Puister M, Wu J, Ruyken M, van Strijp JA, Rooijakkers SH. Staphylococcal complement inhibitor modulates phagocyte responses by dimerization of convertases. J Immunol. 2010;184:420–425. doi: 10.4049/jimmunol.0902865. [DOI] [PubMed] [Google Scholar]
- 27.Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren Campagne M. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Wu YQ, Ricklin D, Janssen BJ, Lambris JD, Gros P. Structure of Complement Fragment C3b-factor H and Implications for Host Protection by Complement Regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rooijakkers SH, van Kessel KP, van Strijp JA. Staphylococcal Innate Immune Evasion. Trends Microbiol. 2005;13:596–601. doi: 10.1016/j.tim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Janssen BJ, Gomes L, Koning RI, Svergun DI, Koster AJ, Fritzinger DC, Vogel CW, Gros P. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28:2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harboe M, Ulvund G, Vien L, Fung M, Mollnes TE. The Quantitative Role of Alternative Pathway Amplification in Classical Pathway Induced Terminal Complement Activation. Clin Exp Immunol. 2004;138:439–446. doi: 10.1111/j.1365-2249.2004.02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chavakis T, Wiechmann K, Preissner KT, Herrmann M. Staphylococcus aureus Interactions with the Endothelium: the Role of Bacterial “Secreteable Expanded Repertoire Adhesive Molecules” (SERAM) in Disturbing Host Defense Systems. Thromb Haemost. 2005;94:278–285. doi: 10.1160/TH05-05-0306. [DOI] [PubMed] [Google Scholar]
- 33.Palma M, Wade D, Flock M, Flock JI. Multiple binding sites in the interaction between an extracellular fibrinogen-binding protein from Staphylococcus aureus and fibrinogen. J Biol Chem. 1998;273:13177–13181. doi: 10.1074/jbc.273.21.13177. [DOI] [PubMed] [Google Scholar]
- 34.Lee LYL, Liang X, Hook M, Brown EL. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb) J Biol Chem. 2004;279:50710–50716. doi: 10.1074/jbc.M408570200. [DOI] [PubMed] [Google Scholar]
- 35.Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Geisbrecht BV, Lambris JD. Cutting Edge: Members of the Staphylococcus aureus Extracellular Fibrinogen-binding Protein Family Inhibit the Interaction of C3d with Complement Receptor 2. J Immunol. 2008;181:7463–7467. doi: 10.4049/jimmunol.181.11.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mollnes TE. Therapeutic Manipulation of the Complement System. In: Szebeni J, editor. The Complement System: Novel Roles in Health and Disease. Kluwer Academic Publishers; Boston, MA: 2004. [Google Scholar]
- 37.Lambris JD, Ross GD. Assay of Membrane Complement Receptors (CR1 and CR2) with C3b- and C3d-coated Fluorescent Microspheres. J Immunol. 1982;128:186–189. [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: Building New Software for Automated Crystallographic Structure Determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated Structure Solution with the PHENIX Suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 43.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Crystal structure of C3c-SCIN (monoclinic form, RCSB code 3L3O; Table 1), where two copies of SCIN (blue to red rainbow from amino to carboxy terminus) and C3c (sand) are found in the asymmetric unit. (b) Electron density map of the same structure generated by refinement of the model without the SCIN molecules. 2Fo−Fc density (blue cage; 1.5σ contour) both copies of C3c, while the Fo−Fc map (green cage; 2.0σ contour) shows clear positive density corresponding to the location of the SCIN polypeptides. Note that the viewing plane of this panel is inclined slightly relative to panel a for clarity. (c) Crystal structure of C3c-SCIN (orthorhombic form 2, RCSB code 3NMS; Table 1), where a single copy of SCIN (blue to red rainbow from amino to carboxy terminus) and C3c (sand) is present in the asymmetric unit. (d) Electron density map of the same structure generated by refinement of the model without the SCIN polypeptide. Map parameters are identical to those in panel b. (e) Generation of the symmetry mate (−x,−y+1, z; drawn at the left) for orthorhombic form 2 indicates a mode of tetramerization similar to that seen for orthorhombic form 1 (Figure 1b), and for the monoclinic form of C3c-SCIN (panel a above). (f) Least-squares superposition of the refined coordinates for orthorhombic form 1 of C3c-SCIN (red), the monoclinic form (silver), and the symmetry-generated tetramer of orthorhombic form 2 (blue). Subtle differences in the orientation of C345C domains in each structure are readily visible.
Refined coordinates of the C3c-SCIN and C3b-SCIN structures were superimposed with those of the SCIN-inhibited convertase (C3b-SCIN-Bb) (RCSB code 2WIN) by least-squares methods. (a) Cartoon representations analogous to those in Figure 1 depicting C3c-SCIN, C3b-SCIN, and C3b-SCIN-Bb pseudo-dimers are shown. The C345C domain in the C3b-SCIN depiction is not colored to reflect its occupancy of zero in this structure. (b) Ribbon diagram showing overlays of C3c-SCIN (silver; drawn from RCSB code 3NSA), C3b-SCIN (blue), and the SCIN-inhibited convertase (red). SCIN molecules are colored in green for all three structures. The C345C domain has been omitted as this region is not visible in the C3b-SCIN structure, while Bb has been removed from the convertase structure for clarity. (c) RMSD values for mainchain atoms relative to C3bBb-SCIN (2WIN). Due to the poorly ordered nature of the various C345C domains, residues corresponding to this region were removed from all structures prior to calculation of RMSD values.
The refined coordinates for the C3c-SCIN structure (drawn from RCSB code 3L3O) were analyzed using the protein interfaces, surfaces, and assemblies (PISA) 25 server to identify likely hydrogen bonds and salt bridges within each SCIN “footprint”. Polar interactions atoms derived from sidechains are shown as dashed red lines, while interactions involving mainchain atoms are drawn as dashed orange lines. (a) Magnified view of the first SCIN binding site on C3c. SCIN is shown as a blue cartoon, with its interacting residues in green according to ball-and-stick convention. C3c (sand) is also shown as a cartoon, however its interacting residues are colored in cyan. (b) Magnified view of the second SCIN binding site on C3c. The color scheme is identical to panel a, except that interacting SCIN residues are shown in purple. Note that the C3c-SCIN structure was analyzed instead of C3b-SCIN because it was determined and refined at significantly higher resolution.