Abstract
Background
Depression has been associated with several circadian rhythm perturbations, suggesting a disruption of the circadian clock system in affective disorders. The interaction of several circadian clock genes generates these daily circadian rhythms.
Methods
This cross-sectional study evaluated whether circadian gene expression differed between individuals with a history of depression and participants without a similar history. The participants were 60 healthy older adults. Half of the participants had a history of depression. Real-time quantitative polymerase chain reaction was used to measure the circadian gene Clock, BMAL1, Period1, and Period2 messenger RNA levels in peripheral blood leukocytes.
Results
Individuals with a history of depression had higher Clock, Period1, and Bmal1 mRNA levels, compared to non-depressed participants.
Limitations
Although circadian gene expression fluctuates throughout the day, clock genes mRNA levels were evaluated only in the morning.
Conclusions
These results suggest that disruptions of the molecular mechanisms underlying the circadian clock system may be associated with depression.
Keywords: depression, circadian genes, clock genes, gene expression, circadian rhythms
Introduction
Depression has been associated with several circadian rhythm perturbations (Wirz-Justice, 2006). Insomnia and a shorter REM stage latency after sleep onset are frequently observed among depressed patients (Kupfer and Foster, 1972; Riemann et al., 2001). Depression has also been related to greater nocturnal elevation in body temperature (Rausch et al., 2003). Moreover, depressed patients display an earlier morning cortisol spike and have higher overall cortisol output than non-depressed patients (Yehuda et al., 1996). Furthermore, some studies have reported lower blood concentrations of melatonin, and delayed melatonin release in depressed patients, compared to controls (Parry and Newton, 2001). Collectively, these results suggest a disruption of the circadian clock system related to major depression (Monteleone and Maj, 2008).
The suprachiasmatic nucleus (SCN) of the hypothalamus is the internal clock entraining several physiological systems to a cycle of about 24 hours (Hastings, 1997). The interaction of several circadian clock genes is thought to be responsible for the circadian fluctuation in protein expression (Zheng and Sehgal, 2008). These circadian genes have been found not only in the SCN, but also in several central and peripheral tissues such as extra-SCN brain regions, eye, heart, kidney, lung, liver, skeletal muscle, oral mucosa, and peripheral blood leukocytes (PBLs), indicating the presence of peripheral circadian oscillators throughout the body (Buijs and Kalsbeek, 2001).
Altered circadian gene expression may represent a vulnerability factor associated with increased risk for depression (Mendlewicz, 2009; Turek, 2007). The generation of circadian rhythms is instantiated by the transcriptional and translational feedback loops of several clock genes (Shearman et al., 2000). Perturbations in one or several of these genes may disrupt the circadian fluctuation in mRNA and protein expression and possibly also increase risk for depression (Turek, 2007; Zheng and Sehgal, 2008).
Clock gene polymorphisms have been associated with increased risk of recurrence of bipolar depression (Benedetti et al., 2003), but this result has not been replicated in all studies (Desan et al., 2000; Johansson et al., 2003). However, several other circadian genes polymorphisms have been related to risk for affective illness. Furthermore, the interaction of several clock genes polymorphisms may be necessary to disrupt circadian gene function (Kripke et al., 2009; Lavebratt et al., 2009). Indeed, the interaction of 3 clock genes polymorphisms predicted bipolar affective disorder (Shi et al., 2008). The current study therefore evaluated differences in four circadian gene mRNAs expressed in PBLs of individuals with and without a history of depression.
Methods
Participants
Participants were part of a larger study of caregiving stress and health. The 60 participants were recruited via notices placed in community and university newspapers, senior citizen centers, the Alzheimer’s Disease Association, and from neurologists’ referrals. Caregivers (n =25) were providing at least 5 hours of care per week for a family member with a progressive dementia. Noncaregiving controls (n=35) were demographically similar to caregivers but without caregiving responsibilities. Subjects with immunologically-related health problems (e.g. cancer or recent surgeries), or those taking medications with broad immunological consequences, were excluded from the study. From the larger sample, participants were included in the present study if they had their blood drawn between 9 and 11 AM, were between the ages of 45 and 85, had a history of depression, or were demographically similar to a participant with a history of depression. Participants were studied between September 2007 and February 2008. The Ohio State University Biomedical Research Review Committee approved the project; all subjects gave written informed consent prior to participation.
Measures
Diagnostic Interview for Genetic Studies (DIGS (Nurnberger et al., 1994)). This semi-structured interview provided data on both current and lifetime history of affective disorders using DSM-IV criteria. Participants were classified as having had a history of depression if they had a past or current diagnosis of major depressive disorder, dysthymia, or major depressive disorder not otherwise specified. Interviews were administered by well-trained graduate psychology or nursing students who were supervised by a clinical psychologist with extensive experience with structured interviews.
The Center for Epidemiological Studies Depression Scale (CES-D) assessed the severity of depressive symptoms (Basco et al., 1997; Radloff, 1977). Studies have shown acceptable test-retest reliability and excellent construct validity (Basco et al., 1997). Widely used, the CES-D has distinguished depressed from non-depressed participants in community and clinical samples (Basco et al., 1997).
The Pittsburgh Sleep Quality Index (PSQI) (Buysse et al., 1989), a self-rated questionnaire, provided data on sleep quality and disturbances over a one-month interval. The PSQI has good diagnostic sensitivity and specificity in distinguishing poor and good sleepers (Buysse et al., 1989).
Health-related data were collected to assess the possibility that relationships between depression and circadian gene expression might reflect the contribution of other variables. Health questions from the Older Adults Resources Survey (OARS) (Fillenbaum and Smyer, 1981) assessed underlying diseases. Assessment of health-related behaviors included body mass index (BMI), smoking status, medication use, and alcohol intake (Kiecolt-Glaser and Glaser, 1988). Two questions assessed exercise (Washburn et al., 1987).
Determination of mRNA levels
All blood samples were drawn between 9 AM and 11 AM to control for diurnal variation in circadian gene expression. Peripheral blood leukocytes were isolated from heparinized blood samples using an accuspin (Sigma) and Histopaque-1077 (Sigma) separation solution. RNA was isolated from human PBLs using Trizol Reagent (Invitrogen). RNA concentration was determined in a nanodrop spectrophotometer. Complementary DNA was produced by reverse transcription using Superscript III Reverse Transcriptase (Invitrogen) and random hexamers. The messenger RNA (mRNA) levels of 4 circadian genes were determined by real-time quantitative polymerase chain reaction using the Power Sybr Green method (Applied Biosystems). The primer pairs were: CCAGAGGCCCCTAACTCCTC/ TGGTCTGCCATTGGATGATCT for brain and muscle Arnt-like protein 1 (Bmal1), CAGTGCTCCTGTTCCTGCA TC/ CCCGCCAACTGCAGAATCT for Period1, ACTGCCAAAATCTTACTCTGC/ AGCAAGGCTCAACAAATCATC for Period2, TTGGCAAAATGTCATGAGCAC/ TTGCCCCTTAGTCAGGAACCT, and for Clock. The results for the circadian genes were normalized against the housekeeping gene, G3PDH, Glyceraldehyde 3 Phosphate Dehydrogenase.
Statistical Analysis
Analysis of variance and chi-square tests assessed group differences in age, sex, caregiving status, health, and self-reported depressive symptoms and sleep disturbances. Circadian gene variables had non-normal, highly skewed distributions that were not normalized even after data transformation. Spearman’s Rho correlations evaluated the relationships among the four circadian genes and depressive symptoms and mood disturbances. Given the non-normal distributions of the gene variables, the nonparametric Mann-Whitney U test was used to examine group differences in circadian gene expression among individuals with and without a history of depression. A logistic regression model evaluated whether circadian gene expression was associated with history of depression over and above differences in sex, age, caregiving status, and health and assessed the unique contribution of each circadian gene. Logistic regression models were fitted with the circadian gene mRNA levels as predictors and history of depression as a dependent variable. Statistical significance was evaluated using 2-sided tests at the .05 level of significance.
Results
Sociodemographic and Clinical Characteristics of Study Participants
Participants’ mean age was 71.02 (SD = 10.11). The sample was comprised of 46 women and 14 men. Fifty-four participants were Caucasian and 6 were African-American. About one-third of the sample had a high school education and 68.3 % of the participants were college graduate. Twenty-five participants were caring for a relative with dementia.
Thirty participants had a lifetime history of depression, and 30 were never clinically depressed. Among the 30 participants with a history of depression, 16 had a diagnosis of a single major depressive episode, 13 had recurrent major depressive episodes, and 1 had a diagnosis of depressive disorder not otherwise specified. Among the 30 participants with a history of depression, 4 were currently clinically depressed. No participant had dysthymia, bipolar disorder, or seasonal affective disorder. Individuals with a history of depression did not significantly differ from participants without a history of depression on age, F(1,59) = .01, p = .91, sex, χ2 (1) = 1.49, p = .22, and caregiving status, χ2 (1) = 1.71, p = .19.
History of depression and Circadian Gene mRNA Expression
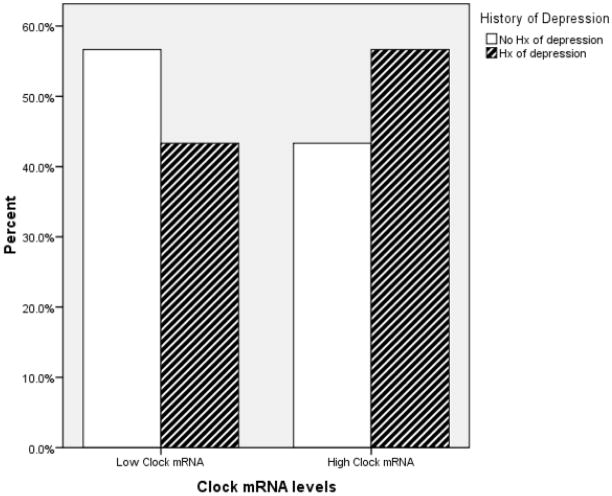
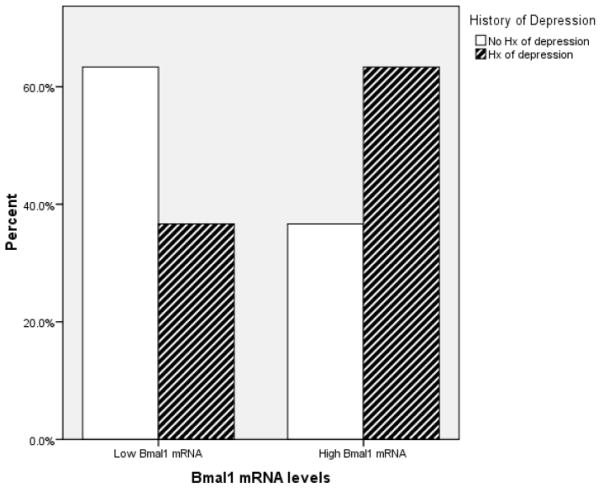
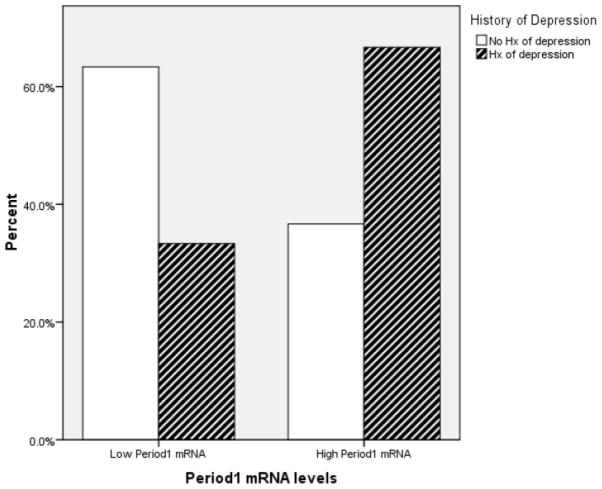
The Mann-Whitney U test revealed that there was a significant difference between individuals with and without a history of depression in circadian gene mRNA expression. Individuals with a history of depression had greater Clock (U = 320.00, z = −1.92, p = .05), Period 1 (U = 292.50, z = −2.33, p = .02), and Bmal1 (U = 308.00, z = −2.1, p = .04). However, Period2 (U = 419.00, z = −.46, p = .65) mRNA expression did not significantly differ between individuals with and without a history of depression. Figure 1–3 depict the relationships between Clock, Period1, and Bmal1 mRNA expression and history of depression.
Figure 1.
The graph depicts the proportion of individuals having high- and low- Clock mRNA expression as a function of their history of depression status. For illustration purposes, the Clock mRNA expression variable was dichotomized using a median split.
Figure 3.
The graph depicts the proportion of individuals having high- and low- Bmal1 mRNA expression as a function of their history of depression status. For illustration purposes, the Bmal1 mRNA expression variable was dichotomized using a median split.
When the four circadian gene variables were entered simultaneously in a logistic regression model, only Clock mRNA levels was an independent predictor of history of depression, z = 3.77, p = .05, while Period1, z = .71, p = .41, Period2, z=.06, p=.80, and Bmal1, z = 1.48, p = .23 did not uniquely predict history of depression. The association between Clock mRNA levels and history of depression persisted even after adjusting for differences in age, sex and caregiving status.
History of Depression, Current Depression and Insomnia Symptoms, and Circadian Gene Expression
Adjusting for caregiving status, individuals with a history of depression did not have greater current depressive symptomatology than participants without such history, F (3,57) = .25, p = .62. However, individuals with a history of depression reported greater sleep disturbances, compared to participants without a history of affective illness, F (3,57) = 7.44, p = .008. No significant Spearman’s rho correlations were found between circadian gene expression and depressive and insomnia symptoms, all ps > .12.
Potentially Confounding Health Variables
Differences in health and health behaviors between participants with and without a history of depression were evaluated as potential confounding variables. When body mass index, alcohol and tobacco use, smoking status, amount of exercise in the past week, and number of self-reported medical conditions were added to the logistic regression model, Clock mRNA expression was still an independent predictor of history of depression, z = 3.91, p = .05.
The most common medications taken by participants were statins (N=24), non-steroidal anti-inflammatory drugs (N=23), diuretics (N=14), angiotensin II receptor antagonists (N=17), beta-blockers (N=17), antidepressants (N=13), thyroid supplements (N=13), calcium-channel blockers (N=13), and sedative/anti-anxiety medications (N=6). Individuals with a history of depression were more likely to use non-steroidal anti-inflammatory drugs, χ2= 5.71, p = .02, compared to participants without a history of affective illness. Furthermore, antidepressant medication has been associated with altered circadian gene expression in animal models (Uz et al., 2005; Ogden et al., 2004). When antidepressant, anti-anxiety, and non-steroidal anti-inflammatory drugs were entered in the model, Clock mRNA expression remained an independent predictor of history of depression, z = 3.89, p = .05.
Discussion
Individuals with a history of depression displayed a different pattern of circadian gene expression than individuals without a history of unipolar affective disorder. Specifically, the presence of a history of depression was associated with higher Clock, Bmal1, and Period1 mRNA expression. When the circadian gene variables were analyzed as a set, only Clock mRNA levels independently predicted history of depression.
This over-expression of circadian genes mRNA may represent a biomarker of vulnerability to unipolar affective disorder. The current findings parallels data showing that Clock gene polymorphism was related to the number of lifetime episodes of depression (Benedetti et al., 2003). However, other human genetics studies have failed to establish an association between Clock gene polymorphisms and depression (Desan et al., 2000; Johansson et al., 2003). Several studies have reported associations between different circadian clock genes polymorphisms and mood disorders (Lavebratt et al., 2009; Soria et al.). However no specific clock genes have been reliably related to depression (Kripke et al., 2009). The lack of consistent findings may be due to the fact that the circadian clock system comprises several genes forming redundant transcriptional and translational feedback loops (Zheng and Sehgal, 2008). Therefore, the redundant processes inherent to this system might mitigate the effect of one polymorphism on circadian gene function. Furthermore, the intercellular coupling of circadian oscillatory may compensate for the impact of one component of the circadian clock gene system (Benca et al., 2009). Indeed, several polymorphisms might be necessary to actually disrupt the functioning of the circadian clock genes system (Shi et al., 2008). In the current study, we assessed disruption of the circadian clock system at another levels of analysis, namely mRNA expression. The fact that we found an over-expression of both positive and negative regulators of the circadian clock system at the mRNA levels reinforces the idea of a link between disruptions of the molecular circadian oscillators and depression.
Several authors have proposed that disorganization of the circadian clock system at the molecular level may be directly related to affective disturbances (Bunney and Potkin, 2008; Bunney and Bunney, 2000; McClung, 2007; Mendlewicz, 2009; Monteleone and Maj, 2008; Turek, 2007). Moreover, altered circadian gene expression may also indirectly influence vulnerability to depression by impacting the occurrence of depression risk factors such as sleep disturbances (Jackson et al., 2003; Turek, 2007). For example, the C variant of the clock gene polymorphism has been associated with insomnia symptoms in individuals with recurrent major depression (Benedetti et al., 2003; Serretti et al., 2005). In accord with this theory, individuals with a history of depression reported more sleep disturbances than non-depressed controls. However, sleep disturbances per se were not associated with circadian gene expression. Alternatively, some theorists have suggested that depression may induced sensitization processes and epigenetic changes that lead to long term alterations in gene expression (Anisman et al., 2008).
One of the limitations of this study is that circadian genes’ mRNA expression was assessed in PBLs. It is unclear whether peripheral expression of circadian genes reflects the functioning of the master clock, the SCN of the hypothalamus. There is some evidence that mood disturbances affect the functioning of the central circadian clock system. Chronic administration of fluoxetine impacted Clock, Bmal1, and Npas2 mRNA expression in the mouse hippocampus (Uz et al., 2005). Furthermore, the mood stabilizer, valproate, decreased the expression of CK1δ and Cry2 in the amygdala (Ogden et al., 2004). The efficacy of these drugs in the treatment of affective disorders suggests a possible central disorganization of the circadian clock system among patients with mood disorders. Our results raise the possibility that this disorganization of circadian oscillators extends to peripheral tissues as well.
The potential clinical implications of the present findings are the possibility of identifying individuals at risk of developing depression based on the functioning of their circadian clock gene system. These individuals may be particularly responsive to chronobiological treatments of depression such as sleep deprivation, light therapy, interpersonal and social rhythm therapy or agomelatine treatment, a melatonergic antidepressant drug. Furthermore, the treatment of insomnia symptoms among depressed patients is associated with improved depression outcomes (Manber et al., 2008). Targeting disturbances of circadian rhythms may then be especially important among individuals presenting altered circadian gene expression.
The present sample included only individuals with a history of unipolar depression. Replication with individuals diagnosed with bipolar disorder and seasonal affective disorder are warranted, given that these two disorders have been associated with dysregulation of the circadian gene system (McClung, 2007). Moreover, circadian gene expression fluctuates throughout the day. In the current study, circadian gene expression was evaluated in the morning between 9 and 11 AM. It would be of interest to compare circadian gene expression between individuals with and without a history of depression during a 24-hour period in a controlled environment in which exposure to light and social zeitgebers will be systematically regulated.
In summary, this study suggests that individuals with a history of depression had altered expression of circadian genes in PBLs, compared to participants without a history of affective illness. This suggests a disruption of peripheral circadian oscillators among depression-prone individuals. Prospective, longitudinal studies are needed to explore whether individuals presenting altered circadian gene expression are at increased risk for mood disorders or whether persistent sleep disturbances lead to disruption of the circadian clock gene function and increased risk for depression.
Figure 2.
The graph depicts the proportion of individuals having high- and low- Period1 mRNA expression as a function of their history of depression status. For illustration purposes, the Period 1 mRNA expression variable was dichotomized using a median split.
Acknowledgments
Role of Funding Source
Funding for this study was provided by NIH Grants AG025732, AI059089, and K23-NS43222, General Clinical Research Center Grant MO1-RR0034, Comprehensive Cancer Center Grant CA16058, and a Fonds de la Recherche en Santé du Québec (FRSQ) Doctoral Training Award; the NIH and FRSQ had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
We thank Ms Liisa Hantsoo, who assisted with the preparation and proof-reading of the manuscript, and Ms. Carrie Houts, who provided advice for the statistical analysis of the data.
Footnotes
Contributors
Author Gouin managed the literature searches, undertook the statistical analyses, and wrote the first draft of the manuscript. Author Connors performed the gene expression analyses. Authors Kiecolt-Glaser, Glaser, Beversdorf and Malarkey designed the study and wrote the protocol. Author Atkinson contributed to the design of the study and oversaw the data collection. All authors contributed to and have approved the final manuscript.
Conflict of Interest
Dr. Beversdorf reported speaking for Novartis, Eisai, and Pfizer within the past year on topics unrelated to the content of this manuscript. None of the other authors reported biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anisman H, Merali Z, Stead JD. Experiential and genetic contributions to depressive- and anxiety-like disorders: clinical and experimental studies. Neurosci Biobehav Rev. 2008;32:1185–1206. doi: 10.1016/j.neubiorev.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Basco MR, Krebaum SR, Rush AJ. Outcome measures of depression. In: Strupp HH, Horowitz LM, Lambert MJ, editors. Measuring patient changes in mood, anxiety, and personality disorders. American Psychological Association; Washington D. C: 1997. pp. 207–245. [Google Scholar]
- Benca R, Duncan MJ, Frank E, McClung C, Nelson RJ, Vicentic A. Biological rhythms, higher brain function, and behavior: Gaps, opportunities, and challenges. Brain Res Rev. 2009;62:57–70. doi: 10.1016/j.brainresrev.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, Smeraldi E. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet B Neuropsychiatr Genet. 2003;123B:23–26. doi: 10.1002/ajmg.b.20038. [DOI] [PubMed] [Google Scholar]
- Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- Bunney JN, Potkin SG. Circadian abnormalities, molecular clock genes and chronobiological treatments in depression. Br Med Bull. 2008;86:23–32. doi: 10.1093/bmb/ldn019. [DOI] [PubMed] [Google Scholar]
- Bunney WE, Bunney BG. Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression. Neuropsychopharmacology. 2000;22:335–345. doi: 10.1016/S0893-133X(99)00145-1. [DOI] [PubMed] [Google Scholar]
- Buysse DJ, Reynolds CF, Monk TH, Berman SR, Kupfer DJ. Pittsburgh Sleep Quality Index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989;28:193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- Fillenbaum GG, Smyer MA. The development, validity, and reliability of the OARS Multidimensional Functional Assessment Questionnaire. J Gerontol. 1981;36:428–434. doi: 10.1093/geronj/36.4.428. [DOI] [PubMed] [Google Scholar]
- Hastings MH. Central clocking. Trends Neurosci. 1997;20:459–464. doi: 10.1016/s0166-2236(97)01087-4. [DOI] [PubMed] [Google Scholar]
- Jackson A, Cavanagh J, Scott J. A systematic review of manic and depressive prodromes. J Affect Disord. 2003;74:209–217. doi: 10.1016/s0165-0327(02)00266-5. [DOI] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Glaser R. Methodological issues in behavioral immunology research with humans. Brain Behav Immun. 1988;2:67–78. doi: 10.1016/0889-1591(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms. 2009;7:2. doi: 10.1186/1740-3391-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer DJ, Foster FG. Interval between onset of sleep and rapid-eye-movement sleep as an indicator of depression. Lancet. 1972;2:684–686. doi: 10.1016/s0140-6736(72)92090-9. [DOI] [PubMed] [Google Scholar]
- Lavebratt C, Sjoholm LK, Partonen T, Schalling M, Forsell Y. PER2 variantion is associated with depression vulnerability. Am J Med Genet B Neuropsychiatr Genet. 2009;153B:570–581. doi: 10.1002/ajmg.b.31021. [DOI] [PubMed] [Google Scholar]
- Manber R, Edinger JD, Gress JL, San Pedro-Salcedo MG, Kuo TF, Kalista T. Cognitive behavioral therapy for insomnia enhances depression outcome in patients with comorbid major depressive disorder and insomnia. Sleep. 2008;31:489–495. doi: 10.1093/sleep/31.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendlewicz J. Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS drugs. 2009;23(Suppl 2):15–26. doi: 10.2165/11318630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Monteleone P, Maj M. The circadian basis of mood disorders: recent developments and treatment implications. Eur Neuropsychopharmacol. 2008;18:701–711. doi: 10.1016/j.euroneuro.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, Kuczenski R, Niculescu AB. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Parry BL, Newton RP. Chronobiological basis of female-specific mood disorders. Neuropsychopharmacology. 2001;25:S102–108. doi: 10.1016/S0893-133X(01)00340-2. [DOI] [PubMed] [Google Scholar]
- Radloff LS. The CES-D scale: A self-report depression scale for research in the general population. Applied Psychological Measurement. 1977;1:385–401. [Google Scholar]
- Rausch JL, Johnson ME, Corley KM, Hobby HM, Shendarkar N, Fei Y, Ganapathy V, Leibach FH. Depressed patients have higher body temperature: 5-HT transporter long promoter region effects. Neuropsychobiology. 2003;47:120–127. doi: 10.1159/000070579. [DOI] [PubMed] [Google Scholar]
- Riemann D, Berger M, Voderholzer U. Sleep and depression--results from psychobiological studies: an overview. Biol Psychol. 2001;57:67–103. doi: 10.1016/s0301-0511(01)00090-4. [DOI] [PubMed] [Google Scholar]
- Serretti A, Cusin C, Benedetti F, Mandelli L, Pirovano A, Zanardi R, Colombo C, Smeraldi E. Insomnia improvement during antidepressant treatment and CLOCK gene polymorphism. Am J Med Genet B Neuropsychiatr Genet. 2005;137B:36–39. doi: 10.1002/ajmg.b.30130. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shi J, Wittke-Thompson JK, Badner JA, Hattori E, Potash JB, Willour VL, McMahon FJ, Gershon ES, Liu C. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1047–1055. doi: 10.1002/ajmg.b.30714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, Gutierrez-Zotes A, Puigdemont D, Bayes M, Crespo JM, Martorell L, Vilella E, Labad A, Vallejo J, Perez V, Menchon JM, Estivill X, Gratacos M, Urretavizcaya M. Differential Association of Circadian Genes with Mood Disorders: CRY1 and NPAS2 are Associated with Unipolar Major Depression and CLOCK and VIP with Bipolar Disorder. Neuropsychopharmacology. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW. From circadian rhythms to clock genes in depression. Int Clin Psychopharmacol. 2007;22(Suppl 2):S1–8. doi: 10.1097/01.yic.0000277956.93777.6a. [DOI] [PubMed] [Google Scholar]
- Uz T, Ahmed R, Akhisaroglu M, Kurtuncu M, Imbesi M, Dirim Arslan A, Manev H. Effect of fluoxetine and cocaine on the expression of clock genes in the mouse hippocampus and striatum. Neuroscience. 2005;134:1309–1316. doi: 10.1016/j.neuroscience.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Washburn RA, Adams LL, Hale GT. Physical activity assessment for epidemiologic research: The utility of two simplified approaches. Prev Med. 1987;16:626–646. doi: 10.1016/0091-7435(87)90047-8. [DOI] [PubMed] [Google Scholar]
- Wirz-Justice A. Biological rhythm disturbances in mood disorders. Int Clin Psychopharmacol. 2006;21:S11–S15. doi: 10.1097/01.yic.0000195660.37267.cf. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Teicher MH, Trestman RL, Levengood RA, Siever LJ. Cortisol regulation in posttraumatic stress disorder and major depression: a chronobiological analysis. Biol Psychiatry. 1996;40:79–88. doi: 10.1016/0006-3223(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]