Abstract
Via high-throughput screening of a natural compound library, we have identified a lipopeptide aldehyde, fellutamide B (1), as the most potent inhibitor of the Mycobacterium tuberculosis (Mtb) proteasome tested to date. Kinetic studies reveal that 1 inhibits both Mtb and human proteasomes in a time-dependent manner under steady-state condition. Remarkably, 1 inhibits the Mtb proteasome in a single-step binding mechanism with Ki = 6.8 nM, whereas it inhibits the human proteasome β5 active site following a two-step mechanism with Ki = 11.5 nM and Ki* = 0.93 nM. Co-crystallization of 1 bound to the Mtb proteasome revealed a structural basis for the tight binding of 1 to the active sites of the Mtb proteasome. The hemiacetal group of 1 in the Mtb proteasome takes the (R) - configuration, whereas in the yeast proteasome it takes the (S) - configuration, indicating that the pre-chiral CHO group of 1 binds to the active site Thr1 in a different orientation. Re-examination of the structure of the yeast proteasome in complex with 1 showed significant conformational changes at the substrate-binding cleft along the active site. These structural differences are consistent with the different kinetic mechanisms of 1 against Mtb and human proteasomes.
Keywords: Mycobacterium tuberculosis, proteasome, slow-binding inhibition, peptide aldehyde, fellutamide B, enzyme conformational change
Mycobacterium tuberculosis (Mtb) is estimated to have caused around 1 billion deaths in the past two centuries [1]. During this time, tuberculosis became curable. However, alarming rates of drug resistance are now putting cure out of reach for increasing numbers of patients [2]. Even with effective drugs, tuberculosis kills nearly 2 million each year. Mtb resides in macrophages of its human host. Despite triggering a vigorous immune response, Mtb resists elimination by the macrophage [3, 4].
Mtb expresses an active proteasome that plays an important role in Mtb’s defense against nitrosative stress imposed by the host [5, 6]. An Mtb mutant strain whose expression of the proteasome was suppressed was susceptible to nitric oxide stress in vitro, and was attenuated in the lungs and spleens of mice following the establishment of infection [5, 7]. Consistent with the results of genetic suppression, chemical inhibition of the Mtb proteasome rendered Mtb susceptible to reactive nitrogen intermedites in non-replicating conditions that were designed to mimic some of the physiological settings that Mtb encounters in the host [5, 8]. Recently, a prokaryotic version of the eukaryotic ubiquitin-proteasome system, the Pup-proteasome system, was demonstrated to degrade certain proteins in Mtb [9–11].
Proteasomes are a family of N-terminal nucleophile hydrolases consisting of 2 sets of 7 copies of α and β subunits that assemble into a barrel-shaped complex in 7 7 7 7 fashion [12]. The active site N-terminal Thr-1 of the -subunit is exposed to the interior chamber after the autocatalytic removal of the propeptide in association with complex assembly. Eukaryotic proteasomes play pivotal roles in the cell cycle, signal transduction and degradation of irreparably damaged proteins [13]. Inhibiting the proteasome can lead to apoptosis, making the human proteasome a target for cancer chemotherapy [14]. The peptidyl boronate bortezomib is in clinical use and other proteasome inhibitors are in clinical trials [15]. However, due to the inherent cytotoxicity of proteasome inhibitors, chemical compounds targeting the Mtb proteasome must exhibit high selectivity for Mtb over human proteasomes in order to be considered for development as chemotherapeutics for tuberculosis.
Previously, we screened a diverse collection of synthetic compounds and discovered that certain oxathiazol-2-ones are highly selective Mtb proteasome inhibitors [8]. This finding demonstrated that a protein degradation pathway could serve as a novel target for anti-infectives. The crystal structure of the inhibitor-treated Mtb proteasome revealed conformational plasticity of the Mtb core particle that appeared to be responsible for the species selectivity [8]. In a separate effort to exploit the distinct oligopeptide substrate preferences of the Mtb proteasome, we obtained several bortezomib analogs that varied in the P1 residues. However, the most selective of these showed only 8-fold more potent inhibition of the Mtb proteasome than a mammalian proteasome [16]. In the current study, we screened a natural product library and discovered a potent lipopeptide aldehyde with nM Ki against Mtb proteasome. Peptide aldehydes are among the first identified proteasome inhibitors, and have been widely used in biological studies involving ubiquitin-proteasome pathway. Peptide aldehydes typically inhibit the proteasome by forming a reversible bond with the α-OH of the active site Thr-1 [6, 17]. 1 did not show better inhibitory activity than reported peptide aldehydes against mammalian proteasomes. However, it was a thousand-fold more potent against the Mtb proteasome than the peptide aldehydes previously tested. Herein we describe kinetic and structural characterizations of inhibition of Mtb and human proteasomes by 1.
EXPERIMENTAL
Materials and Methods
Wild type (Mtb20SWT) and open-gate mutant (Mtb20SOG) Mtb proteasomes were over-expressed and purified as reported [6]. A library containing 1,100 natural products purified from bacteria and 979 from plants was purchased from Analyticon Discovery GmbH (Germany). Suc-LLVY-AMC and human proteasomes were purchased from Boston Biochem (Cambridge, MA). Buffer materials were purchased from Sigma-Aldrich (St. Louis, MO). Screening was carried out at the High-Throughput Screening Resource Facility jointly maintained by Rockefeller University and Weill Cornell Medical College (New York, NY).
High-throughput screen
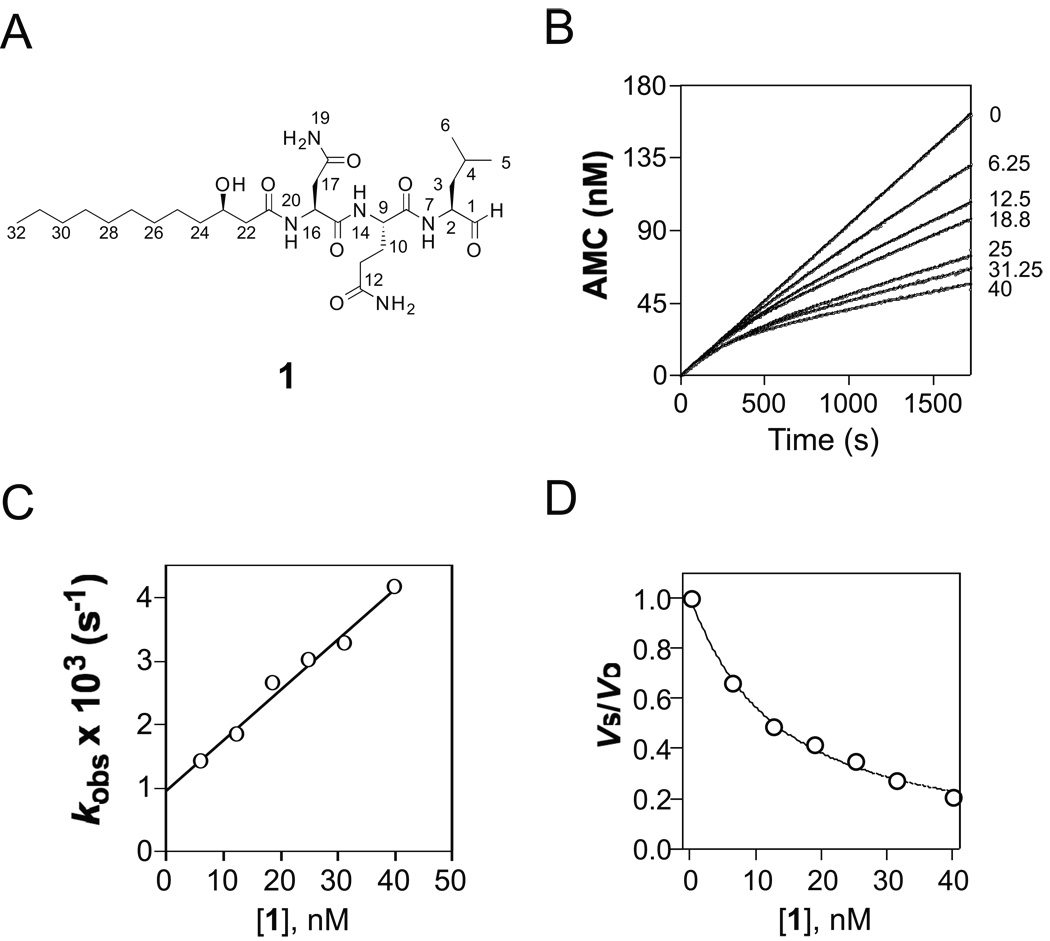
Compounds were dissolved in dimethyl sulfoxide (DMSO) and robotically dispensed into the wells of Falcon Microtest 384-well plates, with DMSO as vehicle control. After a 45-min preincubation with Mtb20SOG (10 µl) at 37 °C, reaction buffer containing substrate (5 µl) was added. Final concentrations were as follows: test compounds, 33 µM; Suc-LLVY-AMC, 50 µM; Mtb20SOG, 1 nM; HEPES, 20 mM; EDTA, 0.5 mM; pH 7.5. Plates were placed on an orbital shaker in a humidified incubator at 37 °C for 45 minutes. End point fluorescence was recorded at excitation 360 nm, emission 460 nm. Z’ values were > 0.5. Three compounds affording > 60% inhibition were retested using re-supplied compounds. Only one, NP-007173, whose structure was determined by Analyticon Discovery GmbH as 1 (figure 1a), showed dose-dependent inhibition of Mtb20SOG.
Figure 1.
Inhibition of Mtb proteasome by fellutamide B (1)
A) Structure of 1. B) Reaction progress curves of cleavage of Suc-LLVY-AMC by Mtb proteasome in the absence and presence of 1 at 6.25 nM – 40 nM; the curves were fit by nonlinear regression to equation (1) to determine the apparent first-order rate constant kobs values, which were corrected by equation (2) to yield real kobs. C) Dependence of kobs on inhibitor concentration. The solid line was drawn by fitting the data to equation (3), yielding kon = (129 ± 0.8) × 103 M−1s−1, and koff = (0.96 ± 0.13) × 10−3 s−1. D) Plot of vs/vo versus inhibitor concentration. The data were fitted to equation (4), yielding Ki = 6.8 ± 0.2 nM.
Kinetics of inhibition
Kinetic measurements were conducted on a Hitachi F-2500 fluorimeter equipped with a water jacket and magnetic stirrer. Progress curves were obtained by monitoring the cleavage of the substrate Suc-LLVY-AMC (50 µM) by Mtb20SOG (0.234 nM) in 2 mL assay buffer (20 mM HEPES, 0.5 mM EDTA, pH 7.5) at 37 °C by fluorimetry. When the velocity of the substrate hydrolysis by the Mtb20SOG reached steady-state, NP-007173 at varying concentrations was added, and the progress curve was recorded till the equilibrium was well established (less than 30 minutes). The AMC fluorescence was recorded at λex = 360 nm and λem = 460 nm with 10 nm slits. In the case of human 20S inhibition by NP-007173, the kinetic reactions were conducted using a miniature assay based on a 96-well plate due to the limited supply of materials. Briefly, a black 96-well plate was prepared by spotting an appropriate amount of NP-007173 in DMSO at the bottom of the wells. 200 µL of preincubated buffer containing human 20S (0.4 nM), proteasomal activator PA28 (2 nM), and the chymotryptic β5 substrate suc-LLVY-AMC (25 µM) was dispensed into each well at 37°C and the reaction progress was immediately monitored for 2 hours on a Molecular Devices SpectraMax M5 Plate-Reader.
Data analysis
Progress curves of Mtb20SOG and human proteasome inhibition by 1 were fitted
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
to a non-linear least square equation (1) to estimate the kobs values with the Prism (Graphpad Software Inc., San Diego, CA). Here vo and vs are the initial and steady-state velocities of the reaction in the presence of inhibitor, respectively, and kobs is the pseudo first-order rate constant describing the rate of inter-conversion from the initial to the steady-state velocities, when no appreciable substrate depletion occurs and the concentration of total active sites is much less than that of inhibitor [18]. Because the concentration of substrate used in the assays was similar to its KM, the kobs values obtained were apparent values. The kobs values at all inhibitor concentrations were then plotted against inhibitor concentrations. A linear relationship allowed use of equation (2) to estimate k3 (i.e. kon) and k4 (i.e. koff). Here, k3 is the apparent rate of association of enzyme and inhibitor if the apparent kobs values were used for the plot and k4 is the rate of disassociation of enzyme inhibitor complex. The disassociation constant Ki was either obtained as k4/k3 or by fitting vs/vo against inhibitor concentrations with the Morrison quadratic equation 3 [18], where ET is the total concentration of the active sites of the enzyme (3.3 nM for Mtb20SOG and 0.8 nM for human proteasome). Because 1 is a competitive inhibitor of the Mtb proteasome, the apparent values of kobs, and k3 are corrected by multiplying (1+ [S]/KM), and Ki by dividing (1+ [S]/KM). Both vi and vs of 1 diminished with the progression of inhibition of h20S β5, indicating that the inhibition of h20S by 1 was via a two-step enzyme isomerization mechanism. The plot of kobs values as a function of [1] yielded a rectangular hyperbola, fitting of which to equation 5 yielded , k5 and k6. The Ki* was calculated by equation 6.
Crystallization of the Mtb 20S proteasome
The purified and concentrated Mtb proteasome was dialyzed against 10 mM HEPES buffer at pH 7.5 for 12 hours. Then the sample was set up in crystallization plates with the sitting drop method at 4 °C. The crystallization droplet contained 3 µL of 20S proteasome at a concentration of 10 mg/mL, mixed with 3 µL of well solution containing 60 mM Na citrate (pH 5.8) and 13% PEG 6000 as the precipitant. Crystals with approximate dimensions of 200 µm × 60 µm × 50 µm grew after 5 to 7 days. For cryo-crystallography, the crystal solution was replaced in several concentration steps with a cryo-protecting solution containing the original mother liquor and 35% dimethylformamide. For inhibitor soaking, the crystals were moved into the cryo-protectant solution containing 1 mM NP-007173, and incubated at 4°C for 10 to 14 hours before the crystals were flash frozen.
Diffraction data collection and structure solution
X-ray diffraction data were collected at Beamline X25 of National Synchrotron Light Source in Brookhaven National Laboratory. The wavelength for the data collection was 1.000 Å. The diffraction data were processed and scaled with the HKL2000 package [19].
Structure refinement
The Mtb proteasome structure (PDB ID: 2FHG) was used as the starting model for molecular replacement [20]. Conventional crystallographic rigid body refinement, simulated annealing, positional and temperature factor refinement were carried out with CNS [21]. Fourteen non-crystallographic symmetry (NCS) restraints were applied during the refinements. After each refinement, the model was manually rebuilt and adjusted in Coot [22]. After Rfree of the model had been reduced to below 26%, the inhibitor structure was built into the electron density. The topology and parameter files were generated for 1 with idealized geometry on HI-CUP server. The electron density map at all 14 active sites fit well with the inhibitor structure, except for the alkyl tail. The refined structural coordinates have been deposited in the Protein Data Bank with access ID: 3KRD.
Results
1. Identification of 1 as a potent Mtb proteasome inhibitor
Screening of 2079 natural products identified 3 compounds that afforded > 60% inhibition of the Mtb proteasome. Only one, NP-007173, displayed reproducible inhibitory activity against the Mtb proteasome upon testing with freshly purchased samples. Its structure was assigned as a lipopeptide aldehyde, (R)-3-hydroxydodecanoyl-Asn-Gln-Leu-CHO (Fig. 1a), by NMR and mass spectral data provided by the supplier. Peptide aldehydes were among the first proteasome inhibitors identified. They inhibit proteasomes by forming a tetrahedral intermediate with the β-hydroxyl of the active site Thr-1 [23]. SciFinder® search identified this compound as fellutamide B, which was isolated from the marine fungus Penicillium fellutanum and found to be cytotoxic to cancer cells [24]. Recently, fellutamide B (1) was shown to induce the release of nerve growth factor from fibroblasts and glial-derived cells through inhibition of proteasome activity, with IC50 values 9.6 nM for β5, 1.2 µM for β2, 2.0 µM for β2 [25, 26]. However, the kinetic mechanism of proteasome inhibition was not provided.
2. 1 inhibits the Mtb proteasome via a one-step slow binding mechanism
Peptide aldehydes have been reported to be classical reversible inhibitors of the proteasome with Ki values ranging from nM to µM [17, 27]. We set out to determine the kinetic mechanism of the inhibition of Mtb20SOG by 1. Progress curves of proteasomal hydrolysis of Suc-LLVY-AMC showed time-dependent inhibition at all concentrations of 1 (Fig. 1B). Each time-course curve was fitted to a nonlinear least squares equation 1 to estimate the kobs values.
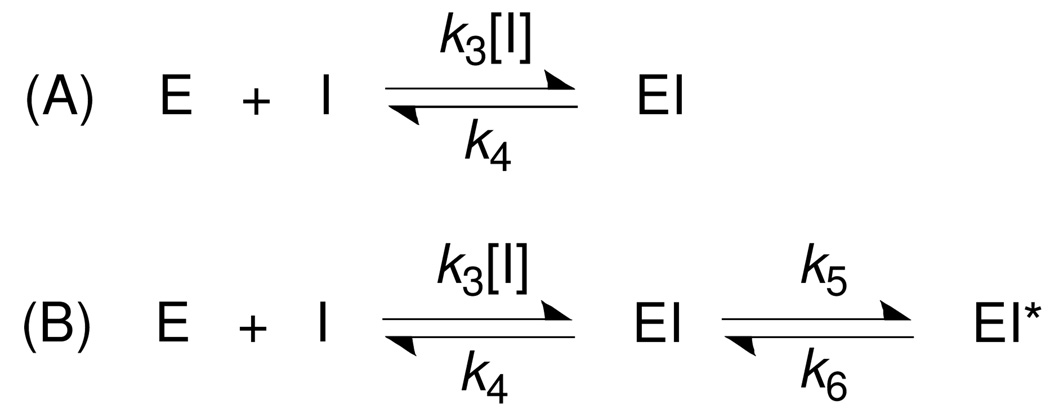
Two basic kinetic mechanisms can account for the slow-binding inhibition, as illustrated in Scheme 1. Scheme (A) shows a simple equilibrium between an enzyme and inhibitor that is governed by association rate constant k3 and dissociation rate constant k4. Scheme (B) shows a two-step mechanism by which an enzyme isomerization step forms a tight EI* complex following the formation of an initial relatively loose EI complex.
Scheme 1.
Two types of time-dependent inhibition mechanism. (A) One-step binding mode and (B) two-step binding mode.
Plotting the kobs values against the concentrations of fellutamide B (10–50 nM) yielded a straight line (Fig. 1C), indicating that inhibition of the proteasome by fellutamide B follows a simple reversible slow-binding inhibition mechanism (Fig. 1E) under the experimental conditions [18]. The slope of the fit, konapp, was (79.5 ± 0.5) × 103 M−1s−1, which was corrected by multiplying (1+[S]/KM) to yield a corrected kon of (129 ± 0.8) × 103 M−1s−1 (Table 1). The intercept on the Y-axis, koff, was (0.96 ± 0.13) × 10−3 s−1, from which the half-life of the enzyme inhibitor complex was calculated to be 12 minutes. The of fellutamide B against Mtb20SOG was determined to be (11.1 ± 0.4) nM (Fig. 1D) by fitting vs/vo versus [1] to the Morrison equ 3 or by koff/kon, which was also corrected to yield a real Ki of (6.8 ± 0.2) nM, in good agreement with the Ki (7.4 nM) calculated from koff/kon. So far, 1 is the most potent Mtb proteasome peptide aldehyde inhibitor of the Mtb proteasome that we have tested [6].
Table 1.
Kinetic parameters of Mtb and human proteasomes by 1
| K3 (M−1s−1) | K4 (s−1) | k5 (s−1) | k6 (s−1) | Ki (nM) | Ki* (nM) |
t1/2 minutes |
|
|---|---|---|---|---|---|---|---|
| Mtb20SOG | 129,000 | 0.00096 | NA | NA | 6.8 | NA | 12.0 |
| Hu20S β5 | NA | NA | 0.0052 | 0.00046 | 11.5 | 0.93 | 25.2 |
3. 1 inhibits the human proteasome β5 via a two-step mechanism
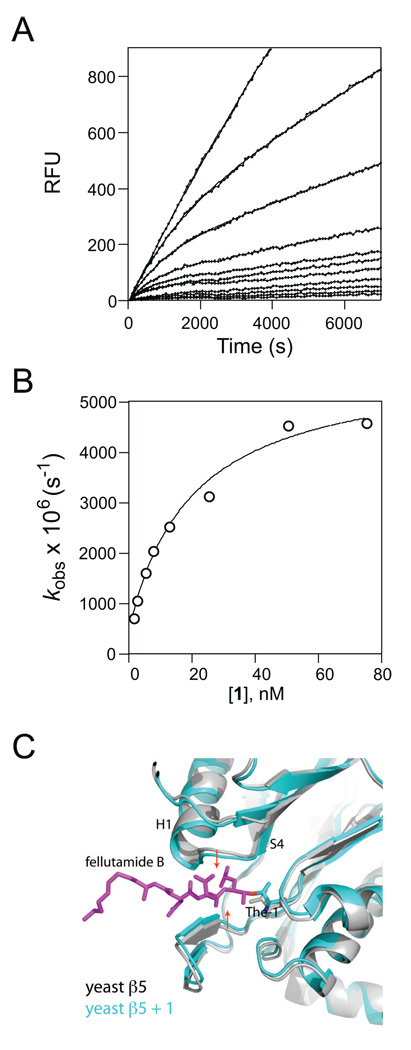
Similar to inhibition of Mtb20SOG, 1 also inhibited human proteasome (hu20S) in a time-dependent manner (Fig. 2a). At all concentrations, 1 decreased both initial and steady-state velocities, indicating a two-step slow binding inhibition. The values of kobs were obtained by fitting the progression curves to equation 1. The plot of kobs as a function of [1] yielded a rectangular hyperbola (Fig. 2b), in agreement with abovementioned two-step slow binding inhibition, where the encounter complex EI isomerizes to tighter EI*. Fitting the curve to equ. 5 yielded k5 = 5.3 × 10−3 s−1, k6 = 0.48 × 10−3 s−1, and apparent Ki 20.1 nM, which is corrected to Ki (11.6 nM) by equation 4 (Table 1). The Ki*, which is defined as the dissociation constant for the high-affinity complex of enzyme-inhibitor, is calculated to be 0.96 nM using equation (6). The k5/k6 ratio = 11 indicates a 91% distribution of enzyme into EI*, reflecting a 1.5 kcal/mol (ΔG = −RTlnK) of advantage on binding affinity over EI.
Figure 2.
1 inhibits the human proteasome β5 via a two-step mechanism that likely involves enzyme conformational changes. (A) Reaction progress curve of hydrolysis of Suc-LLVY-AMC by hu proteasome in the presence of 1 at 0–75 nM; the curves were fit to equation (1) to determine the apparent kobs. (B) The plot of kobs values to [1] demonstrates a two-step inhibition of human 20S β5 by 1. Fitting to equation (6) yields Ki, k5 and k6. (C) Binding of 1 induces significant substrate pocket narrowing of the yeast proteasome β5. The native yeast proteasome β5 structure (gray) is superimposed with β5 bound to 1 (cyan). 1 is shown as sticks in magenta. The two red arrows indicate tightening of the substrate pocket upon binding of 1.
Since the kinetic mechanism indicated an enzyme isomerization, it would be ideal to examine the possible conformation changes in the structure of mammalian proteasome bound by 1. There is no available structure of the human proteasome alone or in complex with 1. However, the human proteasome 5 and the yeast 5 are 66% identical and 80% similar, whereas the human proteasome 5 and the mycobacterial proteasome are only 28% identical and 44% similar. We therefore examined the structure reported for the yeast proteasome co-crystallized with 1 [26]. Superimposition of structures of native yeast β5 and 1-bound β5 reveals significant conformational changes around the active site: The upper loop and bottom β-sheet move inward by 0.5 Å and 1 Å, respectively, as indicated in Fig. 2C. This conformational change increases the contact between the enzyme and inhibitor, likely accounting for the enhanced binding affinity from EI to EI* as reflected by the 1.5 kcal/mol decrease in free energy. This marked conformational change is in good agreement with the prediction by the kinetic studies.
4. Structural analysis of fellutamide-bound Mtb proteasome
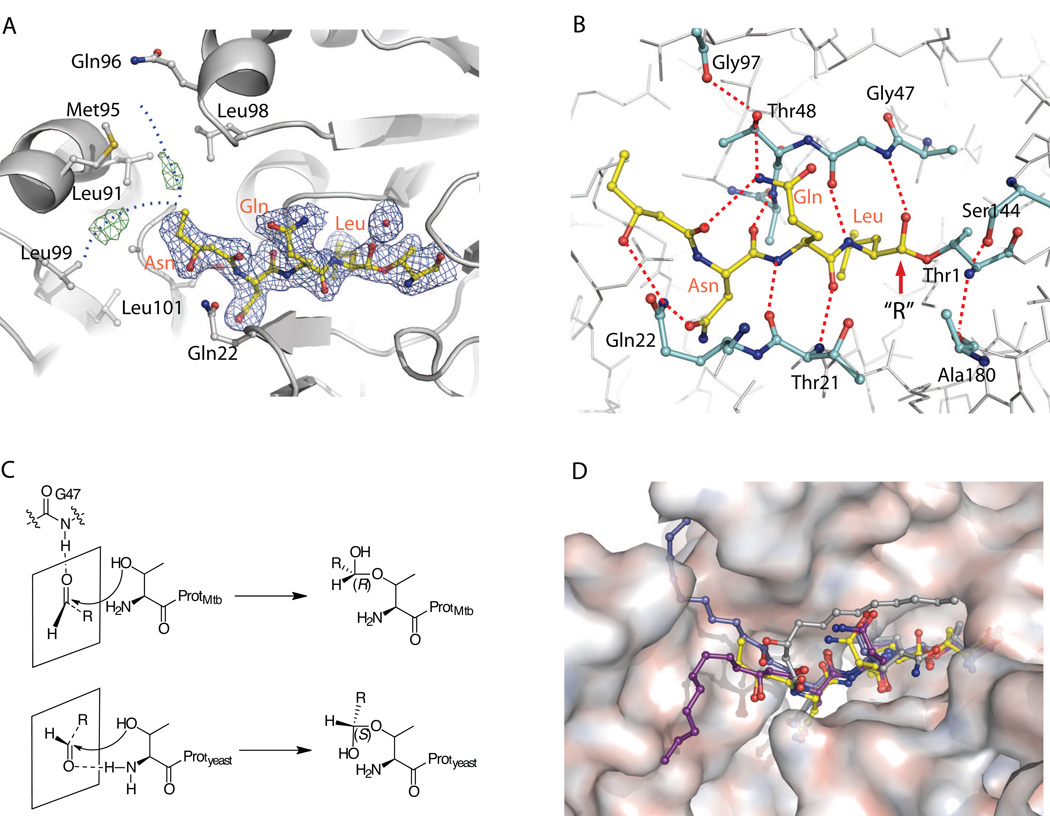
To gain structural insight into the differential inhibition mechanisms of 1 against the Mtb and eukaryotic proteasomes, we determined the X-ray co-crystal structure of the Mtb proteasome with 1 to 2.5 Å resolution (Fig. 3). Statistics for the co-crystal structure determination are listed in Supplemental Table 1. Consistent with the one-step mechanism observed above, there is no significant structural change at the Cα backbone level in the Mtb proteasome. In the crystal structure, 1 has strong electron density at the tripeptide and the head group of the 3-hydroxydodecanoic acid regions nested within the substrate-binding pocket, but only weak and discontinuous densities at the aliphatic tail region (C24–C32) of the dodecanoic acid (Fig. 3A). As expected, the aldehyde of the fellutamide B forms a tetrahedral intermediate with the β-hydroxyl group of the active site Thr-1, with well-defined electron density for the covalent linkage between the aldehyde C1 and γ-Oxygen of Thr-1 (Fig. 3B). This mode of inhibition, mimicking the intermediate in peptide substrate hydrolysis, has been observed for all previously characterized aldehyde inhibitors, including 1, in the yeast proteasome [26, 28]. In the Mtb proteasome, the main chain atoms of the inhibitor’s tripeptide interact with the main chain atoms of Thr-21 at the lower substrate-binding surface, and with that of Gly-47 and Ala-49 at the upper surface through several H-bonds, forming a short anti-parallel β-sheet structure. These interactions presumably keep the inhibitor inside the substrate pocket long enough to stabilize the tetrahedral intermediate between the inhibitor and the enzyme [28].
Figure 3.
Crystal structure of fellutamide B (1) bound to the Mtb proteasome
A) The 2Fo-Fc electron density map of fellutamide B shown at 1σ level is shown in blue mesh and the Fo-Fc map at 3σ in green mesh. The likely positions of the disordered alkyl tail from C24 to C32 of 1 are sketched by two dashed blue curves. B) The hydrogen-bonding network that stabilizes 1 in the substrate-binding pocket. The identities of selected proteasome residues are labeled in black, and the inhibitor residues are shown in orange. C) Scheme of the different stereochemistry of fellutamide B in the Mtb and yeast proteasomes. Upper panel, the carbonyl group takes trans-configuration, is hydrogen bonded with Gly-47 when approaching the Thr-1, and subsequently yields a (R)-hemiacetal; lower panel, the carbonyl group takes cis-configuration, is hydrogen bonded with NH2-Thr-1, and subsequently yields a (S)-hemiacetal. D) A surface view of the substrate-binding pocket of Mtb proteasome with the resolved structure of the fellutamide B (1) (yellow). The three structures of 1 bound to yeast proteasome β1, β2, and β5 are superimposed and shown in gray, blue, and purple, respectively (PDB ID 3D29).
An interesting and perhaps important distinction in the way 1 binds the Mtb proteasome is that the carbonyl-C of the hemiacetal group formed from the nucleophilic attack of aldehyde group by the β-OH of the Thr-1 takes an (R)-conformation as with other peptide aldehyde inhibitors [23], whereas in all 6 active sites of the yeast proteasome it takes the (S)-conformation (Fig. 3B) [26]. In the yeast proteasome, the cis configuration is stabilized by a H-bond with the α-amine of Thr-1. In the Mtb proteasome, the carbonyl oxygen forms a H-bond (3.1 Å) with the amide of Gly-47 and is further stabilized by another H-bond with a nearby water (Fig. 3B). It has been established that the amide backbone of Gly-47 plays an important role in the hydrolysis of protein substrates by serving as the oxyanion hole, which stabilizes the transition state of the intermediate through hydrogen bonding with the oxygen atom of the carbonyl group of the peptide bond that is undergoing hydrolysis [26]. From our results, the amide back-bone of Gly-47 not only stabilizes the hydrolysis intermediates, but also appears to orient the substrate to approach the active site, insofar as the orientation of the pre-chiral CHO prior to the attack by the γO determines the chirality of the hemiacetal as illustrated in Fig. 3C.
While the aliphatic chain of 1 is fully stabilized and well resolved in each of the three active β-subunits of the yeast proteasome [26], the alkyl tail (C24 to C32) of the aliphatic chain does not have well-defined electron density in the Mtb proteasome, except for two discontinuous difference densities (Fig. 3A). These densities suggest that the aliphatic tail might adopt two alternative conformations: either extending upward into a groove between two neighboring β-subunits (similar to its binding in the yeast β2), or bending downward (similar to the yeast β5). Attempts to build such alternative configurations with partial occupancies did not result in improved density in this region, indicating the flexibility and the possibility of more than two configurations of the aliphatic tail. Thus we left out the alkyl tail (C24–C32) in the refined structure.
The short anti-parallel β-sheet formed between many peptide aldehyde inhibitors and proteasomes is maintained by several backbone H-bonds and is not very stable by itself [28]. In the Mtb proteasome, however, the C21 carbonyl of 1 forms an intra-molecular H-bond with the amine of P2-Gln, orienting the Gln side chain to form an additional H-bond with Thr-48 of the enzyme (Fig. 3B). The C23 hydroxyl and the Asn side chain of 1 both form H-bonds with the amine of Gln-22 of the enzyme (Fig. 3B). These additional H-bonds, enabled by the C21 carbonyl and the C23 hydroxyl, likely account for the high affinity of the inhibitor for the Mtb proteasome, and may further explain the peculiar observation that the tripeptide Ac-NQL-AMC, lacking the enabling hydroxyl and carbonyl groups, could not be hydrolyzed by the Mtb proteasome [16].
In the yeast proteasome, the tail section binds quite differently in the three different proteolytic sites [26]. Fig. 3D provides an overall comparison of the conformations of 1 when bound with the Mtb proteasome and with the active yeast β-subunits. Although the alkyl tail of 1 is not modeled in the Mtb proteasome, the fact that we see discontinuous densities in the experimental density map indicates that the 1 alkyl tail may be able to insert into two regions such as found in yeast β5 and β2. Albeit less specifically or less tightly in Mtb than in yeast, the alkyl tail of 1 likely contributes to its high affinity for the Mtb proteasome.
Discussion
Peptide aldehydes are widely used as proteasome inhibitors in studies with cells in vitro. N-terminal Z-capped tripeptide aldehydes typically inhibit the Mtb proteasome with IC50 values ranging from 10 – 70 µM [6]. In unpublished work, we tested 20 N-Ac-Leu-Leu-Xaa-aldehydes against the Mtb proteasome (both wild type and open-gate mutant), where Xaa varied from small hydrophobic amino acids to aromatic amino acids. The IC50 values were similar and ranged from 25.8 µM to over 100 µM. They are classical Michaelis-Menton inhibitors. Surprisingly, the peptidomimetic 1 displayed time-dependent inhibition against both Mtb and human proteasomes. Compared to other peptide aldehydes, such as Z-Leu-Leu-Leu-Cho (ki = 4 nM) [29], 1 did not inhibit the mammalian proteasome any more potently, indicating that the elongated N-terminal capping group did not improve the overall binding affinity, but instead changed its kinetic behavior against the mammalian proteasome as seen in its slow onset of inhibition. However, 1 is over 1,000 fold more potent against the Mtb proteasome than the other peptide aldehydes that we have tested, indicating that elongation at the N-terminus dramatically changes the behavior of peptidyl aldehyde inhibitors, and may compensate for the unfavorable residue at the P1 position as well.
It is also surprising that 1 inhibits Mtb20S and hu20S with different kinetic mechanisms. Inhibition of Mtb20S by 1 follows a simple reversible slow-binding mechanism, whereas inhibition of hu20S β5 by 1 follows a two-step slow-binding mechanism. The one-step inhibition mechanism of 1 on the Mtb proteasome is supported by our crystal structure of the complex, which revealed no significant changes in the proteasome. The two-step mechanism of 1 on β5 of hu20S is consistent with the previously published structure of 1-bound yeast proteasome, which revealed conformational changes in the core particle (Fig. 2C) [26]. A Change in yeast proteasome structure upon inhibitor binding is interesting, because in our earlier report on oxathiazolones, the non-peptidic Mtb proteasome specific inhibitors, we observed marked conformational changes in Mtb proteasome following binding of oxathiazolones in the active site, whereas oxathiazolones did not inhibit the human proteasome [8]. These inhibitor-induced structural changes hint at the potential plasticity of proteasomes. Indeed, structural flexibility might be required for proteasomes to achieve affinity for their diverse substrates in a cell. The conformational changes seen to date in eukaryotic and Mtb proteasomes are triggered by different small molecular scaffolds. More work is needed to better understand the differential structural responses to small molecule inhibitors between the seemingly well-conserved structures of Mtb and eukaryotic proteasomes.
In summary, we have identified fellutamide B as so far the most potent Mtb proteasome inhibitor. Fellutamide B inhibits Mtb 20S in a single step mechanism, while it inhibits the human proteasome in a two-step mechanism. The partial flexibility of the hydrophobic tail of 1 when complexed with the Mtb proteasome suggests that a shorter tail might further improve its binding for Mtb proteasome. Alternatively, the possible two binding pockets in the Mtb proteasome for the alky tail might allow for construction of more species-selective inhibitors with a branched tail that occupies both pockets simultaneously. A branched tail, while potentially advantageous for inhibition of the Mtb proteasome, might interfere with binding to human proteasomes. Studies in this direction are ongoing.
Table 2.
Data collection and model refinement statistics.
| WT 20S + Fellutamide B | |
|---|---|
| Crystal form | |
| Space group | P21 |
| Unit cell dimensions | |
| a , b, c, (Å) | 170 × 118 × 194 |
| (°) | 112.6 |
| Data collection | |
| Wavelength (Å) | 1.000 |
| Temperature (K) | 100 |
| Resolution (Å) | 50 - 2.5 (2.59-2.5) a |
| Unique reflections | 241959 |
| Redundancy | 4.7 |
| Rsym | 0.107 (0.485) |
| Completeness (%) | 96.6 (71.8) |
| I/σ(I) | 9.7 (1.8) |
| Model statistics | |
| Model composition: | |
| nonhydrogen atoms | 48845 |
| Protein residues | 6118 |
| Ligands | 14 |
| Refinement: | |
| Resolution (Å) | 50 - 2.5 |
| Rcryst / Rfree | 0.200 / 0.240 |
| Test reflections (%) | 5 |
| Average B-factor | 45.0 |
| R.m.s deviations: | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.207 |
| Ramachandran plot | |
| Most favored (%) | 95.5 |
| Allowed (%) | 4.1 |
| Disallowed (%) | 0.4 |
Values in parentheses refer to the highest resolution shell.
ACKNOWLEDGMENTS
Supported by NIH PO1-AI056293, NIH R01AI070285 and the Milstein Program in Chemical Biology of Infectious Diseases. A TB Drug Accelerator grant from the Bill and Melinda Gates Foundation supported the purchase of the natural product library. We thank Dr. F. Glickman and R. Realubit of the High-Throughput Screening Resource Facility for their help and Drs. L. Dick and C. Tsu (Millennium Pharmaceuticals Inc., Boston, MA) for donation of N-Ac-Leu-Leu-Xaa-aldehydes. X-ray diffraction data were collected at beamlines X6A, X25, and X29 in the National Synchrotron Light Source, a facility supported by US DOE and NIH. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ryan F. How the battle against tuberculosis was won – and lost. Boston, MA: Lttle, Brown; 1993. The Forgotten Plague. [Google Scholar]
- 2.Murray JF. Am J Respir Crit Care Med. 2004;169:1181–1186. doi: 10.1164/rccm.200402-140OE. [DOI] [PubMed] [Google Scholar]
- 3.Young D, Hussell T, Dougan G. Nat Immunol. 2002;3:1026–1032. doi: 10.1038/ni1102-1026. [DOI] [PubMed] [Google Scholar]
- 4.Tischler AD, McKinney JD. Curr Opin Microbiol. 2010;13:93–99. doi: 10.1016/j.mib.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 6.Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C. Mol Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 7.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, Nathan C. Nature. 2009;461:621–626. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Striebel F, Hunkeler M, Summer H, Weber-Ban E. EMBO J. 2010 doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Nat Struct Mol Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 12.Baumeister W, Walz J, Zuhl F, Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg AL. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg AL. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 15.Jackson G, Einsele H, Moreau P, Miguel JS. Cancer Treat Rev. 2005;31:591–602. doi: 10.1016/j.ctrv.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Lin G, Tsu C, Dick L, Zhou XK, Nathan C. J Biol Chem. 2008;283:34423–34431. doi: 10.1074/jbc.M805324200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormack TA, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Plamondon L, Stein RL, Dick LR. Biochemistry. 1998;37:7792–7800. doi: 10.1021/bi980097q. [DOI] [PubMed] [Google Scholar]
- 18.Copeland RA. Evaluation of enzyme inhibitors in drug discovery: A guide for medicinal chemists and pharmacologists. Hoboken: John Wiley & Sons, Inc.; 2005. pp. 214–248. [PubMed] [Google Scholar]
- 19.Otwinoswki Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 20.Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. Mol Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 24.Shigemori H, Wakuri S, Yazawa K, Nakamura T, Sasaki T, Kobayashi J. Tetrahedron. 1991;47:8529–8534. [Google Scholar]
- 25.Schneekloth JS, Sanders JL, Jr, Hines J, Crews CM. Bioorg Med Chem Lett. 2006;16:3855–3858. doi: 10.1016/j.bmcl.2006.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hines J, Groll M, Fahnestock M, Crews CM. Chem Biol. 2008;15:501–512. doi: 10.1016/j.chembiol.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein RL, Melandri F, Dick L. Biochemistry. 1996;35:3899–3908. doi: 10.1021/bi952262x. [DOI] [PubMed] [Google Scholar]
- 28.Borissenko L, Groll M. Chem Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 29.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]