Abstract
Background
Cell lines are invaluable model systems for the investigation of cancer. Knowledge of the molecular alterations that exist within cell models is required to define the mechanisms governing cellular phenotypes.
Methods
Five tongue squamous cell carcinomas cell lines and one submaxillary salivary gland epidermoid carcinoma cell line were analyzed for copy number and mRNA expression by tiling-path DNA microarrays and Agilent Whole Human Genome Oligoarrays, respectively.
Results
Integrative analysis of genetic and expression alterations revealed the molecular landscape of each cell line. Molecular results for individual cell lines and across all samples have been summarized and made available for easy reference.
Conclusion
Our integrative genomic analyses have defined the DNA and RNA alterations for each individual line. These data will be useful to anyone modelling oral cancer behaviour, providing a molecular context that will be useful for deciphering cell phenotypes.
Keywords: oral cancer, head and neck cancer, cell models, copy number alteration, RNA expression alteration
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the eighth most common cancer in the world.(1) It is a heterogeneous disease primarily affecting the oral cavity, salivary glands, oropharynx, hypopharynx, and larynx. Model systems, such as cell lines are often used as research tools to study many cancer types, including HNSCC.(2, 3) They allow for manipulation of tumour cells in a laboratory setting. However, tumourigenic cell lines in such studies have often not been fully characterized at the molecular level. Like tumours themselves, cancer cell lines can vary greatly with respect to their genetic background. Analysis of cell phenotypes without concurrent analysis of underlying molecular changes represents a lost opportunity to understand the mechanistic basis for disease.
One characteristic of most solid tumours is genomic instability. This is demonstrated by the numerous DNA gains and deletions present in tumour genomes.(4) DNA alterations can lead to aberrant expression of oncogenes and tumour suppressor genes that drive tumourigenesis. For HNSCC development, gene alterations such as loss of chromosomes 3p and 9p, mutation of p53, loss of 13q, and high-level amplification of CCND1 have been associated with premalignant disease stages.(5–8) Loss of chromosomes 4q and 8p, on the other hand, are believed to occur as later events in tumourigenesis.(9) Such DNA alterations, including high-level amplification of CCND1,(10) often leads to overexpression, while homozygous deletion of CDKN2A leads to silencing of p16INK4A in oral tumours.(11)
Established HNSCC cell lines — particularly SCC-4, SCC-9, SCC-15, SCC-25, A253, and Cal27 — are widely used as models of head and neck cancer. A preliminary search in PubMed reveals 407 publications that use at least one of these lines, demonstrating their importance as tools for modeling biochemical, immunological, and pharmacological behaviours. These various behaviours could be influenced by the molecular alterations that exist in each line, yet few efforts have been made to characterize such changes. Previously, genetic alterations of four of the OSCC cell lines (SCC-4, SCC-9, SCC-15, and SCC-25) have been characterized by cDNA microarrays with low resolution and with no data on intragenic regions.(12) Thus, comprehensive characterization of genomic alterations in the most commonly used cell models are needed. In addition, a priori knowledge of both genomic and gene expression changes for a given cell line would facilitate selection of the most appropriate model system for a given in vitro analysis. In this report we provide full characterization of genome and transcriptome alterations in six commonly used HNSCC cell lines. These data will serve as an excellent resource when designing future experiments that attempt to model HNSCC behaviour.
MATERIALS AND METHODS
Cell lines
Five tongue squamous cell carcinomas cell lines (SCC-4,(13) SCC-9,(13) SCC-25,(13) SCC-15,(13) and Cal27(14)) and one submaxillary salivary gland epidermoid carcinoma cell line (A-253)(15) were purchased from the American Type Culture Collection (ATCC). All cell lines were cultured according to ATCC recommendations. Cells were harvested for DNA and RNA extractions when they reached ~ 80% confluency. Genomic DNA was extracted by the standard phenol-chloroform extraction protocol, and total RNA was extracted using the TRIzol (Invitrogen, Carlsbad, CA) protocol followed by DNase I treatment.
Tiling-path DNA microarray
Extracted DNA from each cell line was analyzed by whole genome tiling-path microarrays (SMRT v.2) developed at the British Columbia Cancer Research Centre Array Laboratory.(16) For this array, the whole genome is represented as more than 26,000 overlapping bacterial artificial chromosome (BAC) clones spotted in duplicate with tiling coverage of the human genome. Pooled normal male DNA was used as reference for all microarray experiments. Labelling, hybridization, scanning, and washing of the slides were performed as previously described.(8)
Imaging and analysis of genomic data
Array images were analyzed with GenePix Pro 6.1. A three-step normalization procedure, including locally weighted linear regression (LOWESS) fitting, spatial, and median normalization was used to remove systematic biases.(17) SeeGH software was used to combine duplicate spot data and display log2 signal intensity ratios in relation to genomic locations in the hg18 assembly (NCBI Build 36.1).(18) Data points with standard deviation >0.075 and signal to noise ratio <3 in either channel were removed from each sample.
Two separate algorithms, aCGH-Smooth and DNACopy, were used to detect copy number gains and losses.(19, 20) An alteration was only called when detected concurrently by both algorithms. aCGH-Smooth employs a local search algorithm that uses a maximum likelihood estimation to smooth the observed array CGH values between consecutive breakpoints to a common value.(19) The Lambda and the maximum number of breakpoints in initial pool were set to 6.75 and 100 respectively.(8) DNAcopy employs a circular binary segmentation algorithm that uses a random permutation test to determine the statistical significance of change points.(20) Default settings in DNAcopy were used for our data.
A third algorithm based on moving-average was used for the identification of high-level DNA amplification and presumptive homozygous deletions.(21) The threshold was set at log2 signal intensity ratio >0.8 for high-level amplification or <−0.8 for homozygous deletions. Only regions containing ≥3 overlapping clones with such calls were identified in order to avoid false-positives due to hybridization artefacts.
Gene expression profiling
Total RNA collected from each cell line was analyzed by Agilent Whole Human Genome Microarray 4×44K. This array represents more than 41,000 unique human transcripts. Labelling and hybridizations were performed according to manufacturer’s instructions (Agilent Technologies). Hybridized arrays were scanned using Axon GenePix 4000B and 4200A scanner.
Data analysis of expression profiles
Array images were analyzed using GenePix Pro 6.1. For normalization processing, each background-subtracted intensity value was divided by the median array intensity of each microarray. The median array intensity was calculated based on the background-subtracted intensity value for all spots excluding control type spots on the array. Genes within regions of high-level copy number change were extracted for each cell line. Oral cell lines with no high-level copy number change serve as the baseline for the genes in each alteration region and the mean of each median-normalized intensity value of each probe was compared with those of the cell lines with high-level change.
RESULTS AND DISCUSSION
Cancer cell lines are important model systems for investigating the biology of head and neck cancers. They allow for characterization of a variety of disease phenotypes and also serve as a tool for functional screens. When using such models, it is imperative to determine how closely the model resembles clinical disease. However, genome-wide characterization of most cell lines has not previously been attempted. Characterizing DNA alterations in these cells and determining the contribution of these changes to mRNA expression would provide a very valuable reference point for interpreting experimental results generated using these cell lines. The set of six genomic and six expression profiles has been deposited to Gene Expression Omnibus (GEO) database at NCBI, series accession number GSE16872.
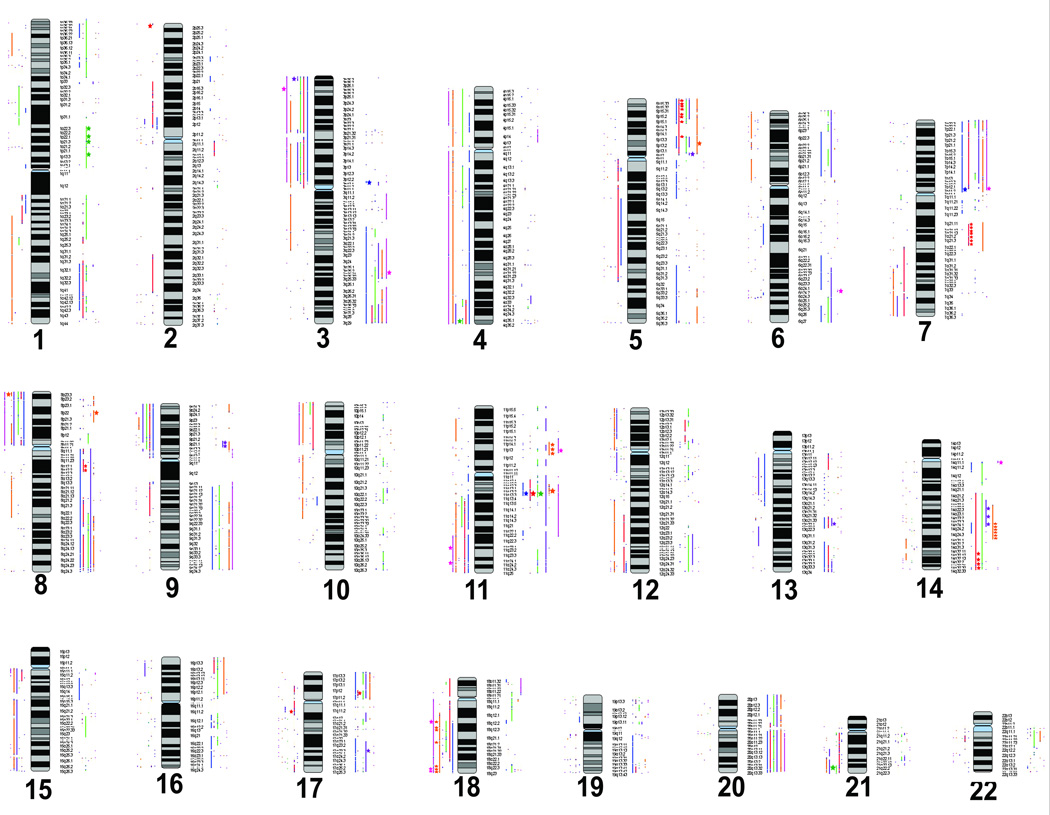
High-level DNA amplification is a common mechanism for driving overexpression of oncogenes, while homozygous deletion is known to inactivate tumour suppressor genes and also contribute to cancer processes.(21–23) High resolution genomic analysis delineates the precise boundaries of amplified or deleted regions, thus defining genes that warrant further investigation. Parallel analysis of gene expression data from the same cells helps to differentiate between "driver" and "passenger" genes in a given segmental DNA change, the former being genes that contribute to the cancer phenotype, the latter being altered simply due to its proximity to a "driver" gene.(21, 24) For example, any candidate gene in an amplicon that is not overexpressed is unlikely to represent a "driver" gene. Figure 1 displays a summary of altered regions detected in each oral cell line, while the specific base pair positions are listed in Supplemental tables S1 to S6. Furthermore, as this paper serves as a reference, the copy number status of well known cancer genes obtained from the Cancer Gene Census is catalogued in Table 1.(25) Whole genome copy number karyogram of each cell line and description of each cell line is represented in Supplemental figure S1.
Figure 1.
Summary of copy number alteration in each cell line. Copy number gain is presented as vertical lines on the right side of the chromosome, and vertical lines on the left side indicate copy number loss. High-level copy number change is represented by a star on the right (DNA amplification) or left (homozygous deletion) of the chromosome. Genetic alteration of each cell line is labelled with different colours, SCC-15 (blue), SCC-4 (red), SCC-25 (green), SCC-9 (purple), A-253 (orange), and Cal27 (pink).
Table 1.
Copy number status of cancer genes in the six head and neck cancer cell lines.
| Gene | Gene Name | Gene ID |
Locus | SCC- 15 |
SCC- 4 |
SCC- 25 |
SCC- 9 |
A- 253 |
Cal27 |
|---|---|---|---|---|---|---|---|---|---|
| ABI1 | abl-interactor 1 | 10006 | 10p12.1 | − | − | − | |||
| ABL1 | v-abl Abelson murine leukemia viral oncogene homolog 1 |
25 | 9q34.12 | + | + | + | + | ||
| ABL2 | v-abl Abelson murine leukemia viral oncogene homolog 2 |
27 | 1q24-q25 | + | |||||
| ACSL6 | acyl-CoA synthetase long-chain family member 6 |
23305 | 5q23.3 | − | − | ||||
| AF15Q14 | AF15q14 protein | 57082 | 15q15.1 | ||||||
| AF1Q | ALL1-fused gene from chromosome 1q |
10962 | 1q21 .2 | − | + | ||||
| AF5q31 | ALL1 fused gene from 5q31 |
27125 | 5q31.1 | − | − | ||||
| AKAP9 | A kinase (PRKA) anchor protein (yotiao) 9 |
10142 | 7q21.2 | + | + | ||||
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 |
207 | 14q32.33 | + | + | ||||
| AKT2 | v-akt murine thymoma viral oncogene homolog 2 |
208 | 19q13.2 | + | |||||
| ALK | anaplastic lymphoma kinase (Ki-1) |
238 | 2p23.2 | ||||||
| ALO17 | KIAA1618 protein | 57714 | 17q25.3 | + | − | + | |||
| APC | adenomatous polyposis of the colon gene |
324 | 5q22.2 | − | − | ||||
| ARHGAP26 | GTPase regulator associated with focal adhesion kinase pp125(FAK) |
23092 | 5q31.3 | + | − | ||||
| ARHGEF12 | RHO guanine nucleotide exchange factor (GEF) 12 (LARG) |
23365 | 11q23.3 | − | − | + | + | − | − |
| ARNT | aryl hydrocarbon receptor nuclear translocator |
405 | 1q21 | ||||||
| ASPSCR1 | alveolar soft part sarcoma chromosome region, candidate 1 |
79058 | 17q25.3 | + | − | + | |||
| ATF1 | activating transcription factor 1 |
466 | 12q13.12 | + | − | ||||
| ATIC | 5-aminoimidazole-4- carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
471 | 2q35 | − | |||||
| ATM | ataxia telangiectasia mutated |
472 | 11q22.3 | − | − | + | − | − | |
| BCL10 | B-cell CLL/lymphoma 10 |
8915 | 1p22.3 | − | |||||
| BCL11A | B-cell CLL/lymphoma 11A |
53335 | 2p16.1 | ||||||
| BCL11B | B-cell CLL/lymphoma 11B (CTIP2) |
64919 | 14q32.2 | ++ | + | ||||
| BCL2 | B-cell CLL/lymphoma 2 |
596 | 18q21.33 | − | − | − | − | − | |
| BCL3 | B-cell CLL/lymphoma 3 |
602 | 19q13.31 | + | |||||
| BCL5 | B-cell CLL/lymphoma 5 |
603 | 17q23.2 | ||||||
| BCL6 | B-cell CLL/lymphoma 6 |
604 | 3q27.3 | + | + | + | + | + | + |
| BCL7A | B-cell CLL/lymphoma 7A |
605 | 12q24.31 | − | + | + | |||
| BCL9 | B-cell CLL/lymphoma 9 |
607 | 1q21 | − | |||||
| BCR | breakpoint cluster region |
613 | 22q11.23 | − | + | + | |||
| BIRC3 | baculoviral IAP repeat- containing 3 |
330 | 11q22.2 | + | − | + | − | + | |
| BLM | Bloom Syndrome | 641 | 15q26.1 | ||||||
| BMPR1A | bone morphogenetic protein receptor, type IA |
657 | 10q23.2 | − | |||||
| BRAF | v-raf murine sarcoma viral oncogene homolog B1 |
673 | 7q34 | − | − | ||||
| BRCA1 | familial breast/ovarian cancer gene 1 |
672 | 17q21.31 | + | |||||
| BRCA2 | familial breast/ovarian cancer gene 2 |
675 | 13q13.1 | − | + | − | |||
| BRD4 | bromodomain containing 4 |
23476 | 19p13.12 | ||||||
| BRIP1 | BRCA1 interacting protein C-terminal helicase 1 |
83990 | 17q23.2 | ||||||
| BTG1 | B-cell translocation gene 1, anti- proliferative |
694 | 12q22 | − | − | ||||
| BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) |
701 | 15q15.1 | − | |||||
| C12orf9 | chromosome 12 open reading frame 9 |
93669 | 12q14.3 | ||||||
| C15orf21 | chromosome 15 open reading frame 21 |
283651 | 15q21.1 | − | − | ||||
| CARD11 | caspase recruitment domain family, member 11 |
84433 | 7p22.2 | + | + | + | + | + | + |
| CARS | cysteinyl-tRNA synthetase |
833 | 11p15.4 | ||||||
| CBFA2T1 | core-binding factor, runt domain, alpha subunit 2;translocated to, 1 (ETO) |
862 | 8q21.3 | + | + | + | − | ||
| CBFA2T3 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 (MTG-16) |
863 | 16q24.3 | ||||||
| CBFB | core-binding factor, beta subunit |
865 | 16q22.1 | + | |||||
| CBL | Cas-Br-M (murine) ecotropic retroviral transforming |
867 | 11q23.3 | − | − | + | + | − | − |
| CCDC6 | coiled-coil domain containing 6 |
8030 | 10q21.2 | − | |||||
| CCNB1IP1 | enhancer of invasion 10 - fused to HMGA2 |
57820 | 14q11.2 | + | + | ++ | |||
| CCND1 | cyclin D1 | 595 | 11q13.3 | ++ | ++ | ++ | + | ++ | + |
| CCND2 | cyclin D2 | 894 | 12p13.32 | + | − | − | |||
| CCND3 | cyclin D3 | 896 | 6p21.1 | + | + | + | − | ||
| CDH1 | cadherin 1, type 1, E- cadherin (epithelial) (ECAD) |
999 | 16q22.1 | + | |||||
| CDH11 | cadherin 11, type 2, OB-cadherin (osteoblast) |
1009 | 16q21 | ||||||
| CDK4 | cyclin-dependent kinase 4 |
1019 | 12q14.1 | + | |||||
| CDK6 | cyclin-dependent kinase 6 |
1021 | 7q12 | ++ | + | ||||
| CDKN2A- p14ARF |
cyclin-dependent kinase inhibitor 2A-- p14ARF protein |
1029 | 9p21.3 | − | − | − | − | − | |
| CDKN2A - p16(INK4a) |
cyclin-dependent kinase inhibitor 2A (p16(INK4a)) gene |
1029 | 9p21.3 | − | − | − | − | − | |
| CDX2 | caudal type homeo box transcription factor 2 |
1045 | 13q12.2 | − | + | − | |||
| CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha |
1050 | 19q13.11 | + | |||||
| CEP1 | centrosomal protein 1 | 11064 | 9q33.2 | + | + | + | + | ||
| CHEK2 | CHK2 checkpoint homolog (S. pombe) |
11200 | 22q12.1 | + | |||||
| CHIC2 | cysteine-rich hydrophobic domain 2 |
26511 | 4q12 | − | − | ||||
| CHN1 | chimerin (chimaerin) 1 | 1123 | 2q31.1 | ||||||
| CIC | capicua homolog (Drosophila) |
23152 | 19q13.2 | + | |||||
| CLTC | clathrin, heavy polypeptide (Hc) |
1213 | 17q23.2 | ||||||
| CLTCL1 | clathrin, heavy polypeptide-like 1 |
8218 | 22q11.21 | + | + | ||||
| CMKOR1 | chemokine orphan receptor 1 |
57007 | 2q37.3 | ||||||
| COL1A1 | collagen, type I, alpha 1 |
1277 | 17q21.33 | ||||||
| COPEB | core promoter element binding protein (KLF6) |
1316 | 10p15.1 | − | − | − | − | ||
| COX6C | cytochrome c oxidase subunit VIc |
1345 | 8q22.2 | + | + | + | + | ||
| CREB1 | cAMP responsive element binding protein 1 |
1385 | 2q33.3 | − | |||||
| CREB3L2 | cAMP responsive element binding protein 3-like 2 |
64764 | 7q32-q34 | − | − | ||||
| CREBBP | CREB binding protein (CBP) |
1387 | 16p13.3 | + | + | + | |||
| CTNNB1 | catenin (cadherin- associated protein), beta 1 |
1499 | 3p22.1 | − | − | − | − | − | |
| CXXC6 | Leukemia-associated protein with a CXXC domain |
80312 | 10q21.3 | + | − | ||||
| CYLD | familial cylindromatosis gene |
1540 | 16q12.1 | + | |||||
| DDB2 | damage-specific DNA binding protein 2 |
1643 | 11p11.2 | + | + | ||||
| DDIT3 | DNA-damage- inducible transcript 3 |
1649 | 12q13.3 | + | + | ||||
| DDX10 | DEAD (Asp-Glu-Ala- Asp) box polypeptide 10 |
1662 | 11q22.3 | − | − | + | − | − | |
| DDX6 | DEAD (Asp-Glu-Ala- Asp) box polypeptide 6 |
1656 | 11q23.3 | − | − | + | + | − | − |
| DEK | DEK oncogene (DNA binding) |
7913 | 6p22.3 | + | + | − | |||
| DUX4 | double homeobox, 4 | 22947 | 4q35.2 | − | − | − | − | − | − |
| EGFR | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
1956 | 7p12.3- p12.1 |
++ | + | + | + | ++ | |
| EIF4A2 | eukaryotic translation initiation factor 4A, isoform 2 |
1974 | 3q27.3 | + | + | + | + | + | + |
| ELKS | ELKS protein | 23085 | 12p13.33 | + | − | − | |||
| ELL | ELL gene (11–19 lysine-rich leukemia gene) |
8178 | 19p13.11 | ||||||
| ELN | elastin | 2006 | 7q11.23 | + | |||||
| EML4 | echinoderm microtubule associated protein like 4 |
27436 | 2p21 | ||||||
| EP300 | 300 kd E1A-Binding protein gene |
2033 | 22q13.2 | ||||||
| EPS15 | epidermal growth factor receptor pathway substrate 15 (AF1p) |
2060 | 1p32 | ||||||
| ERBB2 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) |
2064 | 17q12 | ||||||
| ERCC2 | excision repair cross- complementing rodent repair deficiency, complementation group 2 (xeroderma pigmentosum D) |
2068 | 19q13.32 | + | |||||
| ERCC3 | excision repair cross- complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) |
2071 | 2q14.3 | ||||||
| ERCC4 | excision repair cross- complementing rodent repair deficiency, complementation group 4 |
2072 | 16p13.12 | + | − | + | |||
| ERCC5 | excision repair cross- complementing rodent repair deficiency, complementation group 5 (xeroderma pigmentosum, complementation group G (Cockayne syndrome)) |
2073 | 13q33.1 | + | + | ||||
| ERG | v-ets erythroblastosis virus E26 oncogene like (avian) |
2078 | 21q22.2 | − | − | − | |||
| ETV1 | ets variant gene 1 | 2115 | 7p21.2 | + | + | + | + | ||
| ETV4 | ets variant gene 4 (E1A enhancer binding protein, E1AF) |
2118 | 17q21.31 | ||||||
| ETV5 | ets variant gene 5 | 2119 | 3q37.2 | + | + | + | + | + | + |
| ETV6 | ets variant gene 6 (TEL oncogene) |
2120 | 12p13.2 | + | + | − | − | ||
| EVI1 | ecotropic viral integration site 1 |
2122 | 3q26.2 | + | + | + | + | ||
| EWSR1 | Ewing sarcoma breakpoint region 1 (EWS) |
2130 | 22q12.2 | + | |||||
| EXT1 | multiple exostoses type 1 gene |
2131 | 8q24.11 | + | + | + | + | + | |
| EXT2 | multiple exostoses type 2 gene |
2132 | 11p11.2 | + | + | ||||
| FANCA | Fanconi anemia, complementation group A |
2175 | 16q24.3 | ||||||
| FANCC | Fanconi anemia, complementation group C |
2176 | 9q22.32 | + | + | + | + | ||
| FANCD2 | Fanconi anemia, complementation group D2 |
2177 | 3p25.3 | − | − | − | − | ||
| FANCE | Fanconi anemia, complementation group E |
2178 | 6p21.31 | + | + | + | |||
| FANCF | Fanconi anemia, complementation group F |
2188 | 11p14.3 | − | |||||
| FANCG | Fanconi anemia, complementation group G |
2189 | 9p13.3 | − | − | − | ++ | + | |
| FBXW7 | F-box and WD-40 domain protein 7 (archipelago homolog, Drosophila) |
55294 | 4q31.3 | − | − | − | |||
| FCGR2B | Fc fragment of IgG, low affinity IIb, receptor for (CD32) |
2213 | 1q23 | − | + | ||||
| FEV | FEV protein - (HSRNAFEV) |
54738 | 2q35 | ||||||
| FGFR1 | fibroblast growth factor receptor 1 |
2260 | 8p12 | − | − | − | − | ||
| FGFR1OP | FGFR1 oncogene partner (FOP) |
11116 | 6q27 | + | + | + | |||
| FGFR2 | fibroblast growth factor receptor 2 |
2263 | 10q26.12- q26.13 |
||||||
| FGFR3 | fibroblast growth factor receptor 3 |
2261 | 4p16.3 | ||||||
| FH | fumarate hydratase | 2271 | 1q42.1 | + | − | ||||
| FIP1L1 | FIP1 like 1 (S. cerevisiae) |
81608 | 4q12 | − | − | ||||
| FLCN | folliculin, Birt-Hogg- Dube syndrome |
201163 | 17p11.2 | + | + | + | + | + | |
| FLI1 | Friend leukemia virus integration 1 |
2313 | 11q24.3 | − | − | + | + | − | − |
| FLT3 | fms-related tyrosine kinase 3 |
2322 | 13q12.2 | − | + | − | |||
| FNBP1 | formin binding protein 1 (FBP17) |
23048 | 9q34.11 | + | + | + | + | ||
| FOXO1A | forkhead box O1A (FKHR) |
2308 | 13q14.11 | − | − | ||||
| FOXO3A | forkhead box O3A | 2309 | 6q21 | ||||||
| FOXP1 | forkhead box P1 | 27086 | 3p13 | − | − | − | − | − | − |
| FSTL3 | follistatin-like 3 (secreted glycoprotein) |
10272 | 19p13.3 | ||||||
| FUS | fusion, derived from t(12;16) malignant liposarcoma |
2521 | 16p11.2 | + | |||||
| FVT1 | follicular lymphoma variant translocation 1 |
2531 | 18q21.33 | − | − | − | − | − | |
| GAS7 | growth arrest-specific 7 |
8522 | 17p13.1 | + | + | + | |||
| GATA2 | GATA binding protein 2 |
2624 | 3q21.3 | − | |||||
| GMPS | guanine monphosphate synthetase |
8833 | 3q25.31 | + | + | + | + | ++ | |
| GNAQ | guanine nucleotide binding protein (G protein), q polypeptide |
2776 | 9q21.2 | + | − | + | + | ||
| GNAS | guanine nucleotide binding protein (G protein), alpha stimulating activity polypeptide 1 |
2778 | 20q13.32 | + | + | + | + | + | + |
| GOLGA5 | golgi autoantigen, golgin subfamily a, 5 (PTC5) |
9950 | 14q32.12 | ++ | + | ||||
| GOPC | golgi associated PDZ and coiled-coil motif containing |
57120 | 6q22.1 | + | |||||
| GPHN | gephyrin (GPH) | 10243 | 14q23.3 | + | + | + | + | ||
| HCMOGT-1 | sperm antigen HCMOGT-1 |
92521 | 17p11.2 | + | + | + | + | ||
| HEAB | ATP_GTP binding protein |
10978 | 11q12.1 | − | + | + | + | ||
| HIP1 | huntingtin interacting protein 1 |
3092 | 7q11.23 | + | |||||
| HIST1H4I | histone 1, H4i (H4FM) | 8294 | 6p22.1 | + | + | − | |||
| HLF | hepatic leukemia factor | 3131 | 17q22 | ||||||
| HLXB9 | homeo box HB9 | 3110 | 7q36 | − | − | − | |||
| HMGA1 | high mobility group AT- hook 1 |
3159 | 6p21.31 | + | + | + | |||
| HMGA2 | high mobility group AT- hook 2 (HMGIC) |
8091 | 12q14.3 | ||||||
| HNRNPA2B 1 |
heterogeneous nuclear ribonucleoprotein A2/B1 |
3181 | 7p15.2 | + | + | + | + | + | |
| HOXA11 | homeo box A11 | 3207 | 7p15.2 | + | + | + | + | + | |
| HOXA13 | homeo box A13 | 3209 | 7p15.2 | + | + | + | + | + | |
| HOXA9 | homeo box A9 | 3205 | 7p15.2 | + | + | + | + | + | |
| HOXC11 | homeo box C11 | 3227 | 12q13.13 | + | + | ||||
| HOXC13 | homeo box C13 | 3229 | 12q13.13 | + | + | ||||
| HOXD11 | homeo box D11 | 3237 | 2q31.1 | ||||||
| HOXD13 | homeo box D13 | 3239 | 2q31.1 | ||||||
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog |
3265 | 11p15.5 | ||||||
| HRPT2 | hyperparathyroidism 2 | 3279 | 1q21-q31 | − | |||||
| HSPCA | heat shock 90kDa protein 1, alpha |
3320 | 14q32.31 | ||||||
| HSPCB | heat shock 90kDa protein 1, beta |
3326 | 6p21.1 | + | + | + | − | ||
| IDH1 | isocitrate dehydrogenase 1 (NADP+), soluble |
3417 | 2q33.3 | − | |||||
| IGH@ | immunoglobulin heavy locus |
3492 | 14q32.33 | + | + | ||||
| IGKC | immunoglobulin kappa locus |
50802 | 2p11.2 | ||||||
| IGL@ | immunoglobulin lambda locus |
3535 | 22q11.1- q11.2 |
− | + | + | |||
| IKZF1 | IKAROS family zinc finger 1 |
10320 | 7p12.2 | + | + | + | + | + | |
| IL2 | interleukin 2 | 3558 | 4q27 | − | − | − | |||
| IL21R | interleukin 21 receptor | 50615 | 16p12.1 | + | + | ||||
| IL6ST | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
3572 | 5q11.2 | + | − | ||||
| IRF4 | interferon regulatory factor 4 |
3662 | 6p25.3 | − | + | + | − | ||
| IRTA1 | immunoglobulin superfamily receptor translocation associated 1 |
83417 | 1q23.1 | − | − | + | |||
| ITK | IL2-inducible T-cell kinase |
3702 | 5q33.3 | ||||||
| JAK2 | Janus kinase 2 | 3717 | 9p24.1 | − | − | − | − | + | |
| JAK3 | Janus kinase 3 | 3718 | 19p13.11 | ||||||
| JAZF1 | juxtaposed with another zinc finger gene 1 |
221895 | 7p15.2- p15.1 |
+ | + | + | + | + | |
| KIAA1549 | KIAA1549 | 57670 | 7q34 | − | − | ||||
| KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
3815 | 4q12 | − | − | ||||
| KRAS2 | v-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene homolog |
3845 | 12p12.1 | + | − | − | |||
| KTN1 | kinectin 1 (kinesin receptor) |
3895 | 14q22.3 | + | + | ++ | + | ||
| LAF4 | lymphoid nuclear protein related to AF4 |
3899 | 2q11.2 | + | |||||
| LASP1 | LIM and SH3 protein 1 | 3927 | 17q12 | + | |||||
| LCK | lymphocyte-specific protein tyrosine kinase |
3932 | 1p35- p34.3 |
+ | |||||
| LCP1 | lymphocyte cytosolic protein 1 (L-plastin) |
3936 | 13q14.13 | − | − | ||||
| LHFP | lipoma HMGIC fusion partner |
10186 | 13q13.3 | − | − | ||||
| LIFR | leukemia inhibitory factor receptor |
3977 | 5p13.1 | + | + | + | + | + | |
| LMO1 | LIM domain only 1 (rhombotin 1) (RBTN1) |
4004 | 11p15.4 | ||||||
| LMO2 | LIM domain only 2 (rhombotin-like 1) (RBTN2) |
4005 | 11p13 | + | ++ | ++ | |||
| LPP | LIM domain containing preferred translocation partner in lipoma |
4026 | 3q27.3- q28 |
+ | + | + | + | + | + |
| LYL1 | lymphoblastic leukemia derived sequence 1 |
4066 | 19p13.13 | + | |||||
| MAF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog |
4094 | 16q23.1 | ||||||
| MAFB | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) |
9935 | 20q12 | + | + | + | + | + | + |
| MALT1 | mucosa associated lymphoid tissue lymphoma translocation gene 1 |
10892 | 18q21.31 | − | − | − | − | − | |
| MAML2 | mastermind-like 2 (Drosophila) |
84441 | 11q21 | − | − | − | + | − | + |
| MAP2K4 | mitogen-activated protein kinase kinase 4 |
6416 | 17p11.2 | + | − | + | + | ||
| MDM2 | Mdm2 p53 binding protein homolog |
4193 | 12q15 | ||||||
| MDS1 | myelodysplasia syndrome 1 |
4197 | 3q26.2 | + | + | + | |||
| MDS2 | myelodysplastic syndrome 2 |
259283 | 1p36.12- p36.11 |
− | |||||
| MECT1 | mucoepidermoid translocated 1 |
94159 | 19p13.11 | + | |||||
| MEN1 | multiple endocrine neoplasia type 1 gene |
4221 | 11q13.1 | + | + | + | + | + | + |
| MET | met proto-oncogene (hepatocyte growth factor receptor) |
4233 | 7q31.2 | − | |||||
| MHC2TA | MHC class II transactivator |
4261 | 16p13.13 | + | + | − | + | ||
| MITF | microphthalmia- associated transcription factor |
4286 | 3p13 | − | − | − | − | − | − |
| MKL1 | megakaryoblastic leukemia (translocation) 1 |
57591 | 22q13.1- q13.2 |
||||||
| MLF1 | myeloid leukemia factor 1 |
4291 | 3q25.32 | + | + | + | + | + | |
| MLH1 | E.coli MutL homolog gene |
4292 | 3p22.3 | − | − | − | − | − | |
| MLL | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) |
4297 | 11q23.3 | − | − | + | + | − | − |
| MLLT1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 (ENL) |
4298 | 19p13.3 | + | |||||
| MLLT10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 (AF10) |
8028 | 10p12.31 | − | − | ||||
| MLLT2 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 2 (AF4) |
4299 | 4q21.3 | − | − | − | − | ||
| MLLT3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (AF9) |
4300 | 9p21.3 | − | − | − | − | − | |
| MLLT4 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to,4 (AF6) |
4301 | 6q27 | + | + | + | |||
| MLLT6 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 (AF17) |
4302 | 17q21 | + | |||||
| MN1 | meningioma (disrupted in balanced translocation) 1 |
4330 | 22q12.1 | + | |||||
| MPL | myeloproliferative leukemia virus oncogene, thrombopoietin receptor |
4352 | 1p35.1 | + | |||||
| MSF | MLL septin-like fusion | 10801 | 17q25 | + | + | ||||
| MSH2 | mutS homolog 2 (E. coli) |
4436 | 2p21 | − | |||||
| MSH6 | mutS homolog 6 (E. coli) |
2956 | 2p16.3 | − | |||||
| MSI2 | musashi homolog 2 (Drosophila) |
124540 | 17q23.2 | ||||||
| MUC1 | mucin 1, transmembrane |
4582 | 1q22 | − | − | + | |||
| MUTYH | mutY homolog (E. coli) | 4595 | 1p34.1 | + | |||||
| MYC | v-myc myelocytomatosis viral oncogene homolog (avian) |
4609 | 8q24.21 | + | + | + | + | + | |
| MYCL1 | v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian) |
4610 | 1p34.2 | + | |||||
| MYCN | v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
4613 | 2p24.3 | ||||||
| MYH11 | myosin, heavy polypeptide 11, smooth muscle |
4629 | 16p13.11 | + | − | + | |||
| MYH9 | myosin, heavy polypeptide 9, non- muscle |
4627 | 22q12.3 | ||||||
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 (MORF) |
23522 | 10q22.2 | + | − | ||||
| NACA | nascent-polypeptide- associated complex alpha polypeptide |
4666 | 12q13.3 | + | |||||
| NBS1 | Nijmegen breakage syndrome 1 (nibrin) |
4683 | 8q21.3 | + | + | + | − | ||
| NCKIPSD | SH3 protein interacting with Nck, 90 kDa (ALL1 fused gene from 3p21) |
51517 | 3p21.31 | − | |||||
| NCOA1 | nuclear receptor coactivator 1 |
8648 | 2p23.3 | ||||||
| NCOA2 | nuclear receptor coactivator 2 (TIF2) |
10499 | 8q13.3 | + | + | − | |||
| NCOA4 | nuclear receptor coactivator 4 - PTC3 (ELE1) |
8031 | 10q11.23 | − | |||||
| NF1 | neurofibromatosis type 1 gene |
4763 | 17q11.2 | − | |||||
| NF2 | neurofibromatosis type 2 gene |
4771 | 22q12.2 | + | |||||
| NFKB2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
4791 | 10q24.32 | + | |||||
| NIN | ninein (GSK3B interacting protein) |
51199 | 14q22.1 | + | + | + | |||
| NOTCH1 | Notch homolog 1, translocation- associated (Drosophila) (TAN1) |
4851 | 9q34.3 | + | + | + | + | ||
| NPM1 | nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
4869 | 5q35.1 | + | + | ||||
| NR4A3 | nuclear receptor subfamily 4, group A, member 3 (NOR1) |
8013 | 9q31.1 | + | + | + | |||
| NRAS | neuroblastoma RAS viral (v-ras) oncogene homolog |
4893 | 1p13.2 | + | |||||
| NSD1 | nuclear receptor binding SET domain protein 1 |
64324 | 5q35.2- q35.3 |
+ | + | ||||
| NTRK1 | neurotrophic tyrosine kinase, receptor, type 1 |
4914 | 1q23.1 | − | − | + | |||
| NTRK3 | neurotrophic tyrosine kinase, receptor, type 3 |
4916 | 15q25.3 | − | − | ||||
| NUMA1 | nuclear mitotic apparatus protein 1 |
4926 | 11q13.4 | + | ++ | ++ | + | − | + |
| NUP214 | nucleoporin 214kDa (CAN) |
8021 | 9q34.13 | + | + | + | + | ||
| NUP98 | nucleoporin 98kDa | 4928 | 11p15.4 | ||||||
| NUT | nuclear protien in testis | 256646 | 15q14 | − | − | ||||
| OLIG2 | oligodendrocyte lineage transcription factor 2 (BHLHB1) |
10215 | 21q22.11 | − | + | ||||
| OMD | osteomodulin | 4958 | 9q22.31 | + | + | + | + | ||
| PAFAH1B2 | platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa |
5049 | 11q23.3 | − | − | + | + | − | − |
| PALB2 | partner and localizer of BRCA2 |
79728 | 16p12.1 | + | + | ||||
| PAX3 | paired box gene 3 | 5077 | 2q36.1 | ||||||
| PAX5 | paired box gene 5 (B- cell lineage specific activator protein) |
5079 | 9p13.2 | − | − | − | − | + | |
| PAX7 | paired box gene 7 | 5081 | 1p36.13 | + | − | ||||
| PAX8 | paired box gene 8 | 7849 | 2q13 | ||||||
| PBX1 | pre-B-cell leukemia transcription factor 1 |
5087 | 1q23.3 | − | + | − | |||
| PCM1 | pericentriolar material 1 (PTC4) |
5108 | 8p22- p21.3 |
− | − | − | ++ | − | |
| PCSK7 | proprotein convertase subtilisin/kexin type 7 |
9159 | 11q23.3 | − | − | + | + | − | − |
| PDE4DIP | phosphodiesterase 4D interacting protein (myomegalin) |
9659 | 1q21.1 | − | − | + | |||
| PDGFB | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) |
5155 | 22q13.1 | ||||||
| PDGFRA | platelet-derived growth factor, alpha-receptor |
5156 | 4q12 | − | − | ||||
| PDGFRB | platelet-derived growth factor receptor, beta polypeptide |
5159 | 5q32 | + | − | ||||
| PER1 | period homolog 1 (Drosophila) |
5187 | 17p13.1 | + | + | + | |||
| PHOX2B | paired-like homeobox 2b |
8929 | 4p13 | − | − | + | |||
| PICALM | phosphatidylinositol binding clathrin assembly protein (CALM) |
8301 | 11q14.2 | − | − | − | + | − | + |
| PIK3CA | phosphoinositide-3- kinase, catalytic, alpha polypeptide |
5290 | 3q26.32 | + | + | + | + | ||
| PIK3R1 | phosphoinositide-3- kinase, regulatory subunit 1 (alpha) |
5295 | 5q13.1 | ||||||
| PIM1 | pim-1 oncogene | 5292 | 6p21.2 | + | + | + | − | ||
| PLAG1 | pleiomorphic adenoma gene 1 |
5324 | 8q12.1 | + | + | + | |||
| PML | promyelocytic leukemia |
5371 | 15q24.1 | + | − | − | |||
| PMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
5378 | 2q32.2 | − | |||||
| PMS2 | PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
5395 | 7p22.1 | + | + | + | + | + | + |
| PNUTL1 | peanut-like 1 (Drosophila) |
5413 | 22q11.21 | + | + | ||||
| POU2AF1 | POU domain, class 2, associating factor 1 (OBF1) |
5450 | 11q23.1 | − | − | + | − | − | |
| POU5F1 | POU domain, class 5, transcription factor 1 |
5460 | 6p21.33 | + | + | ||||
| PPARG | peroxisome proliferative activated receptor, gamma |
5468 | 3p25.2 | − | − | − | − | ||
| PRCC | papillary renal cell carcinoma (translocation- associated) |
5546 | 1q23.1 | − | − | + | |||
| PRDM16 | PR domain containing 16 |
63976 | 1p36.32 | + | |||||
| PRKAR1A | protein kinase, cAMP- dependent, regulatory, type I, alpha (tissue specific extinguisher 1) |
5573 | 17q24.2 | + | |||||
| PRO1073 | PRO1073 protein (ALPHA) |
29005 | 11q13.1 | + | + | + | + | + | + |
| PRRX1 | paired mesoderm homeo box 1 |
5396 | 1q24 .2 | + | + | − | |||
| PSIP1 | PC4 and SFRS1 interacting protein 1 (LEDGF) |
11168 | 9p22.3 | − | − | − | − | − | |
| PTCH | Homolog of Drosophila Patched gene |
5727 | 9q22.32 | + | + | + | + | ||
| PTEN | phosphatase and tensin homolog gene |
5728 | 10q23.31 | − | |||||
| PTPN11 | protein tyrosine phosphatase, non- receptor type 11 |
5781 | 12q24.13 | + | + | − | |||
| RABEP1 | rabaptin, RAB GTPase binding effector protein 1 (RABPT5) |
9135 | 17p13.2 | + | + | + | |||
| RAD51L1 | RAD51-like 1 (S. cerevisiae) (RAD51B) |
5890 | 14q24.1 | + | + | + | + | + | |
| RANBP17 | RAN binding protein 17 |
64901 | 5q35.1 | + | + | ||||
| RAP1GDS1 | RAP1, GTP-GDP dissociation stimulator 1 |
5910 | 4q23 | − | − | − | |||
| RARA | retinoic acid receptor, alpha |
5914 | 17q21.2 | + | |||||
| RB1 | retinoblastoma gene | 5925 | 13q14.2 | − | − | ||||
| RBM15 | RNA binding motif protein 15 |
64783 | 1p13.3 | ++ | |||||
| RECQL4 | RecQ protein-like 4 | 9401 | 8q24.3 | + | + | + | + | + | |
| REL | v-rel reticuloendotheliosis viral oncogene homolog (avian) |
5966 | 2p16.1 | ||||||
| RET | ret proto-oncogene | 5979 | 10q11.21 | − | |||||
| RHOH | RAS homolog gene family, member H (TTF) |
399 | 4p14 | − | − | + | |||
| ROS1 | v-ros UR2 sarcoma virus oncogene homolog 1 (avian) |
6098 | 6q22.1 | ||||||
| RPL22 | ribosomal protein L22 (EAP) |
6146 | 1p36.31 | + | |||||
| RPN1 | ribophorin I | 6184 | 3q21.3 | − | |||||
| RUNX1 | runt-related transcription factor 1 (AML1) |
861 | 21q22.12 | − | + | − | |||
| RUNXBP2 | runt-related transcription factor binding protein 2 (MOZ/ZNF220) |
7994 | 8p11.21 | − | − | + | − | − | |
| SBDS | Shwachman-Bodian- Diamond syndrome protein |
51119 | 7q11.21 | + | + | ||||
| SDHB | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
6390 | 1p36.13 | + | − | ||||
| SDHC | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
6391 | 1q23.3 | − | + | ||||
| SDHD | succinate dehydrogenase complex, subunit D, integral membrane protein |
6392 | 11q23.1 | − | − | + | − | − | |
| SET | SET translocation | 6418 | 9q33.2 | + | + | + | + | ||
| SFPQ | splicing factor proline/glutamine rich(polypyrimidine tract binding protein associated) |
6421 | 1p34.3 | + | |||||
| SFRS3 | splicing factor, arginine/serine-rich 3 |
6428 | 6p21.31 | + | + | + | |||
| SH3GL1 | SH3-domain GRB2- like 1 (EEN) |
6455 | 19p13.3 | ||||||
| SIL | TAL1 (SCL) interrupting locus |
6491 | 1p33 | + | |||||
| SLC45A3 | solute carrier family 45, member 3 |
85414 | 1q32.1 | + | + | + | − | ||
| SMAD4 | Homolog of Drosophila Mothers Against Decapentaplegic 4 gene |
4089 | 18q21.1 | − | − | − | − | − | |
| SMARCB1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
6598 | 22q11.23 | − | + | + | |||
| SMO | smoothened homolog (Drosophila) |
6608 | 7q32.1 | − | |||||
| SOCS1 | suppressor of cytokine signaling 1 |
8651 | 16p13.13 | + | + | − | + | ||
| SS18 | synovial sarcoma translocation, chromosome 18 |
6760 | 18q11.2 | − | + | − | |||
| SS18L1 | synovial sarcoma translocation gene on chromosome 18-like 1 |
26039 | 20q13.33 | + | + | + | + | + | + |
| STK11 | serine/threonine kinase 11 gene (LKB1) |
6794 | 19p13.3 | ||||||
| STL | Six-twelve leukemia gene |
7955 | 6q22.31 | + | |||||
| SUFU | suppressor of fused homolog (Drosophila) |
51684 | 10q24.32 | + | |||||
| SUZ12 | suppressor of zeste 12 homolog (Drosophila) |
23512 | 17q11.2 | − | − | ||||
| SYK | spleen tyrosine kinase | 6850 | 9q22.2 | + | − | + | + | + | |
| TAF15 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
8148 | 17q12 | − | − | ||||
| TAL1 | T-cell acute lymphocytic leukemia 1 (SCL) |
6886 | 1p33 | + | |||||
| TAL2 | T-cell acute lymphocytic leukemia 2 |
6887 | 9q31.2 | + | + | + | + | ||
| TCEA1 | transcription elongation factor A (SII), 1 |
6917 | 8q11.23 | + | + | + | |||
| TCF1 | transcription factor 1, hepatic (HNF1) |
6927 | 12q24.31 | − | + | + | |||
| TCF12 | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) |
6938 | 15q21.3 | − | − | ||||
| TCF3 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) |
6929 | 19p13.3 | ||||||
| TCL1A | T-cell leukemia/lymphoma 1A |
8115 | 14q32.13 | ++ | + | − | |||
| TCL6 | T-cell leukemia/lymphoma 6 |
27004 | 14q32.13 | ++ | + | − | |||
| TFEB | transcription factor EB | 7942 | 6p21.1 | + | + | + | − | ||
| TFG | TRK-fused gene | 10342 | 3q12.2 | + | |||||
| TFPT | TCF3 (E2A) fusion partner (in childhood Leukemia) |
29844 | 19q13.42 | + | + | + | |||
| TFRC | transferrin receptor (p90, CD71) |
7037 | 3q29 | + | + | + | + | + | + |
| THRAP3 | thyroid hormone receptor associated protein 3 (TRAP150) |
9967 | 1p34.3 | + | |||||
| TIF1 | transcriptional intermediary factor 1 (PTC6,TIF1A) |
8805 | 7q34 | − | |||||
| TLX1 | T-cell leukemia, homeobox 1 (HOX11) |
3195 | 10q24.31 | + | |||||
| TLX3 | T-cell leukemia, homeobox 3 (HOX11L2) |
30012 | 5q35.1 | + | + | ||||
| TMPRSS2 | transmembrane protease, serine 2 |
7113 | 21q22.3 | − | −− | − | − | ||
| TNFRSF17 | tumor necrosis factor receptor superfamily, member 17 |
608 | 16p13.13 | + | + | − | + | ||
| TNFRSF6 | tumor necrosis factor receptor superfamily, member 6 (FAS) |
355 | 10q23.31 | − | |||||
| TOP1 | topoisomerase (DNA) I | 7150 | 20q12 | + | + | + | + | + | + |
| TP53 | tumor protein p53 | 7157 | 17p13.1 | + | + | + | |||
| TPM3 | tropomyosin 3 | 7170 | 1q21.3 | − | − | + | |||
| TPM4 | tropomyosin 4 | 7171 | 19p13.12 | ||||||
| TPR | translocated promoter region |
7175 | 1q31.1 | − | |||||
| TRA@ | T cell receptor alpha locus |
6955 | 14q11.2 | + | + | ||||
| TRBC1 | T cell receptor beta locus |
6957 | 7q34 | − | − | − | |||
| TRD@ | T cell receptor delta locus |
6964 | 14q11.2 | + | + | ||||
| TRIM33 | tripartite motif- containing 33 (PTC7,TIF1G) |
51592 | 1p13.2 | + | |||||
| TRIP11 | thyroid hormone receptor interactor 11 |
9321 | 14q32.12 | + | + | ||||
| TSC1 | tuberous sclerosis 1 gene |
7248 | 9q34.13 | + | + | + | + | ||
| TSC2 | tuberous sclerosis 2 gene |
7249 | 16p13.3 | + | + | + | |||
| TSHR | thyroid stimulating hormone receptor |
7253 | 14q31.1 | + | + | + | |||
| TTL | tubulin tyrosine ligase | 150465 | 2q13 | ||||||
| USP6 | ubiquitin specific peptidase 6 (Tre-2 oncogene) |
9098 | 17p13.2 | + | + | + | |||
| VHL | von Hippel-Lindau syndrome gene |
7428 | 3p25.3 | − | − | − | − | ||
| WHSC1 | Wolf-Hirschhorn syndrome candidate 1(MMSET) |
7468 | 4p16.3 | ||||||
| WHSC1L1 | Wolf-Hirschhorn syndrome candidate 1- like 1 (NSD3) |
54904 | 8p12 | − | − | − | − | ||
| WRN | Werner syndrome (RECQL2) |
7486 | 8p12 | − | − | − | − | − | |
| WT1 | Wilms tumour 1 gene | 7490 | 11p13 | + | ++ | + | |||
| XPA | xeroderma pigmentosum, complementation group A |
7507 | 9q22.33 | + | + | + | + | ||
| XPC | xeroderma pigmentosum, complementation group C |
7508 | 3p25.1 | − | − | − | − | ||
| ZNF145 | zinc finger protein 145 (PLZF) |
7704 | 11q23.2 | − | − | + | − | − | |
| ZNF198 | zinc finger protein 198 | 7750 | 13q12.11 | − | + | ||||
| ZNF278 | zinc finger protein 278 (ZSG) |
23598 | 22q12.2 | + | |||||
| ZNF331 | zinc finger protein 331 | 55422 | 19q13.42 | + | + | ||||
| ZNF384 | zinc finger protein 384 (CIZ/NMP4) |
171017 | 12p13.31 | + | − | − | |||
| ZNF521 | zinc finger protein 521 | 25925 | 18q11.2 | − | + | − | |||
| ZNF9 | zinc finger protein 9 (a cellular retroviral nucleic acid binding protein) |
7555 | 3q21.3 | − | |||||
| ZNFN1A1 | zinc finger protein, subfamily 1A, 1 (Ikaros) |
10320 | 7p12.2 | + | + | + | + | + |
Symbols: +, low-level copy number gain; ++, high-level copy number gain; −, low-level copy number loss; −−, high-level copy number loss.
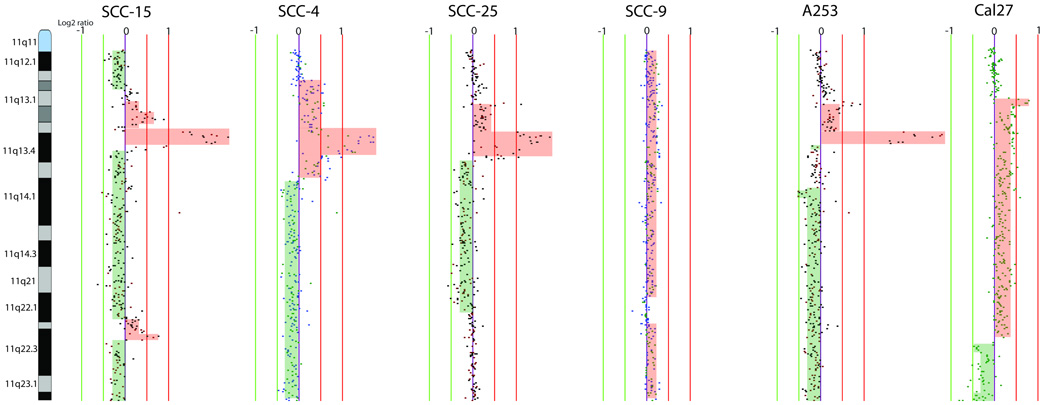
It is known that head and neck cancers, like many solid tumours, are a molecularly heterogeneous group where several regions of alteration and genes have been reported at high frequencies but may occur in various combinations in a given tumour. Genetic alterations frequently occurring in tumours indicate an importance for carcinogenesis. Loss of chromosome 3p and 9p are genetic events frequently documented in head and neck tumours, and both have been implicated as one of the earliest changes in oral premalignant lesions and associated with progression risks.(6, 9) All six cell lines revealed segmental losses of chromosome 3p and 9p (detailed regions of genetic alterations are listed in Supplemental Tables S1 to S6). Cal27 revealed only one region of segmental loss of 9p24.1-p23, while the remaining five cell lines exhibited copy number loss at 9p21.3, which contain the tumour suppressor CDKN2A. Genetic loss of chromosome 8p is also an expected change that has been frequently described in clinical specimens.(26, 27) Whole arm 8p loss was found in SCC-15, SCC-9, Cal27, while A253 exhibited a region of homozygous deletion on 8p23.2-p23.2 and a region of high-level DNA amplification on 8p22. Gain of chromosome 8q is also a frequent genetic alteration in head and neck tumours, harbouring the known oncogene MYC.(26) Cell lines A253, SCC-15, SCC-4, SCC-25, and SCC-9 all showed low level gain of 8q24 (MYC). Regions of high-level amplification, including 3q26, 7p11 and 11q13, occur frequently in head and neck tumours.(26–30) These regions have been associated with PIK3CA, EGFR, and CCND1 respectively. SCC-4 and SCC-9 did not reveal genetic gain of 3q26.32 whereas the lines A253, SCC-15, and SCC-25 showed low level gains. Cell lines SCC-15 and Cal27 exhibited region of high-level amplification at the EGFR locus, and cell lines SCC-4, SCC-9, and A-253 exhibited low-level copy number gain. Cell lines SCC-9 and Cal27 showed genetic gain of the CCND1 locus, whereas four cell lines A253, SCC-15, SCC-4, and SCC-25 displayed high-level amplification of this locus. Complex alterations on chromosome arm 11q were revealed SCC-15, SCC-4, SCC-25, and A253 (Fig. 2).
Figure 2.
Multiple levels of segmental copy number alterations are detected in six cell lines on chromosome 11q. Each BAC clone is displayed as vertical line representing its genomic coverage. Data points to the left and right of the centre line (purple) represent DNA copy number losses and gains, respectively. Regions of gain are highlighted in red and loss is shaded in green.
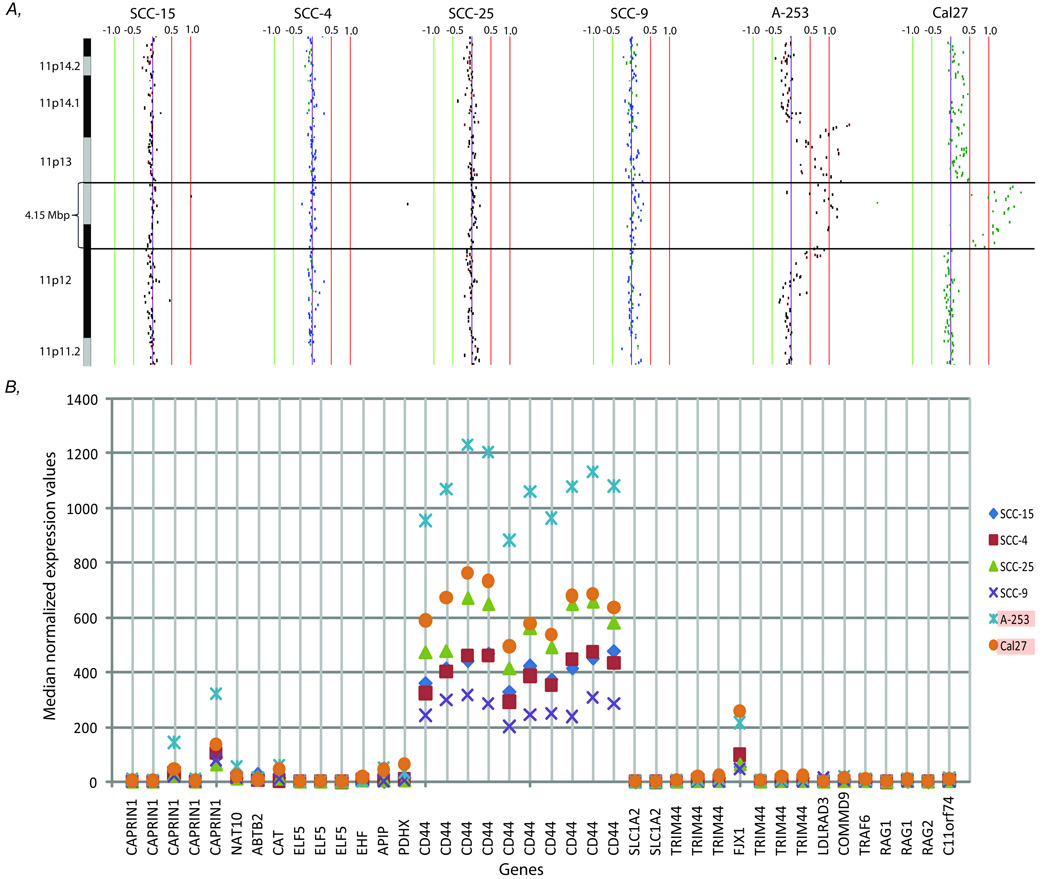
One common characteristic of the cell lines is the presence of high-level amplifications, where many of the lines contained multiple regions of high-level DNA amplification. This is similar to tumour genomes where it has been reported that high-level amplification is observed in ~65% of cases, although usually only one or two regions per tumour.(26) High-level amplifications indicating multiple copy numbers and regions of homozygous deletion, where both copies are lost, often result in gene overexpression and underexpression.(21, 22, 31) These types of alteration often arise under selective pressure of genes important for the growth of cancer cells. Therefore we focused our analysis on the expression of genes within these regions. The expression levels of genes within all the regions of high-level DNA amplification and homozygous deletion detected in the six cell lines are presented in Supplemental figure S2. In general regions of homozygous deletion often resulted in lower expression levels for many of the genes within the region, while higher level of expression was often observed in at least one gene within an amplicon compared to cell lines without such amplicon. For example, a common region of high-level amplification of 4.15 Mbp in size on 11p13-p12 was detected in Cal27 and A-253, which contains 20 RefSeq genes (Fig. 3). The expression of 17 of these genes were analyzed, and the gene CD44 has a two-fold increase in expression compared to the other four cell lines without such high-level amplification. Other genes inside this amplicon, PDHX and CAT, also have a 5-fold increase in expression. The expression of CD44 is also among the top five-percentile of all expressed genes in the six cell lines, suggesting that while high-level DNA amplification causes higher expression of the gene, other mechanisms might also cause its high expression in cell lines without such amplicon. However, as there are many ways to regulate gene expression, some of the amplified regions do not show the expected changes in gene expression. Further molecular examinations of epigenetic alterations and sequence analyses would be essential to fully characterize the different molecular aspects of each cell line.
Figure 3.
Integrative analysis of genetic and expression levels of genes within 11p13-p12 amplicon. A, Alignment of chromosome region 11p for six cell lines. Each BAC clone is displayed as vertical line representing its genomic coverage. Minimal region of DNA amplification of A-253 and Cal27 is marked by two black lines and is 4.15 Mbp in size. B, The expression level of 17 genes within this amplicon is analyzed by expression arrays. For each cell line, median-normalized expression values are represented on the y-axis and the corresponding gene is listed on the x-axis.
CONCLUSIONS
Head and neck cancer cell lines are important model systems for investigating head and neck cancer biology. Comprehensive characterization of their genetic alterations is useful for the selection and interpretation of studies using cell lines. Our data presents the first comprehensive catalogue of copy number and expression alterations in six commonly used and easily obtainable oral cancer cell lines, thus providing a good resource for researchers to select and use these cell lines in experiments.
Supplementary Material
Whole genome copy number karyogram of each cell line. SeeGH software was used to plot the log2 signal intensity ratios of cell line sample versus normal reference male genomic DNA at each BAC. Discussion of each cell line is described underneath the figure.
Scatter plots of median-normalized expression values for genes within regions of high-level copy number change. The median-normalized expression values are plotted on the y-axis, and the corresponding gene for each probe on the Agilent Whole Human Genome Microarray 4×44K microarray are plotted on the x-axis. The regions of high-level copy number alteration are detailed in Supplemental Tables S1 to S6.
Acknowledgements
This work was supported by grants from the National Institute of Dental and Craniofacial Research/National Institutes of Health (R01 DE17013), Canadian Institutes of Health Research, and funds from the Pacific Otolaryngology Foundation. IFLT is supported by scholarships from the Canadian Institutes of Health Research (CIHR) and the Michael Smith Foundation for Health Research (MSFHR). We would like to acknowledge Drs Wan Lam and Timon Buys for helpful discussion.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Lin CJ, Grandis JR, Carey TE, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29(2):163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 3.Martin CL, Reshmi SC, Ried T, et al. Chromosomal imbalances in oral squamous cell carcinoma: examination of 31 cell lines and review of the literature. Oral Oncol. 2008;44(4):369–382. doi: 10.1016/j.oraloncology.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh B. Molecular pathogenesis of head and neck cancers. J Surg Oncol. 2008;97(8):634–639. doi: 10.1002/jso.21024. [DOI] [PubMed] [Google Scholar]
- 5.Califano J, van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56(11):2488–2492. [PubMed] [Google Scholar]
- 6.Mao L, Lee JS, Fan YH, et al. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med. 1996;2(6):682–685. doi: 10.1038/nm0696-682. [DOI] [PubMed] [Google Scholar]
- 7.Rosin MP, Cheng X, Poh C, et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res. 2000;6(2):357–362. [PubMed] [Google Scholar]
- 8.Tsui IF, Rosin MP, Zhang L, Ng RT, Lam WL. Multiple aberrations of chromosome 3p detected in oral premalignant lesions. Cancer Prev Res (Phila Pa) 2008;1(6):424–429. doi: 10.1158/1940-6207.CAPR-08-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell. 2004;5(4):311–316. doi: 10.1016/s1535-6108(04)00090-x. [DOI] [PubMed] [Google Scholar]
- 10.Akervall JA, Michalides RJ, Mineta H, et al. Amplification of cyclin D1 in squamous cell carcinoma of the head and neck and the prognostic value of chromosomal abnormalities and cyclin D1 overexpression. Cancer. 1997;79(2):380–389. [PubMed] [Google Scholar]
- 11.Nakahara Y, Shintani S, Mihara M, Ueyama Y, Matsumura T. High frequency of homozygous deletion and methylation of p16(INK4A) gene in oral squamous cell carcinomas. Cancer Lett. 2001;163(2):221–228. doi: 10.1016/s0304-3835(00)00699-6. [DOI] [PubMed] [Google Scholar]
- 12.Jarvinen AK, Autio R, Kilpinen S, et al. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer. 2008;47(6):500–509. doi: 10.1002/gcc.20551. [DOI] [PubMed] [Google Scholar]
- 13.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41(5):1657–1663. [PubMed] [Google Scholar]
- 14.Gioanni J, Fischel JL, Lambert JC, et al. Two new human tumor cell lines derived from squamous cell carcinomas of the tongue: establishment, characterization and response to cytotoxic treatment. Eur J Cancer Clin Oncol. 1988;24(9):1445–1455. doi: 10.1016/0277-5379(88)90335-5. [DOI] [PubMed] [Google Scholar]
- 15.Giard DJ, Aaronson SA, Todaro GJ, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51(5):1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 16.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 17.Khojasteh M, Lam WL, Ward RK, MacAulay C. A stepwise framework for the normalization of array CGH data. BMC Bioinformatics. 2005;6:274. doi: 10.1186/1471-2105-6-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi B, deLeeuw RJ, Coe BP, Ng RT, MacAulay C, Lam WL. MD-SeeGH: a platform for integrative analysis of multi-dimensional genomic data. BMC Bioinformatics. 2008;9:243. doi: 10.1186/1471-2105-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20(18):3636–3637. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- 20.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 21.Lockwood WW, Chari R, Coe BP, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27(33):4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271(5247):350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 23.Snijders AM, Schmidt BL, Fridlyand J, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24(26):4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 24.Haber DA, Settleman J. Cancer: drivers and passengers. Nature. 2007;446(7132):145–146. doi: 10.1038/446145a. [DOI] [PubMed] [Google Scholar]
- 25.Futreal PA, Coin L, M Marshall, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldwin C, Garnis C, Zhang L, Rosin MP, Lam WL. Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 2005;65(17):7561–7567. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- 27.Smeets SJ, Braakhuis BJ, Abbas S, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25(17):2558–2564. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 28.Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, du Manoir S. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001;61(10):4122–4129. [PubMed] [Google Scholar]
- 29.Huang X, Godfrey TE, Gooding WE, McCarty KS, Jr, Gollin SM. Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer. 2006;45(11):1058–1069. doi: 10.1002/gcc.20371. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Gollin SM, Raja S, Godfrey TE. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci U S A. 2002;99(17):11369–11374. doi: 10.1073/pnas.172285799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22(8):447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole genome copy number karyogram of each cell line. SeeGH software was used to plot the log2 signal intensity ratios of cell line sample versus normal reference male genomic DNA at each BAC. Discussion of each cell line is described underneath the figure.
Scatter plots of median-normalized expression values for genes within regions of high-level copy number change. The median-normalized expression values are plotted on the y-axis, and the corresponding gene for each probe on the Agilent Whole Human Genome Microarray 4×44K microarray are plotted on the x-axis. The regions of high-level copy number alteration are detailed in Supplemental Tables S1 to S6.