Abstract
The ubiquitously expressed basic helix-loop-helix (bHLH) transcription factors E12 and E47, products of alternative splicing of the E2A/TCF3 gene, regulate diverse biological processes including cell growth, differentiation and development. To search for novel protein interactions for E12, we utilized the bHLH domain of E12 as a bait in yeast two-hybrid screening. Yeast two-hybrid, mammalian two-hybrid, and co-immunoprecipitation analyses demonstrate specific interaction of E12 with RANBP17, a novel member of the importin-β superfamily; this interaction maps to the CRM1 homology region of RANBP17. Ectopic expression of RANBP17 leads to a ~3-fold increase in E2A/MyoD mediated transactivation of an E-box regulated luciferase reporter gene. Interaction and transactivation studies also revealed similar functions for RANBP16/XPO7. Furthermore, ectopic expression of either RANBP16 or RANBP17 resulted in increased level of endogenous transcript for the cyclin-dependent kinase inhibitor, p21Waf1/Cip1, a well-characterized E2A target gene. Together, these biochemical and functional data reveal RANBP16 and RANBP17 as novel regulators of E2A protein action.
Keywords: bHLH Transcription Factor, E2A Proteins (E12 and E47), RANBP17, RANBP16, E-box, Yeast Two-Hybrid, Nucleocytoplasmic Transport, CRM1
INTRODUCTION
The basic helix-loop-helix (bHLH) family of transcription factors regulates gene transcription during cell differentiation, proliferation, lineage commitment, and neoplastic transformation in a variety of cell types [Desprez et al., 2003; Norton, 2000; Perry and Soreq, 2002]. Members of this family are defined by the bHLH domain near their C-termini that is comprised of two functionally distinct regions. The basic domain is rich in positively charged amino acids critical for DNA binding while the HLH region governs dimerization with other bHLH family members. The DNA sequence bound by bHLH proteins is the hexanucleotide E-box element (5'-CANNTG-3'), present in a variety of tissue-specific enhancers [Funk et al., 1991; Greenbaum and Zhuang, 2002; Murre et al., 1989; Yutzey and Konieczny, 1992]. The E2A proteins, E12 and E47, are products of the E2A gene generated by alternate splicing. As such, E12 and E47 possess distinct, albeit highly conserved, sequences in their respective bHLH domains. Given their ubiquitous expression, E12 and E47 are classified as class I bHLH transcription factors [Hikima et al., 2005; Massari and Murre, 2000; Murre et al., 1994 ]. E12 and E47 homo- or heterodimerize with other bHLH transcription factors [Lassar et al., 1991; Massari and Murre, 2000; Quong et al., 2002]; cell type selectivity of bHLH protein action is conferred by tissue restricted bHLH factor(s) that serve as dimerization partners for the E2A proteins [Massari and Murre, 2000]. These include the myogenic regulatory factors MyoD, MRF-4, Myf-5, and myogenin [French et al., 1991; Lassar et al., 1991; Venuti and Cserjesi, 1996]; BETA2/NeuroD that function in neurogenesis, pancreatic development and insulin gene expression [Glick et al., 2000; Lee, 1997; Mutoh et al., 1997]; and SCL/Tal1 in hematopoietic lineage development [Gould and Bresnick, 1998; Hsu et al., 1994]. The action of bHLH factors is also controlled by the dominant negative regulatory Id proteins (Id1-Id4), a distinct group of HLH factors that lack a basic region but possess an HLH dimerization domain [Benezra et al., 1990; Langlands et al., 1997]. Furthermore, the physiological specificity of transcriptional activation by bHLH factors is not solely dependent on their intrinsic DNA binding specificities but also involves cooperative interactions with other components of the transcriptional machinery [Eckner et al., 1996; Massari et al., 1999; Turner et al., 2004].
In regard to protein interactions of bHLH factors, bHLH-bHLH dimerization among bHLH protein family members is perhaps the best studied [Massari and Murre, 2000]. However, there is ample evidence that interactions of E2A proteins are not solely with other members of the bHLH family, nor only within their HLH regions. A number of non-bHLH interaction partners for E2A proteins have been described. For example, E2A proteins interacts with p300/CBP and SAGA histone acetyltransferase complex [Bayly et al., 2004; Eckner et al., 1996; Massari et al., 1999]; the MAPK-activated protein kinases 3pK and MK2 interact with E47 to repress E-box-mediated transcriptional activity [Neufeld et al., 2000]; calmodulin binds the basic region of E2A proteins to inhibit DNA binding [Saarikettu et al., 2004]; and a C-terminal 31-kDa caspase-mediated cleavage product of p130cas, which contains high sequence homology with Id proteins, heterodimerizes with E2A proteins to result in inhibition of p21Waf1/Cip1 transcription [Kim et al., 2004]. E2A protein-protein interactions can thus serve as key integration points for a multitude of intracellular signals that reflect and/or control overall cellular status in a variety of biological settings.
Transport between nucleus and cytoplasm through the nuclear pore complex is primarily mediated by importin β-related factors which represent the largest class of nuclear transport receptors [Cook et al., 2007; Kutay et al., 2000]. The common features of importin β-related transport receptors include a molecular weight in the range of ~90 to ~130 kDa and the presence of a conserved region for RanGTP binding in their N-termini [Kutay et al., 2000; Yoneda, 2000]. The closely related proteins RANBP16 (also termed XPO7) and RANBP17 show an overall 67% amino acid identity and phylogenetic analysis has determined that they are the most evolutionarily distant members of the importin β superfamily [Kutay et al., 2000]. The N-terminal importin β domain of RANBP17 is located at amino acids 30-95, which is found within a larger region of homology with the CRM1 nuclear export protein, at amino acids 8-167 of RANBP17. Northern blot analysis of polyA+ RNA has led to RANBP17 transcript(s) expression described as testis-enriched in mouse and human tissue samples [Koch et al., 2000]. However, Northern blot analyses have also detected RANBP17 transcript(s) in human heart, liver, kidney, pancreas and placenta tissues, and in erythroid HEL and the megakaryocyte cell lines Meg01 and M07E [Bernard et al., 2001; MacLeod et al., 2003].
While the role of RANBP17 in nuclear transport has yet to be reported, that of RANBP16/XPO7 has been addressed, albeit in only a few studies [Kutay et al., 2000; Mingot et al., 2004]. Gorlich and colleagues showed that RANBP16 has exportin function, with likely broad substrate specificities [Mingot et al., 2004]. However, Bischoff and coworkers observed an intermediate dissociation constant for RanGTP hydrolysis and could therefore not define RANBP16 specifically as an importin or exportin [Kutay et al., 2000]. They suggested it may have a unique function in bidirectional cargo transport at the nuclear pore [Kutay et al., 2000]. Given that RANBP16 did not recognize proteins that contain the classical type of basic nuclear import signal, but rather interacted with positively charged regions of several types of targets, it has been suggested that RANBP16 may be a component of a novel type of nuclear transport mechanism [Mingot et al., 2004]. The only additional information on RANBP16/XPO7 function is a recent report on its role in the localization of LKB1, a serine/threonine kinase that regulates cell polarity, metabolism, and growth. STRADα induces relocalization of LKB1 from the nucleus to the cytoplasm and stimulates its catalytic activity. RANBP16/XPO7 associates with STRADα to impact LKB1 localization [Dorfman and Macara, 2008]. Direct interaction of STRADα with RANBP16/XPO7 was reported in this study, however RANBP17 was reported as negative for interaction with STRADα.
In a search for novel protein-protein interactions for the bHLH transcription factor E12, we conducted yeast two-hybrid screening and identified RANBP17. Here we carry out studies that demonstrate that RANBP17, and the closely related RANBP16, are novel E2A binding partners that can function to enhance transcriptional activation mediated by E2A proteins.
MATERIALS AND METHODS
Cloning and Expression Constructs
E2A protein expression constructs
The E12 bHLH region bait used in yeast two hybrid screening contains amino acids 508-654 of human E12 in the pAS Gal4 DNA binding domain fusion vector (Clontech Corp.). For mammalian two hybrid assay, the same sequence of E12 and RANBP17 present in the yeast two hybrid constructs were subcloned into the mammalian two hybrid expression vectors, pVP16 and pM (Clontech Corp.), such that they maintained reading frame with the VP16 transactivating and the GAL4-DNA binding domains, respectively. A cDNA clone for full-length human E12 was obtained from Dr. C. Murre (Harvard University) and subcloned into pcDNA3.1+ (Invitrogen Corp.). A full-length cDNA for human E47 (GenBank accession number BC110579) was purchased from Open Biosystems, sequence verified, and the insert subcloned into pcDNA3.1+. PCR-based cloning incorporating EcoRI and Sal I restriction sites was used to generate an expression construct for amino acids 502-654 of E12 in the pCMV-Myc mammalian expression plasmid (Clontech Corp.) such that a start codon was provided with the epitope tag. Primer sequences used were for this were: 5’-GCCGAATTCAAGGAGGACGAGGAGAA CACG-3’ and 5’-GGCGTCGACTCACATGTGCCCGGCGGGGTT-3’
RANBP16 and RANBP17 expression constructs
Mammalian expression constructs for full-length human RANBP16 and full-length human RANBP17 that contained three copies of an HA tag at their respective N-termini, RANBP16-pMT2SM and RANBP17-pMT2SM, referred to herein as HA-RANBP17 and HA-RANBP16, were a kind gift from Dr. W.G. Janssen (University of Heidelberg, Germany). During the sequencing of RANBP17-pMT2SM, we found that a single amino acid differed from the GenBank reference sequence for human RANBP17, NM 022897.2. We corrected the RANBP17 sequence to match the RANBP17 GenBank reference sequence. An I.M.A.G.E. cDNA clone for the short form of human RANBP17 (sRANBP17) in the mammalian expression vector pCMV-SPORT6 (GenBank accession number BI824159) was obtained from the American Type Culture Collection (ATCC). HA-RANBP17-CRM1 was constructed in the pCMV-HA vector (Clontech Corp.) utilizing PCR-based cloning with primers that incorporated EcoRI and Kpn I cloning sites: 5’-GCCCGAATTCTGGCGCTGCACTTCCAGAGTTTG-3’ and 5’-GGCGGTACCTCATGTTCTCCAAGTTGTTGGAATCTG-3’
Luciferase Reporter Constructs
An E-box regulated firefly luciferase reporter construct, termed E-box-pLuc, was generated via insertion of three copies of the consensus E-box element, CAGGTG just 5’ to an SV40 minimal promoter in the pGL3-Promoter vector (Promega Corp.). pGAL4-Luc was utilized as a reporter for protein-protein interaction in mammalian two hybrid assay and was provided by Dr. B. Rowan (Tulane University, New Orleans LA). It contains a GAL4 DNA binding site upstream of a minimal TATA box which governs the expression of the firefly luciferase gene.
Yeast Two-Hybrid Screening and Mating Assays
A DU145 prostate cancer cell two-hybrid library was generated in the pGADT7-Rec vector using the Clontech Matchmaker 3 Library Construction Kit (Clontech Corp.), following manufacturer's instructions. Approximately 2 × 106 library clones were screened by yeast mating with selection by growth for 10 days on agar media lacking Leu, Trp, and His, followed by standard filter-lift β-galactosidase assay of colonies. Plasmid DNA was isolated from β-galactosidase positive yeast colonies, transformed into E. coli DH5α, followed by preparation of E. coli plasmid DNA. DNA plasmids for library cDNA clones were then individually transformed into S. cerevisiae strain AH109 and tested for bait specificity by mating with S. cerevisiae strain Y187 that harbored either the E12 bait construct, a lamin negative control construct (provided in the Matchmaker System 3 Kit), or pGBTK7 empty vector. Mating mixtures were plated on the either double (Leu-, Trp-) dropout (DDO) or triple (Leu-, Trp- , His-) dropout (TDO) nutrient selective media and bait specificity of the interaction was scored by appropriate growth pattern on selective media and by X-α-galactosidase activity. Inserts for yeast two hybrid library cDNA clones that demonstrated bait-specific interaction were sequenced.
Mammalian Two-Hybrid Assay
HeLa cells were plated at 4 × 104 per well in a 24 well plates and the following day transfected with DNA constructs using Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA). Indicated combinations of DNA constructs (0.2 μg for each) were co-transfected with 0.4 μg of pGAL4-Luc and 0.2 μg of pRLTK plasmid DNA per well. 48 h after transfection, cells were lysed with passive lysis buffer and dual luciferase activities were measured with a Turner Systems luminometer in accordance with the manufacturer's instruction (Dual-Luciferase Reporter Assay System, Promega Corp.). Relative luciferase activities were expressed as the ratio of firefly and Renilla luciferase activities. Studies shown in each graph were performed a minimum of three independent times with samples prepared as independent triplicates for each assay. Representative data from a minimum of three independently performed studies are shown. Statistical significance was calculated using single factor ANOVA.
E-Box Reporter Assay
As indicated in the respective figure legend, combinations of the following DNA expression constructs were used in an E-box regulated luciferase reporter assay: E12 or E47 (0.004 μg), MyoD (0.1 μg), RANBP (0.1 μg of HA-RANBP17, HA-RANBP16, sRANBP17, or HA-RANBP17-CRM1), Id1 (0.5 μg), E-box-pLuc (0.2 μg), pRLTK (0.1 μg). Numbers in parenthesis indicate the mass of DNA construct used per well of a 24 well culture plate. Addition of empty vector DNA was included as needed to equalize the total combined mass of DNA per well to 1 μg, or to 1.2 μg when transfections included the Id1 expression construct. Co-transfections were carried out in HeLa cells using Lipofectamine 2000 (Invitrogen Corp), with media change at 4 h post-transfection. Samples were assayed for firefly luciferase at 48 h post-transfection with normalization by standard protein assay (Bio-Rad). Studies presented in each graph were performed a minimum of three independent times with samples prepared as independent triplicates for each assay, with representative data shown. Statistical significance was calculated using single factor ANOVA.
RNA Preparation and Q-PCR Analysis
293T cells were transiently transfected with HA-RANBP17 or HA-RANBP16 DNA expression constructs, or with empty vector DNA as a negative control. Transfections were carried out three independent times with representative data shown. Transfection was via Lipofectamine 2000 with media change 4 hr post-transfection. RNA was isolated from cells using TriZol Reagent (Invitrogen Corp.), according to manufacturer's instruction. First-strand cDNA was synthesized using 5 μg of DNase I-treated total RNA. Transcript levels were analyzed by SYBR Green-based quantitative real-time PCR (Q-PCR) in 25 μl-reactions containing 1X SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 200 nM each forward and reverse primers, and 10 ng of cDNA. Primers used for detection of human p21Waf1/Cip1 transcript were 5’-GTCTTGTACCCTTGTGCCTC-3’ and 5’-GCTTCCTCTTGGAGAAGATCAG-3’ and for human GAPDH 5’-GAAGGTGAAGGTCGGAGTCA-3’ and 5’-TTCACACCCATGACGAACAT-3’. Q-PCR was conducted with an ABI 7500 Real-Time PCR System. PCR was carried out over 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 40 s, with an initial cycle of 50°C for 2 min and 95°C for 10 min to activate AmpliTaq Gold DNA polymerase; a dissociation curve was generated over the range of 60–95°C. For the latter, a single sharp peak was observed in each case, indicative of a single PCR product species. Transcript expression was normalized against GAPDH. The cycle threshold value was generated using ABI Prism 7500 SDS software, version 1.2. Fold changes were determined by the ΔΔCT method and are shown as means ±SD of triplicates. Statistical significance was calculated using single factor ANOVA.
Immunocytochemistry and Intracellular Localization Studies
For immunostaining studies, COS cells were plated on laminin-coated coverslips and transfected with DNA for expression constructs using Lipofectamine 2000. 48 h after transfection, cells on coverslips were washed twice with PBS and fixed with 100% ice-cold methanol for 10 min, washed in PBS, and blocked by incubation in 1% BSA in PBS for 30 min at room temperature. Cells were incubated with polyclonal E2A antibody for detection of E12 (1:100, Santa Cruz Biotech.) and/or monoclonal HA antibody (1:100, Covance Corp.) in 1% BSA in PBS for 1.5 h. Following three washes in 1% BSA in PBS, cells were incubated with Alexafluor 568-conjugated goat anti-mouse (1:800, Invitrogen Corp.) and/or FITC-conjugated goat anti-rabbit (1:200, Bio-Rad) secondary antibodies, as indicated. Following a 1 h incubation, coverslips were washed and mounted on glass slides and cells observed at 400 X or 600 X magnification using a Nikon Eclipse E800 fluorescence microscope equipped with a digital camera. Image acquisition and merging was performed with Image-Pro Plus software (Media Cybernetics, Carlsbad, CA). For quantitative assessment of intracellular localization, signals were scored as predominately cytoplasmic, predominantly nuclear or equally distributed in nucleus and cytoplasm, similar to previously published methodology [Yagita et al., 2002]. For each experiment, 50-100 cells were scored from three independent transfections, with three independent experiments performed. Statistical significance was calculated using single factor ANOVA.
Western Blot Analysis, Subcellular Fractionation and Co-immunoprecipitation
For analysis of total cell lysates, HeLa cells were transfected with 4 μg of HA-RANBP17 or empty vector per well of a 6-well plate using Lipofectamine 2000 (Invitrogen Corp.). Cells were harvested at 48 h post-transfection by lysis in TNN(+) cell lysis buffer (10 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP-40, 1 mM EDTA supplemented with a protease inhibitor cocktail). Lysates were incubated on ice for 30 min with intermittent vortexing and after centrifugation at 20,800 × g for 15 min, supernatant was collected and protein content determined (Bio-Rad, Hercules, CA). For subcellular fractionation, cell lysates were prepared according to the instructions for preparation of nuclear extracts (Active Motif, Carlsbad CA). For Western blot analysis, 30 μg of protein extract was fractionated on 10 % SDS-PAGE gel, followed by electroblotting onto PVDF membrane using 0.025 M Tris/0.192 M glycine transfer buffer with 20% methanol. Membranes were blocked for 1 h in 5% non-fat milk in PBS containing 0.5% Tween 20 (PBS-T) followed by a 1 h incubation at room temperature with a mouse monoclonal HA antibody (1:2000, Covance Research Products, Berkeley, CA), a mouse monoclonal lamin A/C antibody (1:200, Affinity BioReagents) or a rabbit polyclonal β-tubulin antibody (1:10,000, Covance Research Products, Berkeley CA). Secondary antibody was HRP-conjugated goat anti-mouse (1: 2000, Santa Cruz Biotech, Santa Cruz CA.) or anti-rabbit (1: 2000, Bio-Rad) antibody and signals were detected using ECL Plus enhanced chemiluminescence reagent (GE Healthcare). Western blot analyses were conducted a minimum of three times with essentially same results and representative data shown.
For co-immunoprecipitation, COS cells were transfected with the indicated expression constructs and 48 h post-transfection total cell lysates were prepared by sonication in TNN(+) cell lysis buffer (10 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP-40, 1 mM EDTA supplemented with a protease inhibitor cocktail). 50 μg of cell lysate was incubated with E2A antibody by rotation overnight at 4°C followed by incubation for 3 h with 25 μl of 50% (v/v) suspension of Protein-A-agarose beads (Santa Cruz Biotech, Inc.). After centrifugation at 10,800 × g for 30 s, pelleted beads were washed three times with TNN(+) cell lysis buffer. Beads were resuspended in SDS-PAGE loading buffer, boiled for 10 min, centrifuged and the supernatant subjected to Western blot analysis. No signal was detected when immunoprecipitation was performed in the absence of antibody. A slight modification of this approach, found during the course of our studies to reduce non-specific background, was used for the experiment in Figure 6D. In this case lysates were pre-incubated with Protein-A-agarose beads for one hour at 4°C, beads were removed and protocol continued at the primary antibody addition stage, as described above. For the Western blot analysis shown in Figure 6D, we utilized protein-A-conjugated HRP, which recognizes native but not denatured IgG, to minimize signals from immunoglobulin heavy and light chains. All co-immunoprecipitation studies were performed a minimum of three times with essentially the same results and representative data is shown.
RESULTS
Two Hybrid Screening Reveals RANBP17 is a Novel Interaction Partner for E12
Yeast two hybrid screening was employed to search for novel binding partners for E12. The transcriptional activation regions of E12 (AD1 and AD2) that are located within the first 500 amino acids of the protein render a yeast two hybrid bait comprised of full-length E12 unsuitable for screening as it autonomously induced transcriptional activation of the yeast-two hybrid reporter gene(s) (data not shown). We therefore employed a bait that encoded amino acids 508-654 of E12 and that includes the bHLH domain. Figure 1A shows an illustration of the full-length E12 protein consisting of 654 amino acids and the region of E12 used in the bait construct. Following screening of 2 × 106 clones of a DU145 prostate cancer cell yeast two hybrid cDNA library, we identified a clone that evidenced specific interaction with the E12 bait and that maintained reading frame with the GAL4 activation domain encoded by the cDNA library vector. It contained amino acids 1 - 252 of RANBP17 (RANBP171-252).
Figure 1. Yeast and Mammalian Two-Hybrid Analyses Identify RANBP17 as a Binding Partner for E12.
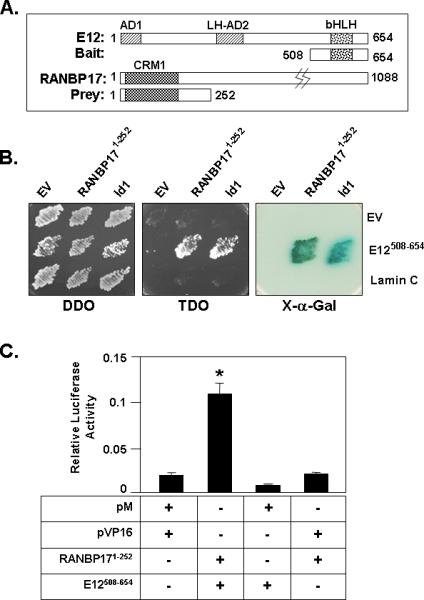
(A) Schematic representation of full-length E12 (E121-654), full-length RANBP17 (RANBP171-1088), the E12 bait (E12508-654) and the RANBP17 library prey clone (RANBP171-252). (B) The indicated pairwise combinations of the bait (indicated at right) and prey constructs (indicated at top) and corresponding empty control vectors were separately transformed into yeast strains Y187 and AH109, mated and protein-protein interaction was assayed as described in Materials and Methods. DDO, media lacking Leu and Trp; TDO media lacking Leu, Trp and His; X-α-Gal, X-α-galactosidase. (C) Mammalian two hybrid analysis. HeLa cells were transfected with the indicated expression constructs for E12508-654 and RANBP171-252 in theVP16 activation domain fusion or the pM binding domain fusions vectors, respectively. Luciferase activities were measured at 48 h post-transfection. Data shown is mean ± S.D., *, p<0.01 for the E12 and RANBP17 co-transfected samples vs. all others shown. Data is representative of three wholly independently conducted experiments, with transfections carried out in triplicate for each such independent experiment.
Figure 1B shows the result of bait specificity testing of the interaction between the E12 bait and RANBP171-252. Constructs for the E12 bait or the RANBP171-252 prey were transformed into Y187 and AH109 strains of S. cerevisiae, respectively. We included a prey for full length Id1, a known E12 interactor, as an E12 interactor positive control. Empty vectors and a bait construct encoding lamin C are negative controls. The left panel of Figure 1B indicates effective mating of yeast harboring the indicated pair-wise combinations of DNA binding and activation domain plasmids to produce diploids that grow on Leu and Trp deficient DDO agar media plates. The middle panel of Figure 1B demonstrates selective growth on Leu, Trp, and His deficient TDO agar media plates; here interaction is demonstrated by growth in the absence of His. RANBP171-252 interaction with E12 is specific in that yeast that harbor both the E12 bait construct and the RANBP171-252 library clone (Figure 1B, middle panel) grow on TDO media, whereas no growth is noted for the respective negative controls. The colorimetric detection of X-α-galactosidase activity, an additional reporter gene for protein-protein interaction in this system, is shown in the right panel of Figure 1B, further confirming bait-specific interaction of RANBP171-252 with the E12 bait. We also find the predicted interaction for the positive control combination of the E12 bait and Id1 prey.
We next employed mammalian two-hybrid analysis to assess protein-protein interaction in mammalian cells. The inserts from the E12 bait and the RANBP171-252 prey constructs used in the yeast two-hybrid study were subcloned into the mammalian two hybrid activation and binding domain vectors pVP16 and pM, respectively. These expression constructs and a pGAL4-Luc reporter construct were co-transfected into HeLa cells and luciferase activity assessed at 48 h post-transfection. Figure 1C shows that, in comparison to empty vector transfectants, a 5.7 fold increase (p<0.01) in luciferase activity was observed in cells co-transfected with fusion constructs for E12508-654 and RANBP171-252, indicative of interaction of these two protein regions in mammalian cells.
As our yeast and mammalian two hybrid studies only assessed interaction of the bHLH region of E12 with amino acids 1-252 of RANBP17, we employed co-immunoprecipitation studies to test for association of full-length E12 with full-length RANBP17. Unfortunately, assessment of multiple commercial antibodies for RANBP17 proved none were of utility for our studies. We first demonstrated that the expression construct for HA-tagged RANBP17 was functional. The Western blot in Figure 2A indicates protein expression from the HA-RANBP17 construct results in a single protein species consistent with the predicted ~120 kDa mass for RANBP17. Figure 2B demonstrates that E12 and RANBP17 are present in the same protein complex and that this signal is dependent on transfection of expression constructs for both proteins. Figure 2C indicates, as anticipated from our yeast two hybrid screening result, that a region of E12 that was used in the yeast two hybrid bait is sufficient for co-immunoprecipitation with full-length RANBP17.
Figure 2. RANBP17 Co-Immunoprecipitates with E12.
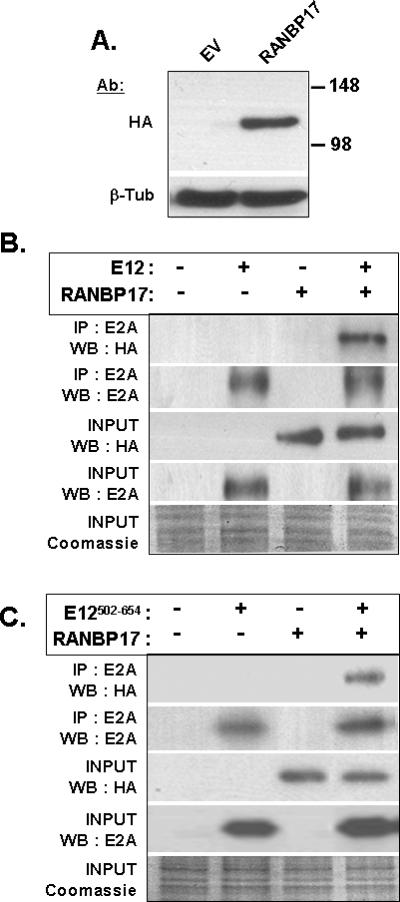
(A) Empty vector pcDNA3.1+ (EV) or the HA-RANBP17 expression construct (RANBP17) was transfected into HeLa and at 48 h post-transfection cell lysates were subjected to Western blot analysis using monoclonal HA primary antibody. β-tubulin (β-Tub) was used as a loading control. (B) HA-RANBP17 and full-length E12 protein was expressed separately or in combination by transfection of COS cells. 50 μg of each cell lysate was subjected to immunoprecipitation and Western blot analysis with the indicated antibodies. 1/20 of total protein (2.5 μg) was analyzed for Western blot (INPUT). Equivalent protein loading for Western blot was verified by Coomassie blue staining. (C) HA-RANBP17 and E12502-654 protein was expressed separately or in combination by transfection of COS cells and analyzed as in B. For B and C, IP: immunoprecipitation, WB: Western blot.
Co-Localization Studies of E12 and RANBP17
The results of the two-hybrid and co-immunoprecipitation interaction studies led us to address whether RANBP17 co-localized with E12. We first assessed the intracellular localization of RANBP17 protein by subcellular fractionation and immunocytochemistry. Nuclear and cytosolic fractions were prepared from cell lysates of HA-RANBP17-transfected HeLa cells and analyzed for RANBP17 protein utilizing Western blot with HA antibody. Figure 3A shows that RANBP17 protein is detected in both the nuclear and cytosolic fractions. Reprobing the membrane for β-tubulin and lamin A/C indicates effective fractionation of cytosolic and nuclear proteins, respectively. Immunocytochemistry of COS cells transfected with full-length HA-RANBP17 reveal three types of cellular localization patterns for RANBP17 protein (Figure 3B). We define these as predominantly cytoplasmic, predominantly nuclear or equally distributed in both nucleus and cytoplasm. Among those cells exhibiting a predominantly nuclear signal for RANBP17 protein, we observed a distinctive pattern of intense nuclear speckles, shown in the middle panel of Figure 3B. A similar nuclear speckled pattern, with an apparent exclusion of signal from nucleoli, had been previously observed by Janssen and coworkers for an eGFP fusion of murine RANBP17, although in that case localization of eGFP-RANBP17 was reported to be nearly exclusively nuclear [Koch et al., 2000]. Figure 3C illustrates that in cells showing a predominately nuclear signal for RANBP17, such signal co-localizes with that for E12; the latter is well-characterized as an exclusively nuclear protein [Lingbeck et al., 2008].
Figure 3. Intracellular Localization of RANBP17 and E12.
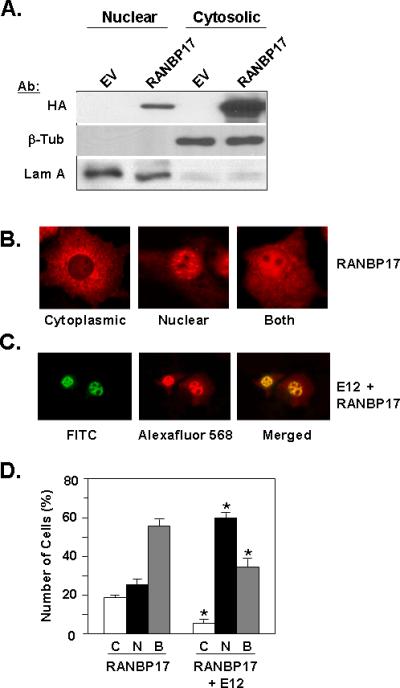
(A) Nuclear and cytoplasmic fractions of HeLa cell lysates transfected with empty vector pcDNA3.1+ (EV) or HA-RANBP17 expression construct were subjected to Western blot analysis with the indicated antibodies. β-tubulin (β-Tub) and lamin A (Lam A) were used as markers for cytoplasmic and nuclear fraction, respectively. (B) Immunocytochemistry for RANBP17 localization. COS cells were transfected with the HA-RANBP17 expression construct and at 48 h post-transfection were processed as described in Materials and Methods. Signal was detected utilizing HA primary antibody and Alexafluor 568 secondary antibody. Images presented are representative of data from five independent experiments and are shown at 400X. (C) Immunocytochemistry for co-localization of RANBP17 and E12. COS cells were co-transfected with HA-RANBP17 and E12 expression constructs and at 48 h post-transfection cells were processed as described in Materials and Methods. Signals were detected utilizing E12 primary antibody with FITC-conjugated secondary antibody (for E12) and HA primary antibody with Alexafluor 568-conjugated secondary antibody (for RANBP17). Images presented are representative of data from five independent experiments and are shown at 200X. (D) Analysis of intracellular localization of RANBP17 in the absence (first three columns) or presence (last three columns) of E12 expression. Immunostaining for RANBP17 and E12 was conducted as described in Materials and Methods. The intracellular localization of RANBP17 protein in individual cells was enumerated. C, cytoplasmic; N, nuclear; and B, both. Data is shown as the percentage of total cells enumerated, with from 50-100 cells enumerated. Localization was assessed in three independent transfections. *; p< 0.01 for pairwise comparisons of C, N, or B data for the RANBP17 plus E12 transfectants vs. RANBP17 only transfectants.
To determine whether the co-expression of E12 altered the subcellular localization of RANBP17, we conducted immunocytochemistry for RANBP17 in COS cells transfected with the HA-RANBP17 expression construct in either the absence or presence of co-transfection of an E12 expression construct. The subcellular localization of RANBP17 was scored as cytoplasmic, nuclear or both. The data in Figure 3D demonstrate that co-expression of E12 results in increased nuclear localization of RANBP17 and decreased signal for RANBP17 protein in the cytoplasm, consistent with interaction between E12 and RANBP17. We also ascertained whether co-expression of RANBP17 could influence E12 intracellular localization. For this we utilized expression constructs for full-length E12 and for an N’-terminally truncated cytoplasmic-localized form of E12 that lacks signal(s) for nuclear localization, termed E12502-654. Co-expression of RANBP17 did not affect the distribution of either full-length E12 or of E12502-654 from their exclusively nuclear and cytoplasmic localization, respectively (data not shown). Thus while nuclear E12 was shown to result in enhanced localization of RANBP17 in the nucleus (Figure 3D), the converse was not observed to occur.
Transcriptional Activation of E12 by RANBP17
To assess the functional consequences of the association of RANBP17 with E12, we determined if RANBP17 affects E12-mediated transcriptional activity. For this assay we generated an E-box responsive luciferase reporter construct, E-box-pLuc that has three copies of an E-box consensus enhancer element (CAGGTG) present just 5’ to an SV40 minimal promoter that drives expression of a firefly luciferase reporter gene (Figure 4A). E12 forms only weakly active homodimers and in most cases exerts its transcriptional action in a heterodimer with a second bHLH protein [Massari and Murre, 2000; Shirakata and Paterson, 1995; Sun and Baltimore, 1991]. We therefore utilized the bHLH protein MyoD, a well-characterized master regulator of skeletal muscle development [Berkes and Tapscott, 2005], as a heterodimerization partner for E12 in these assays. Co-transfection of E-box-pLuc with expression constructs for full-length E12 and MyoD, as anticipated, led to ~ 4-fold increase (p<0.01) in luciferase activity (Figure 4B, columns 1 and 2 ). In contrast, co-transfection of E12 and MyoD did not alter luciferase activity of the pLuc reporter construct, that lacks an E-box elements, was used as a negative control (Figure 4B, columns 5 and 6), indicating that the E12/MyoD-mediated transcriptional activation of E-box-pLuc was E-box dependent. Having validated this assay, we determined the effects of co-transfection of RANBP17 on the ability of E12/MyoD to transactivate the E-box-pLuc luciferase reporter. RANBP17 led to a 2.7-fold (p<0.01) further stimulation of transcriptional activity (Figure 4B, column 2 vs. 3). No significant effects on E-box-pLuc luciferase activity were noted upon co-transfection of RANBP17 alone (Figure 4B, column 4). Importantly, the pLuc plasmid itself (i.e. no E-box) was not regulated by RANBP17 singly or in combination with E12/MyoD (last four columns of Figure 4B), indicating that the transcriptional response to RANBP17 is specifically E-box dependent. This result supports the notion that RANBP17 interacts with E12 in a functionally important manner that enhances the transactivation activity of E12.
Figure 4. Expression of RANBP17 Augments E12-Mediated Transcriptional Activity.
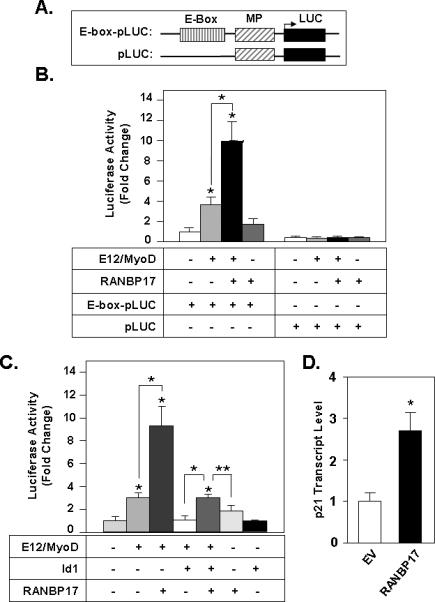
(A) Schematic diagram of luciferase reporter constructs used in the assay. Notation above the boxed regions indicates: E-box, 3 copies of the CAGGTG E-box element; MP, SV40 minimal promoter element; Luc, luciferase reporter gene. (B) The indicated combinations of plasmids including E12, MyoD and full-length RANBP17 and either E-box pLuc or pLuc reporter constructs were transfected into HeLa cells and cell lysates were analyzed for luciferase activity. (C) The effect of RANBP17 on Id1 inhibition of E12/MyoD-mediated transcriptional activity. The combinations of expression constructs (described above) were transfected into HeLa cells with co-transfection of an Id1 expression construct, as indicated. Value for transfection of E-box pLuc only was set to 1 (column 1). *; p<0.01 and **, p<0.05 for the indicated bracketed pairwise comparisons and for the respective data value vs. column 1. For B and C, data is representative of three wholly independently conducted experiments, with transfections carried out in triplicate for each such independent experiment. (D) Regulation of endogenous p21Waf1/Cip1 transcript level by RANBP17. Q-PCR analysis of RNA harvested from transfected 293T cells with *, p< 0.01 compared to empty vector. Representative data from three independent transfection studies is shown.
Id1 is a well-characterized inhibitory member of the HLH protein family. Id1 has an HLH protein interaction interface, but lacks a basic DNA binding domain [Benezra et al., 1990]. As such it functions in a dominant negative manner via the sequestration of E12 or other bHLH factors, into non-functional heterodimers. As a consequence, Id1 can diminish E12-mediated transactivation of E-box regulated genes [Norton et al., 1998; Yokota and Mori, 2002]. We determined whether RANBP17 might alter the ability of Id1 to exert its dominant negative action. Figure 4C shows that, as anticipated, co-transfection of an expression construct for Id1 with that for E12 and MyoD results in abolishment of E-box responsive transcriptional activity of E-boxpLuc (Figure 4C, column 2 vs. 4). In the presence of RANBP17, the dominant negative inhibitory effects of Id1 expression are attenuated (Figure 4C, column 5).
Lastly, we examined the ability of RANBP17 to affect endogenous E2A-regulated gene transcription using transcript level of p21Waf1/Cip1, a key cell cycle regulator important in all cells, as a readout. The direct positive transcriptional regulation of this cyclin-dependent kinase inhibitor by E2A proteins has been highly characterized by multiple investigations [Funato et al., 2001; Funato et al., 2003; Kim et al., 2004; Liu et al., 2004; Prabhu et al., 1997; Semerad et al., 2009; Sun et al., 2009]. To test the ability of RANBP17 to impact levels of endogenous p21Waf1/Cip1 transcript, we expressed RANBP17 in 293T cells using transient transfection, with RNA harvested 48 h later. Q-PCR analysis for p21Waf1/Cip1 shows that expression of RANBP17 led to a 2.7 fold increase in level of endogenous p21Waf1/Cip1 transcript compared with empty vector transfectants.
E12 Transcriptional Activation Function and Protein-Protein Interaction Map to the CRM1 Homology Region of RANBP17
Limited studies of expression of RANBP17 transcript in human and murine cells and tissues have been described [Bernard et al., 2001; Hansen-Hagge et al., 2002; Koch et al., 2000; MacLeod et al., 2003]. A 4.5 kb transcript is presumed to encode full-length RANBP17 protein [Hansen-Hagge et al., 2002; Koch et al., 2000]. Additionally, Northern blots have revealed that transcripts of 2.5, 7.5 and/or 10 kb are expressed in various tissues and cell lines [Bernard et al., 2001; Koch et al., 2000; MacLeod et al., 2003]. The 2.5 kb sized transcript likely encodes a shorter form of RANBP17 protein, as described below. The sequences and protein products of the other RANBP17 transcript sizes are undetermined. Janssen and coworkers identified a naturally occurring variant of RANBP17 transcript, which they cloned from testis [Hansen-Hagge et al., 2002]. This short form of RANBP17, termed herein sRANBP17, is generated via alternate splicing that introduces a premature stop codon to result in a C-terminally truncated protein of 576 amino acids, Figure 5A. We assessed whether sRANBP17 could activate E12/MyoD E-box mediated transcription and determined that it resulted in a ~2-fold increase (p<0.01) in luciferase reporter gene expression (Figure 5B). Given this result and the fact that the RANBP17 sequence present in the two hybrid library cDNA clone was largely comprised of the RANBP17 CRM1 region, we speculated that the transcriptional response and protein interactions of RANBP17 might map to its CRM1 homology region. To test this hypothesis, we generated an HA-tagged expression construct containing the CRM1 homology region of RANBP17 (amino acids 2-167), termed HA-RANBP17-CRM1, and determined its ability to augment E12/MyoD-mediated transcriptional response and to co-immunoprecipitate with E12502-654. Figure 5C demonstrates that expression of HA-RANBP17-CRM1 results in a ~ 2.5-fold increase in E-box mediated transcriptional activity. The co-immunoprecipitation data in Figure 5D reveals that the CRM1 homology region of RANBP17 is sufficient to mediate interaction with E12502-654. For the CRM1 protein its CRM1 region per se governs interaction with RanGTP. Our data indicate that in the case of RANBP17, the CRM1 region also functions as an interface for protein-protein interaction with E12.
Figure 5. The CRM1 Homology Region of RANBP17 is Sufficient for E12 Mediated Transcriptional Activation and Protein Interaction.
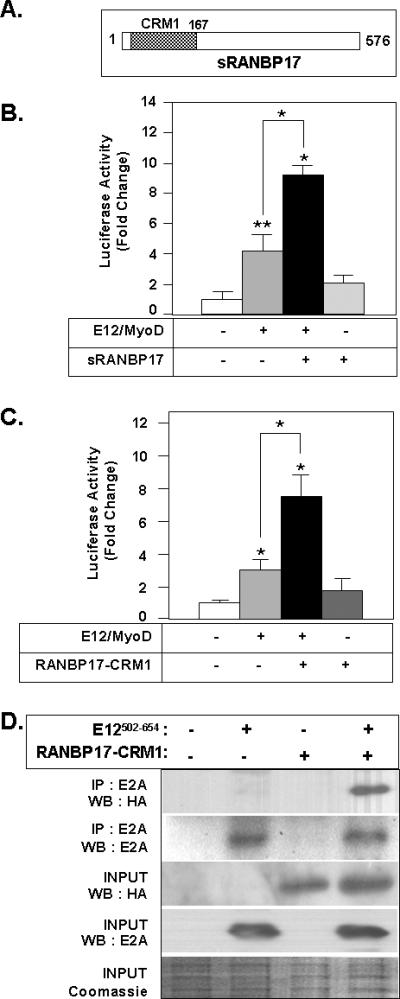
(A) Schematic diagram of sRANBP17 protein used in the expression construct. (B) The indicated combinations of plasmids including E12, MyoD and sRANBP17 and the E-box pLuc reporter construct were transfected into HeLa cells and cell lysates were analyzed for luciferase activity. Value for transfection of E-box pLuc only was set to 1 (column 1). (C) The indicated combinations of plasmids including E12, MyoD and RANBP17-CRM1 and the E-box pLuc reporter construct were transfected into HeLa cells and cell lysates were analyzed for luciferase activity. For B-C, data are shown as mean ± S.D *; p<0.01 and **, p<0.05 for the indicated bracketed pairwise comparisons and for the respective data value vs. that of column 1. Data is representative of three wholly independently conducted experiments, with transfections carried out in triplicate for each such independent experiment. (D) HA- RANBP17-CRM1 and E12502-654 proteins were expressed separately or in combination by transfection of COS cells. 50 μg of each cell lysate was subjected to immunoprecipitation and Western blot analysis with the indicated antibodies. 1/20 of total protein (2.5 μg) was analyzed for Western blot (INPUT). Equivalent protein loading for Western blot was verified by Coomassie blue staining.
Expansion of the E12-RANBP17 Functional Network to Include E47 and RANBP16
We next determined if the functional interaction we identified for E12 and RANBP17 might also occur for proteins that are closely related to E12 or RANBP17, namely E47 and RANBP16, respectively. As previously mentioned, RANBP17 and the closely related RANBP16 are the most evolutionally-distant members of the importin-β protein superfamily [Kutay et al., 2000]. That the CRM1 homology region of RANBP17 was sufficient for protein interaction and transactivation response, taken with the high homology between the respective CRM1 regions of RANBP17 and RANBP16 proteins, led us to test whether RANBP16 might also impact E12-mediated transcriptional function. As shown in Figure 6A, a ~2-fold increase in luciferase activity is observed with RANBP16. Figure 6B shows, similar to as demonstrated in Figure 4D for RANBP17, transient expression of RANBP16 in 293T cells resulted in a 2.2-fold increase in level of endogenous transcript for the E2A target gene p21Waf1/Cip1.
Figure 6. Expansion of the RABP17/E12 Interaction Network to Include Closely Related Proteins.
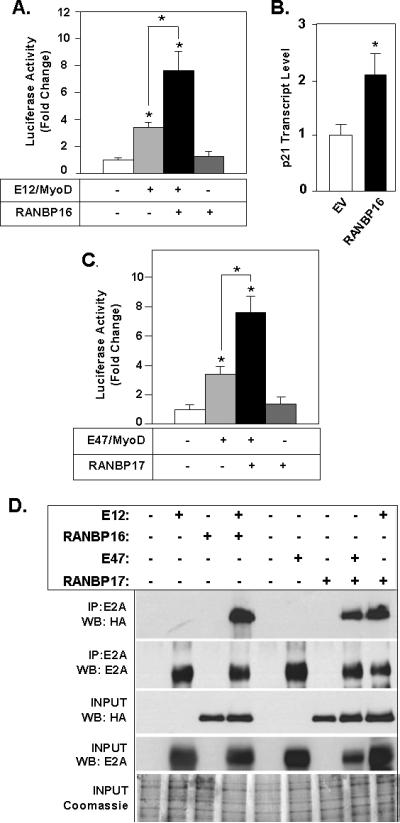
(A) The indicated combinations of plasmids including E12, MyoD and RANBP16 and the E-box pLuc reporter construct were transfected into HeLa cells and cell lysates analyzed for luciferase activity. Value for transfection of E-box pLuc only was set to 1 (column 1). *; p< 0.01 for the indicated bracketed pairwise comparisons and for the respective data value vs. that of column 1. (B) Regulation of endogenous p21Waf1/Cip1 transcript level by RANBP16. Q-PCR analysis of RNA harvested from transfected 293T cells with *, p< 0.01 compared to empty vector. Representative of three independent transfection studies is shown. (C) The indicated combinations of plasmids including E47, MyoD, RANBP17 and the E-box pLuc reporter construct were transfected into HeLa cells and cell lysates analyzed for luciferase activity. For A and C, data is representative of three wholly independently conducted experiments, with transfections carried out in triplicate for each such independent experiment. Data are shown as the mean ± S.D. (D) Co-immunoprecipitation analysis reveals interaction of E12 with RANBP16 and E47 with RANBP17. Various combinations of plasmids as indicated by “+” signs at lane headings were transfected into COS cells. Cell lysates were subjected to immunoprecipitation (IP) and Western blot (WB) analysis with the indicated antibodies. 1/20 of total protein (2.5 μg) was analyzed for Western blot (INPUT). Equivalent protein content per sample was verified by Coomassie blue staining of 20 μg sample aliquots. For comparison purposes, the far right lane shows interaction of RANBP17 with E12, carried out in the same experiment and analyzed on the same Western blot as the other combinations presented in this panel.
E12 and E47 are encoded by alternate splicing of the E2A gene leading to a degree of amino acid distinctions in their respective bHLH domains. However, their respective bHLH regions are nonetheless highly conserved, both in regard to the positively charged basic domain and the HLH interaction interface. Namely, the 61 AA bHLH region of E12 has 12 dispersed AA differences with that of E47, with conserved AA substitutions accounting for 7 of these differences. As such E12 and E47 often utilize the same bHLH heterodimerization partners [Hsu et al., 1994; Langlands et al., 1997; Lassar et al., 1991; Lingbeck et al., 2008]. We therefore asked if the transactivation ability we noted for E12 with RANBP17 would extend to E47. Figure 6C demonstrates the ability of RANBP17 to enhance E-box regulated gene transcription mediated by E47 and MyoD. Here we find that co-transfection of E47 and MyoD results in a 3.4-fold increase (p<0.01) in luciferase activity of E-box-pLuc and that co-transfection the RANBP17 expression construct further augments this response by an additional 2.2-fold (p<0.01). Our data for E12 and RANBP17, or regions thereof, showed that in each instance wherein we observed enhanced E-box mediated transcriptional activity we also demonstrated the respective protein-protein interaction by co-immunoprecipitation. As such, it would be predicted that E12 would co-immunoprecipitate with RANBP16 and E47 with RANBP17. These data for positive interaction are shown in Figure 6D. In this experiment, we also included the combination of E12 and RANBP17 as a positive control. Comparison of co-immunoprecipitation signals for E12 and RANBP16 (fourth lane), E47 and RANBP17 (lane 8), and E12 and RANBP17 (last lane) reveals a similar magnitude of signal intensity for each of these three combinations.
DISCUSSION
Using a yeast two hybrid screening approach we have identified RANBP17, a largely uncharacterized member of the importin-β superfamily, as a novel binding partner of E12. The interaction of RANBP17 and E12 was further demonstrated in mammalian cells by mammalian two hybrid assay and co-immunoprecipitation analyses. Importantly, we show that RANBP17 interacts with E12 such that it confers enhanced transcriptional activity on E12 when examined in mammalian cells. It is the CRM1 domain containing N-terminal region (1-167 AA) of RANBP17 and the C-terminus including the bHLH domain (508-654 amino acids) of E12 that are responsible for this interaction. Interestingly, for the case of the CRM1 protein itself, its CRM1 domain mediates the ability of CRM1 to bind RanGTP [Ossareh-Nazari and Dargemont, 1999; Petosa et al., 2004]. Our study suggests that the CRM1 domain may have a wider role in various protein-protein interactions than has previously been appreciated. It is of interest that we find that transcriptional activity of both E47 and E12 are increased via RANBP17 action and that each co-immunoprecipitate with RANBP17. Thus it might be overall structural similarity, for example the HLH motif, or the enrichment of positively charged amino acids that mediates interaction of RANBP17 with E-proteins. In their studies of RANBP16, Gorlich and coworkers concluded that while cargoes of RANBP16 were structurally diverse, a common theme appeared to be the presence of positively charged domains [Mingot et al., 2004]. It is of interest to note that bHLH regions are typified by a high density of positively charged amino acids [Benezra et al., 1990]; it remains to be seen if the interactions we describe herein will extend to other members of the bHLH protein family. The fact that we demonstrate that E12 transcriptional activity is also augmented by RANBP16, and that RANBP16 and E12 co-immunoprecipitate, reinforces the idea that positively charged motifs are involved in functional interactions for RANBP16 and suggests that this is also the case for RANBP17.
The ubiquitously expressed class I bHLH transcription factors, including E2A proteins, for the most part, impact tissue specific gene regulation via their interaction with cell type specific class II bHLH transcription factors. Proteins that function in the nucleus, including transcription factors, must be regulated in precise temporal and cell type-specific manners. A multitude of studies suggests that the regulation of nucleocytoplasmic shuttling of transcription factors is an important mechanism by which their activities can be regulated. Mutations affecting this process have been identified in various cancers [Fabbro and Henderson, 2003; Kau et al., 2004]. Transport between nucleus and cytoplasm through nuclear pore complex is largely mediated by the superfamily of importin β-related factors which represents the largest class of nuclear transport receptors [Kutay et al., 2000; Mingot et al., 2004]. This type of nucleocytoplasmic transport responds to the nucleocytoplasmic RanGTP gradient [Gorlich et al., 1996; Kutay et al., 1997]. Surprisingly little is known of the machinery specifically involved in E2A protein transport. However given their molecular mass of ~75, they exceed the estimated 20 to 40 kDa upper range of mass for entering the nucleus by diffusion, and thus they are likely to depend on facilitated receptor mediated translocation. Nuclear localization of E2A proteins is required for their proper functioning as transcription factors and E2A proteins contain potential nuclear localization sequence in their N-terminus [Chu and Kohtz, 2001; Huggins et al., 1999; Mitsui et al., 1993; Sloan et al., 1996], however the specific details for the nucleocytoplasmic transport mechanism of E2A proteins are not yet known. It is conceivable that if RANBP17 functions in nucleocytoplasmic transport, which to date has been empirically demonstrated for RANBP16 but not well-examined for RANBP17, then E12 might be a cargo of RANBP17. On the other hand, as RANBP17 and RANBP16 are both very evolutionarily distant members of the importin-β superfamily, they may impact E2A protein function via novel mechanisms.
Interestingly, the RANBP17 gene is the site of recurrent chromosomal 5 breakpoints found in T-cell acute lymphoblastic leukemia (T-ALL) with a (t5;14)(q35;q32) translocation the most frequently observed [Berger et al., 2003; Bernard et al., 2001; Su et al., 2006], and a (t5;14)(q34;q11) translocation also noted [Hansen-Hagge et al., 2002]. It has been suggested that the transcriptional activation of RANBP17, presumed to occur during hematopoietic development, may favor genetic rearrangement by opening up the chromatin in the RANBP17 region of chromosome 5 [Bernard et al., 2001]. To date, the biological effects of these translocations have focused nearly exclusively on the resultant activation of the transcription factor HOX11L2/TLX3 [Bayly et al., 2004; MacLeod et al., 2003; Su et al., 2006]. However, the idea has also been put forth that enhanced expression of RANBP17 transcript could conceivably occur in the (t5;14)(q34;q11) rearrangement, since this generates a head-to-tail juxtaposition of RANBP17 with enhancer elements of the TCR delta gene [Bernard et al., 2001; Hansen-Hagge et al., 2002]. This would be predicted to lead to increased expression of full-length RANBP17 or a C-terminally truncated RANBP17 protein, dependent on the breakpoint site [Bernard et al., 2001; Hansen-Hagge et al., 2002]. It is possible that in these and other settings RANBP17 plays a role in disease via its ability to augment E12 and/or E47-mediated transcriptional signals.
In conclusion, we have identified RANBP16 and RANBP17 as new players in the bHLH protein interaction network. E2A-regulated transcriptional signals have key roles in numerous biological settings. Taken with our finding that RANBP16 and RANBP17 can enhance transcript levels of endogenous p21Waf1/Cip1 , a well-validated E2A transactivation target gene, our data suggests that in certain biological settings RANBP16 or RANBP17 may impact normal and disease processes via functional interaction with E2A proteins.
ACKNOWLEDGMENTS
We thank Dr. W.G. Janssen (University of Heidelberg, Germany) for RANBP17-pMT2SM2 and RANBP16-pMT2SM, Dr. C. Murre (Harvard University) for E12, and Dr. B. Rowan (Tulane University) for the pGAL4-Luc plasmids. We thank Melissa Wilhelm and David Majors for help with yeast two hybrid screening and Kun Liu for technical assistance. This work was supported by a grant from the National Cancer Institute to C.M.S.
Contract grant sponsor: NIH, National Cancer Institute; Contract grant number: 5R01CA098141
REFERENCES
- Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, LeBrun DP. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J Biol Chem. 2004;279:55362–71. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Berger R, Dastugue N, Busson M, Van Den Akker J, Perot C, Ballerini P, Hagemeijer A, Michaux L, Charrin C, Pages MP, Mugneret F, Andrieux J, Talmant P, Helias C, Mauvieux L, Lafage-Pochitaloff M, Mozziconacci MJ, Cornillet-Lefebvre P, Radford I, Asnafi V, Bilhou-Nabera C, Nguyen Khac F, Leonard C, Speleman F, Poppe B, Bastard C, Taviaux S, Quilichini B, Herens C, Gregoire MJ, Cave H, Bernard OA. t(5;14)/HOX11L2-positive T-cell acute lymphoblastic leukemia. A collaborative study of the Groupe Francais de Cytogenetique Hematologique (GFCH). Leukemia. 2003;17:1851–7. doi: 10.1038/sj.leu.2403061. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bernard OA, Busson-LeConiat M, Ballerini P, Mauchauffe M, Della Valle V, Monni R, Nguyen Khac F, Mercher T, Penard-Lacronique V, Pasturaud P, Gressin L, Heilig R, Daniel MT, Lessard M, Berger R. A new recurrent and specific cryptic translocation, t(5;14)(q35;q32), is associated with expression of the Hox11L2 gene in T acute lymphoblastic leukemia. Leukemia. 2001;15:1495–504. doi: 10.1038/sj.leu.2402249. [DOI] [PubMed] [Google Scholar]
- Chu C, Kohtz DS. Identification of the E2A gene products as regulatory targets of the G1 cyclin-dependent kinases. J Biol Chem. 2001;276:8524–34. doi: 10.1074/jbc.M008371200. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–71. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Desprez PY, Sumida T, Coppe JP. Helix-loop-helix proteins in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:225–39. doi: 10.1023/a:1025957025773. [DOI] [PubMed] [Google Scholar]
- Dorfman J, Macara IG. STRADalpha regulates LKB1 localization by blocking access to importin-alpha, and by association with Crm1 and exportin-7. Mol Biol Cell. 2008;19:1614–26. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–90. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- French BA, Chow KL, Olson EN, Schwartz RJ. Heterodimers of myogenic helix-loop-helix regulatory factors and E12 bind a complex element governing myogenic induction of the avian cardiac alpha-actin promoter. Mol Cell Biol. 1991;11:2439–50. doi: 10.1128/mcb.11.5.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato N, Ohtani K, Ohyama K, Kuroda T, Nakamura M. Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Mol Cell Biol. 2001;21:7416–28. doi: 10.1128/MCB.21.21.7416-7428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato N, Ohyama K, Kuroda T, Nakamura M. Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription. J Biol Chem. 2003;278:7486–93. doi: 10.1074/jbc.M212248200. [DOI] [PubMed] [Google Scholar]
- Funk WD, Ouellette M, Wright WE. Molecular biology of myogenic regulatory factors. Mol Biol Med. 1991;8:185–95. [PubMed] [Google Scholar]
- Glick E, Leshkowitz D, Walker MD. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J Biol Chem. 2000;275:2199–204. doi: 10.1074/jbc.275.3.2199. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. Embo J. 1996;15:5584–94. [PMC free article] [PubMed] [Google Scholar]
- Gould KA, Bresnick EH. Sequence determinants of DNA binding by the hematopoietic helix-loop-helix transcription factor TAL1: importance of sequences flanking the E-box core. Gene Expr. 1998;7:87–101. [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S, Zhuang Y. Regulation of early lymphocyte development by E2A family proteins. Semin Immunol. 2002;14:405–14. doi: 10.1016/s1044532302000751. [DOI] [PubMed] [Google Scholar]
- Hansen-Hagge TE, Schafer M, Kiyoi H, Morris SW, Whitlock JA, Koch P, Bohlmann I, Mahotka C, Bartram CR, Janssen JW. Disruption of the RanBP17/Hox11L2 region by recombination with the TCRdelta locus in acute lymphoblastic leukemias with t(5;14)(q34;q11). Leukemia. 2002;16:2205–12. doi: 10.1038/sj.leu.2402671. [DOI] [PubMed] [Google Scholar]
- Hikima J, Middleton DL, Wilson MR, Miller NW, Clem LW, Warr GW. Regulation of immunoglobulin gene transcription in a teleost fish: identification, expression and functional properties of E2A in the channel catfish. Immunogenetics. 2005;57:273–82. doi: 10.1007/s00251-005-0793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Huang L, Tsan JT, Funk W, Wright WE, Hu JS, Kingston RE, Baer R. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol Cell Biol. 1994;14:1256–65. doi: 10.1128/mcb.14.2.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins GS, Chin MT, Sibinga NE, Lee SL, Haber E, Lee ME. Characterization of the mUBC9-binding sites required for E2A protein degradation. J Biol Chem. 1999;274:28690–6. doi: 10.1074/jbc.274.40.28690. [DOI] [PubMed] [Google Scholar]
- Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–17. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- Kim W, Kook S, Kim DJ, Teodorof C, Song WK. The 31-kDa caspase-generated cleavage product of p130cas functions as a transcriptional repressor of E2A in apoptotic cells. J Biol Chem. 2004;279:8333–42. doi: 10.1074/jbc.M312026200. [DOI] [PubMed] [Google Scholar]
- Koch P, Bohlmann I, Schafer M, Hansen-Hagge TE, Kiyoi H, Wilda M, Hameister H, Bartram CR, Janssen JW. Identification of a novel putative Ran-binding protein and its close homologue. Biochem Biophys Res Commun. 2000;278:241–9. doi: 10.1006/bbrc.2000.3788. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–71. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Treichel N, Calado A, Carmo-Fonseca M, Prehn S, Kraft R, Gorlich D, Bischoff FR. Identification of two novel RanGTP-binding proteins belonging to the importin beta superfamily. J Biol Chem. 2000;275:40163–8. doi: 10.1074/jbc.M006242200. [DOI] [PubMed] [Google Scholar]
- Langlands K, Yin X, Anand G, Prochownik EV. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J Biol Chem. 1997;272:19785–93. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–15. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lee JE. NeuroD and neurogenesis. Dev Neurosci. 1997;19:27–32. doi: 10.1159/000111182. [DOI] [PubMed] [Google Scholar]
- Lingbeck JM, Trausch-Azar JS, Ciechanover A, Schwartz AL. In vivo interactions of MyoD, Id1, and E2A proteins determined by acceptor photobleaching fluorescence resonance energy transfer. Faseb J. 2008;22:1694–701. doi: 10.1096/fj.07-095000. [DOI] [PubMed] [Google Scholar]
- Liu Y, Encinas M, Comella JX, Aldea M, Gallego C. Basic helix-loop-helix proteins bind to TrkB and p21(Cip1) promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol Cell Biol. 2004;24:2662–72. doi: 10.1128/MCB.24.7.2662-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod RA, Nagel S, Kaufmann M, Janssen JW, Drexler HG. Activation of HOX11L2 by juxtaposition with 3'-BCL11B in an acute lymphoblastic leukemia cell line (HPB-ALL) with t(5;14)(q35;q32.2). Genes Chromosomes Cancer. 2003;37:84–91. doi: 10.1002/gcc.10194. [DOI] [PubMed] [Google Scholar]
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–40. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot JM, Bohnsack MT, Jakle U, Gorlich D. Exportin 7 defines a novel general nuclear export pathway. Embo J. 2004;23:3227–36. doi: 10.1038/sj.emboj.7600338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Shirakata M, Paterson BM. Phosphorylation inhibits the DNA-binding activity of MyoD homodimers but not MyoD-E12 heterodimers. J Biol Chem. 1993;268:24415–20. [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–35. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–44. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Mutoh H, Fung BP, Naya FJ, Tsai MJ, Nishitani J, Leiter AB. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–4. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld B, Grosse-Wilde A, Hoffmeyer A, Jordan BW, Chen P, Dinev D, Ludwig S, Rapp UR. Serine/Threonine kinases 3pK and MAPK-activated protein kinase 2 interact with the basic helix-loop-helix transcription factor E47 and repress its transcriptional activity. J Biol Chem. 2000;275:20239–42. doi: 10.1074/jbc.C901040199. [DOI] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- Ossareh-Nazari B, Dargemont C. Domains of Crm1 involved in the formation of the Crm1, RanGTP, and leucine-rich nuclear export sequences trimeric complex. Exp Cell Res. 1999;252:236–41. doi: 10.1006/excr.1999.4599. [DOI] [PubMed] [Google Scholar]
- Perry C, Soreq H. Transcriptional regulation of erythropoiesis. Fine tuning of combinatorial multi-domain elements. Eur J Biochem. 2002;269:3607–18. doi: 10.1046/j.1432-1033.2002.02999.x. [DOI] [PubMed] [Google Scholar]
- Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-Lopez M, Baudin F, Mattaj IW, Muller CW. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell. 2004;16:761–75. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–96. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–22. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- Saarikettu J, Sveshnikova N, Grundstrom T. Calcium/calmodulin inhibition of transcriptional activity of E-proteins by prevention of their binding to DNA. J Biol Chem. 2004;279:41004–11. doi: 10.1074/jbc.M408120200. [DOI] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci U S A. 2009;106:1930–5. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakata M, Paterson BM. The E12 inhibitory domain prevents homodimer formation and facilitates selective heterodimerization with the MyoD family of gene regulatory factors. Embo J. 1995;14:1766–72. doi: 10.1002/j.1460-2075.1995.tb07165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SR, Shen CP, McCarrick-Walmsley R, Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol. 1996;16:6900–8. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XY, Della-Valle V, Andre-Schmutz I, Lemercier C, Radford-Weiss I, Ballerini P, Lessard M, Lafage-Pochitaloff M, Mugneret F, Berger R, Romana SP, Bernard OA, Penard-Lacronique V. HOX11L2/TLX3 is transcriptionally activated through T-cell regulatory elements downstream of BCL11B as a result of the t(5;14)(q35;q32). Blood. 2006;108:4198–201. doi: 10.1182/blood-2006-07-032953. [DOI] [PubMed] [Google Scholar]
- Sun R, Su Y, Zhao X, Qi J, Luo X, Yang Z, Yao Y, Xia Z. Human calcium/calmodulin-dependent serine protein kinase regulates the expression of p21 via the E2A transcription factor. Biochem J. 2009;419:457–66. doi: 10.1042/BJ20080515. [DOI] [PubMed] [Google Scholar]
- Sun XH, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–70. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Turner EC, Cureton CH, Weston CJ, Smart OS, Allemann RK. Controlling the DNA binding specificity of bHLH proteins through intramolecular interactions. Chem Biol. 2004;11:69–77. doi: 10.1016/j.chembiol.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Venuti JM, Cserjesi P. Molecular embryology of skeletal myogenesis. Curr Top Dev Biol. 1996;34:169–206. doi: 10.1016/s0070-2153(08)60711-5. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. Embo J. 2002;21:1301–14. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mori S. Role of Id family proteins in growth control. J Cell Physiol. 2002;190:21–8. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000;5:777–87. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- Yutzey KE, Konieczny SF. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic Acids Res. 1992;20:5105–13. doi: 10.1093/nar/20.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]


