Abstract
The checkpoint with Forkhead-associated domain (FHA) and Ring finger domain (CHFR) is a mitotic checkpoint protein with tumor-suppressor functions. In this study, we investigated the epigenetic and genetic mechanisms that regulate CFHR expression in esophageal adenocarcinomas (EACs). Quantitative real-time RT-PCR analysis demonstrated down-regulation of CHFR transcript in 79% of EACs (44/56) as compared to 41 normal samples (P<.001). Immunohistochemical analysis of CHFR protein expression showed absence or weak immunostaining for CHFR in 75% of EACs (56/75), as compared to normal tissue samples. We next examined the promoter DNA hypermethylation of CHFR using quantitative bisulfite pyrosequencing technology. We detected significant CHFR promoter DNA hypermethylation in 31% of tumor samples (18/58), as compared to normal samples (P<.001). Treatment of OE33 cells with 5-Aza-deoxycytidine led to reduction in the promoter DNA methylation levels with restoration of the CHFR mRNA expression, confirming promoter DNA methylation as an epigenetic mechanism regulating CHFR expression. However, we identified several EACs where the CHFR mRNA expression was silenced in absence of notable methylation. Therefore, we examined the relative DNA copy number level of CHFR, as compared to normal samples. The results confirmed a decrease or absence of the relative CHFR DNA copy number levels in 59% of tumor samples. Nine tumors showing loss of CHFR mRNA expression, in absence of promoter DNA hypermethylation, demonstrated a significant loss of relative CHFR DNA copy numbers. Taken together, our findings demonstrate that both epigenetic and genetic mechanisms are involved in silencing CHFR expression in EACs.
Keywords: methylation, copy numbers, CHFR, esophageal, Barrett's, cancer
Introduction
The incidence of adenocarcinoma of the lower esophagus and gastro-esophageal junction has increased at an alarming rate in the Western World in the last few decades 1, 2. Risk factors including obesity, alcohol consumption, smoking, and gastroesophageal reflux disease (GERD) have contributed to the increase in this cancer 3. Chronic GERD is the most common risk factor for the development of Barrett's esophagus (BE) whereby normal squamous mucosa is replaced by a metaplastic specialized columnar epithelium. Patients with BE have a high-risk of progression to dysplasia and subsequent esophageal adenocarcinoma 4, 5.
Although several molecular changes have been demonstrated in tumor initiation and progression 6-8, the contribution of epigenetic mechanisms in Barrett's tumorigenesis is not well characterized. DNA methylation plays an important role in the regulation of gene expression. Aberrant DNA methylation, namely overall DNA hypomethylation and regional DNA hypermethylation has been linked to carcinogenesis of various organs 9. Aberrant DNA hypermethylation of CpG islands in the promoter region has been associated with gene silencing of several genes in cancer such as p16, hMLH1 and CDH1 genes 10-12. Checkpoint with Forkhead-associated domain (FHA) and Ring finger domain (CHFR), located on chromosome 12q24, ubiquitously expressed in normal human tissues, was first identified by Scolnick and Halazonetis in 2000 13. CHFR is a mitotic checkpoint protein with a tumor-suppressor function that has the potential to be a novel biomarker for chemotherapeutic response to microtubule-targeting drugs 14. Under conditions of mitotic stress induced by taxol or nocodazole, the CHFR protein delays nuclear envelope breakdown and chromosome condensation prior to cellular entry into metaphase 13.
In this study, we have examined the epigenetic and genetic mechanism of silencing of CHFR in esophageal adenocarcinomas. We utilized state-of-the-art quantitative bisulfite pyrosequencing technology (Biotage, Uppsala, Sweden) for analysis of promoter DNA methylation and quantitative Real-time PCR for evaluation of relative DNA copy numbers of CHFR.
Materials and Methods
Tissues samples
All tissue samples were obtained from the archives of pathology at Vanderbilt University (Nashville, TN, USA), Otto-von-Guericke University (Magdeburg, Germany), and from the National Cancer Institute Cooperative Human Tissue Network (CHTN). The use of specimens from the archival tissue repository was approved by the Institutional Review Board protocol numbers 03-1078 and 33-2001. All tissue samples included in this study were collected and were coded from tissues that remained after completion of diagnosis, and that are otherwise discarded. All personal identifiers were removed prior to receiving samples. Histopathological diagnosis of the EACs was verified on the basis of H&E-stained sections according to the Vienna classification of gastrointestinalepithelial neoplasia 15. All tissue samples were dissected to obtain ≥70% cell purity. The patients' ages ranged from 34 to 84 years (median at 65 years). The adenocarcinomas ranged from well differentiated to poorly differentiated, stages I-IV, with a mix of intestinal- and diffuse-type tumors.
Quantitative real-time reverse transcription PCR (qRT-PCR) analysis of CHFR
Total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA, USA), and single-stranded cDNA was subsequently synthesized using the Advantage RT-for-PCR Kit (Clontech, Palo Alto, CA, USA). The expression of CHFR was evaluated in a set of 97 frozen primary human samples that included 41 normal mucosa of the oesophagus and the stomach and 56 samples of EACs and GEJs. Seventeen tumor samples had a matching tumor and normal sample from the same patient. The CHFR oligos were designed using the online software, Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The oligos were purchased from Integrated DNA Technologies(Coralville, IA, USA) (CHFR (forward) 5′-CGTAACATCCCGTCCTGACT-3′ and (reverse) 5′-GCTCTCTTCACCTCCAGTGC-3′). The qRT-PCR was performed using an iCycler (Bio-Rad, Hercules, CA, USA), with the threshold cycle number determined by use of iCycler software version 3.0. Reactions were performed in triplicate and the threshold numbers were averaged. The results of the CHFR gene were normalized to HPRT, which had minimal variation in all normal and tumor samples tested 16. Expression fold was calculated according to the formula 2(Rt–Et)/2(Rn–En) 17 where Rt is the threshold cycle number for the reference gene (HPRT) observed in the tumor, Et is the threshold cycle number for the experimental gene (CHFR) observed in the tumor, Rn is the threshold cycle number for the reference gene observed in the normal samples, and En is the threshold cycle number for the reference gene observed in the tumor. Rn and En values were calculated as an average of the 41 normal samples. For all the primary EACs, the gene was considered to be down-regulated if the mRNA expression fold was ≤0.5 in comparison with the normal samples.
DNA Bisulfite treatment and pyrosequencing analysis
DNA was purified using a DNeasy tissue kit (Qiagen). The bisulfite modification of the DNA from cell lines and tissues was performed using an EZ DNA Methylation-Gold Kit (ZYMO Research, Orange, CA, USA), according to the manufacturer's protocol. The CHFR promoter CpG Island was identified using a CpG island online search tool (http://www.uscnorris.com/cpgislands2.cpg.aspx). The criteria used for the definition of CpG islands was; a DNA fragment ≥500 bp with a G+C equal to or greater than 55% with an observed CpG/expected CpG of 0.65. A 20 ng aliquot of modified DNA was subjected to PCR amplification of the specific promoter region containing a CpG island that extends from −46 to −143 bp from the transcription start site and contains 15 CpG sites. The primers were designed using PSQ assay design software (Biotage), where one of the primers was biotin labeled. The forward primer sequence is GAAGTAGTTTGGTTAGGATTAAAGAT, the reverse biotin labeled primer sequence is Bio-ACATTACCACTCCCTCAACTAAT and the sequence primer is GTTTGGTTAGGATTAAAGAT. The Platinum PCR SuperMix High Fidelity enzyme mix (Invitrogen, Carlsbad, CA, USA) was used in the PCR reactions. The PCR products were checked by gel electrophoresis to confirm the size of the product and rule out the formation of primer dimers. The specific PCR products were then subjected to quantitative pyrosequencing analysis using a Biotage PyroMark MD system (Biotage) following the protocol provided by the manufacturer. The results were analysed by Pyro Q-CpG 1.0.9 software (Biotage). Based on the methylation levels in the normal samples, we used 20% methylation as a cut off for identification of promoter DNA hypermethylation of CHFR in tumor samples. Statistical analysis was performed to detect significant changes in the frequencies of DNA methylation of CpG sites between tumor and normal samples.
5-Aza-2′ deoxycytidine (5-Aza) and trichostatin-A (TSA) treatment
For validation of the role of the promoter DNA hypermethylation in transcriptional regulation of CHFR in vitro, esophageal cancer cell line (OE33) was used. OE33 cells were maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and antibiotics (Invitrogen). Cells were seeded at low density for 24 h, and then treated with 5 μM 5-Aza (Sigma-Aldrich, St Louis, MO, USA) for 72 h or 300 nM Trichostatin A (TSA, Wako, Osaka, Japan) for 24 h. Total RNA and DNA were isolated and purified by a Qiagen RNeasy kit and DNeasy tissue kit (Qiagen), as described above. DNA methylation levels of the CpG nucleotides of CHFR promoter were determined by pyrosequencing before and after treatments. The CHFR mRNA expression levels were determined by qRT-PCR, as described above.
Immunohistochemistry
Immunohistochemical (IHC) analysis of CHFR protein expression was performed on a tumor tissue microarray (TMA) that contained 75 adenocarcinomas. All adenocarcinomas were classified according to the recent guidelines of the International Union Against Cancer (UICC) TNM classification system. All EACs originated from the lower esophagus or gastro-oesophageal junction corresponding to the adenocarcinoma of the esophago-gastric junction type 18. Samples from adjacent normal esophageal squamous epithelia, BE, and dysplastic tissues were included when available. All tissue samples were histologically verified and representative regions were selected for inclusion in the TMA. The adenocarcinoma samples ranged from well-differentiated (WD) to poorly-differentiated (PD), stages I to IV, with a mix of intestinal- and diffuse-type tumors. Tissue cores with a diameter of 0.5 mm were retrieved from the selected regions of the donor blocks and punched to the recipient block using a manual tissue array instrument (Beecher Instruments, Silver Spring, MD, USA). Each tissue sample was represented by four tissue cores on the TMA. Sections (5 μm) were transferred to polylysine-coated slides (SuperFrostPlus, Menzel-Gläser, Braunschweig, Germany) and incubated at 37°C for 2 h. The resulting TMA was used for IHC analysis utilizing a CHFR mouse monoclonal antibody (clone BC012072, dilution 1:25, Abcam, Germany) and positive immunohistochemical reactions were revealed using the EnVision+ kit from DAKO (DAKO, Germany). Cores with no evidence of nuclear staining or only rare scattered positive cells, less than 3%, were recorded as negative. The overall intensity of staining was recorded as that for the core with the strongest intensity. Immunohistochemical results were evaluated for intensity and frequency of cell staining. The intensity of staining was graded as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The frequency was graded from 0 to 4 by percentage of positive cells as follows: grade 0, <3%; grade 1, 3-25%; grade 2, 25-50%; grade 3, 50-75%; grade 4, >75%. The index score was the product of multiplication of the intensity and frequency grades, which was then classified into a 4 point scale: index score 0 = product of 0, index score 1 = products 1 and 2, index score 2 = products 3 and 4, index score 3 = products 6 through 12.
Measurement of fold DNA amplification by qPCR
For evaluation of relative DNA copy numbers, we used the quantitative real-time PCR (qPCR) amplifications using iCycler (Bio-Rad, Hercules, CA, USA). PCR reactions were prepared in a total volume of 20 μl containing template DNA (40ng), with the threshold cycle number determined by use of iCycler software version 3.0. Primers were designed using the online software, Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The forward and reverse primers for CHFR genomic DNA were 5′-TGCAGATGCTGTTCCTTACG-3′ and 5′-ACACATCCTCCACGTGACAA-3′, respectively. The oligos were obtained from Integrated DNA Technologies (Coralville, IA, USA). Reactions were performed in triplicate, and the threshold numbers were averaged. The results were normalized to beta-Actin, which had minimal variation in all normal and tumor samples tested. Amplification fold was calculated in the same way as in quantification of qRT-PCR for mRNA expression and the En values were calculated as an average of the 19 normal samples. For all the primary EACs, the gene was considered to have loss of DNA copy number if the relative amplification fold was ≤0.5 in comparison with the normal samples.
Statistical Analysis
The t-test for paired samples using GraphPad prism version 4 software (GraphPad Prism Software, Inc., La Jolla, CA, USA) was used to compare the DNA methylation level between normal and EACs. We also compared the mRNA expression fold between normal and tumor samples and between unmethylated and methylated EACs. The student t-test was used to compare (1) the differences of the DNA methylation level between normal and EACs; (2) the differences of mRNA expression fold between normal and EACs, and (3) the differences of mRNA expression fold between unmethylated and methylated EACs. In addition, we analysed the association between DNA methylation and the clinicopathological factors. The correlation between the DNA methylation level and mRNA expression fold was determined by Spearman correlation. The comparison of IHC scores among normal, EACs and association between IHC score and clinicopathological factors were carried out by Chi-square and Fisher exact tests. All p values were based on two-sided tests, and differences were considered statistically significant when the p value was ≤0.05.
Results
Frequent silencing of CHFR gene expression in EACs
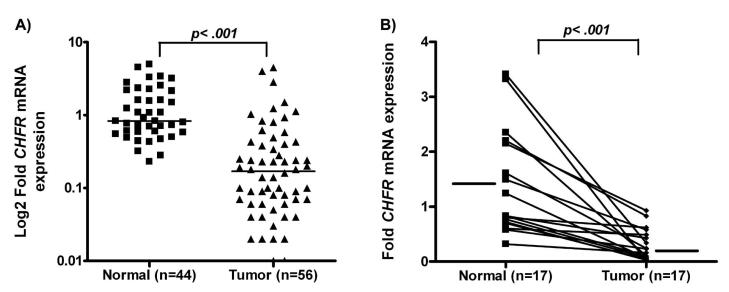
The qRT-PCR analysis revealed frequent down-regulation of CHFR mRNA expression in tumor samples (44/56, 78.6%), as compared to 41 normal samples (p<.0001) (Figure 1A). Further analysis of 34 paired tumor and normal samples demonstrated significant down-regulation of CHFR mRNA expression in tumors compared to their corresponding normal samples (Figure 1B). Of note, several EACs (16/56, 28%) showed complete silencing of CHFR mRNA expression indicated by absence of detectable signal.
Figure 1. Comparison of CHFR mRNA expression levels in Normal vs Tumor.
A) The expression of CHFR in normal mucosa (n=41) and EACs (n=56) was determined by Real-time RT-PCR and normalized to the average value of all the normal samples as described in materials and methods. Black boxes and triangles represent normal and tumor samples, respectively. The horizontal bars represent the mean expression fold. The statistical significance (p<0.0001) was determined by t-test. B) Expression of CHFR in 17 representative tumor samples and their corresponding normal samples from the same patients were analyzed side by side for comparison.
Promoter DNA hypermethylation of CHFR gene correlates with silencing of mRNA expression
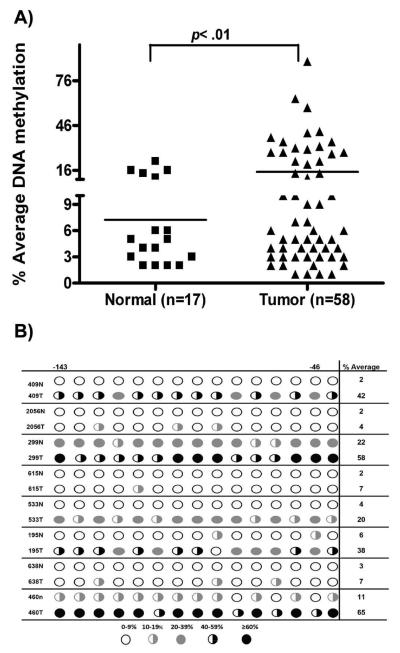
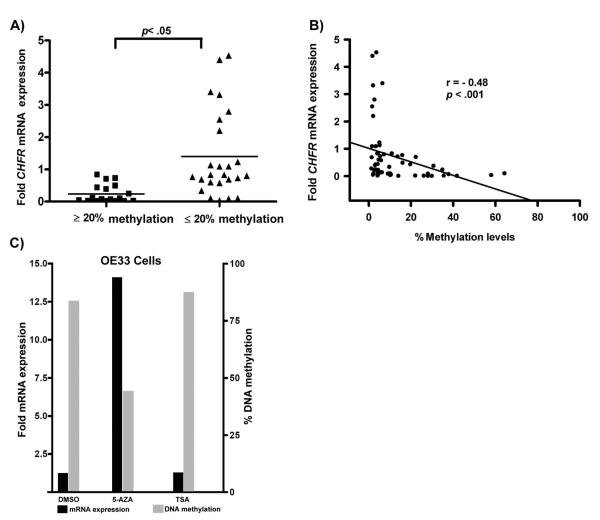
Quantitative analysis of CHFR promoter DNA methylation using state-of-the-art pyrosequencing technology indicated increased promoter DNA methylation levels of all tested CpG nucleotides sequences in tumor samples, as compared to normal samples. The DNA methylation changes were independent of the patients' age. The representative results of the percentage of the average methylation for the CHFR gene promoter in EACs vs normal are shown in Figure 2A. The promoter DNA hypermethylation (≥20%) of the CHFR gene was detected in 31% (18/58) of the EACs. The difference of methylation of CHFR promoter in EACs vs normal tissues was statistically significant (p=0.01). Analysis of DNA methylation in 15 tumor samples, as compared to their matching histologically normal samples from the same patients, demonstrated a significant increase in the level of DNA methylation in tumors compared to their corresponding normal tissue (p= 0.01), a representative summary is illustrated in Figure 2B. We next analyzed the promoter DNA methylation against mRNA expression levels in all samples. As shown in figure 3A, samples with hypermethylation (≥20%) had significant down-regulation for CHFR expression (p≤0.05) as compared to samples with absence or low promoter DNA methylation level (≤20%). Using spearman correlation for analysis we found a significant inverse correlation between promoter DNA methylation and mRNA expression fold for CHFR (coefficient r= −0.48, p<0.001) (Figure 3B). These results suggest that the hypermethylation of the CHFR promoter region is one of the factors involved in suppression of its mRNA expression in EACs.
Figure 2. Comparison of DNA methylation levels.
A) The percentage of promoter DNA methylation of the CHFR gene was determined by quantitative bisulfate pyrosequencing (Biotage). The horizontal bars locate the mean levels of DNA methylation. The statistical analysis of DNA methylation levels were determined by t-test. The tumors (EACs) were compared to normal samples. A p value of 0.05 was considered statistically significant. B) Methylation levels of eight representative matching normal and tumor samples.
Figure 3. A correlation analysis between DNA methylation and gene expression levels.
A) The mRNA expression fold is shown for non methylated (Black box) and methylated (Black triangle) samples. The horizontal bars locate the means expression fold. The statistical difference was determined by t-test for paired samples (p<0.05). B) The Spearman correlation analysis between DNA methylation level and mRNA expression fold in CHFR gene. Significant correlation was found for CHFR (p<0.001). C) Treatment of OE33 esophageal adenocarcinoma cells with 5-Aza led to a decrease in DNA methylation and restoration of CHFR expression. The treatment with TSA or DMSO (control) had no effect on DNA methylation or expression levels of CHFR.
To confirm the role of DNA methylation in silencing CHFR expression, we tested whether interference with the activities of DNA methyltransferases lead to reactivation of CHFR in an esophageal adenocarcinoma cell line, OE33. The OE33 cells express low levels of CHFR and their promoter CpGs are hypermethylated (80%). The 5-Aza treatment but not DMSO or TSA led to reduction in the DNA methylation level from 80% to 45% and a 13-fold increase in mRNA expression (Figure 3C).
Loss of DNA copy numbers interplays with DNA hypermethylation for silencing CHFR mRNA expression in EACs
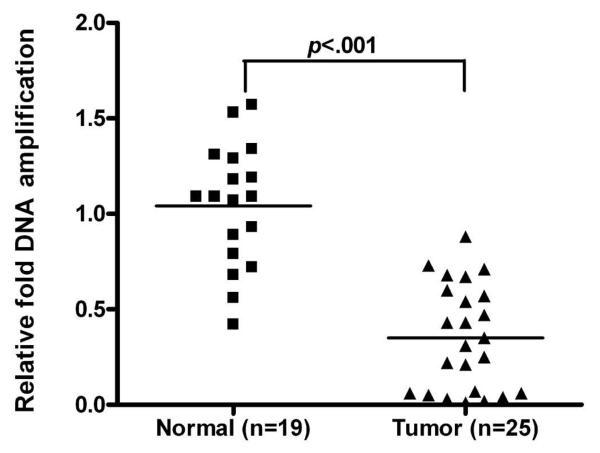
Although promoter DNA hypermethylation correlated statistically with low gene expression levels, we detected silencing of mRNA CHFR expression in 78.6% (44/56) of cases, whereas promoter DNA hypermethylation was only seen in 21.4% (12/56) of EACs. These findings prompted us to find out whether loss of copy numbers could be a contributing factor in silencing CHFR expression. Evaluation of relative DNA copy numbers in 27 tumor samples was compared to the average of eleven histologically normal esophageal samples (adjacent to tumor areas) and eight blood samples from normal individuals (total n=19). There is no significant difference on DNA copy numbers between the normal esophagus and blood samples (data not shown). As shown in Figure 4, there was a loss of DNA copy numbers (≤0.5 fold) in 16 of 27 of the analyzed tumors (59%). Seven cases showed a combination of promoter DNA hypermethylation and loss of DNA copy numbers whereas nine cases showed loss of DNA copy numbers in absence of promoter DNA hypermethylation, (Table 1). This interplay of epigenetic and genetic mechanisms for silencing CHFR suggests that silencing of CHFR is a critical alteration for EACs.
Figure 4. Comparison of DNA copy number amplification (variation is better or not?) in Normal vs Tumor samples.
Quantification of CHFR DNA copy number in 19 normal samples and 25 EACs was determined using Real-time PCR and normalized to the average value of all the normal samples, as described in materials and methods. Black boxes and triangles represent normal and tumor samples respectively. The horizontal bars represent the mean of DNA copy number.
Table 1.
Genetic and epigenetic silencing of CHFR.
| ID | Fold Relative DNA copy numbers |
% Methylation | Fold Relative mRNA expression |
|
|---|---|---|---|---|
| methylation ≤ 20% | 615T | 0.02 | 6.96 | 0 |
| 228T | 0.03 | 1.93 | 0.05 | |
| 533T | 0.05 | 19.61 | 0.43 | |
| 158T | 0.22 | 9.68 | 0.34 | |
| 602T | 0.25 | 4.18 | 0.25 | |
| 602T | 0.31 | 4.18 | 0.58 | |
| 658T | 0.47 | 4.39 | 0.17 | |
| 149T | 0.6 | 1.39 | 0.69 | |
| 101T | 0.68 | 4.14 | 0.02 | |
|
| ||||
| methylation ≥ 20% | 409T | 0.57 | 42 | 0.01 |
| 195T | 0.35 | 38 | 0.07 | |
| 460T | 0.54 | 64 | 0.1 | |
| 299T | 0.07 | 58 | 0.11 | |
| 70T | 0.43 | 35 | 0.24 | |
| 438T | 0.06 | 41 | 0.54 | |
| 1653T | 0.04 | 32 | 0.16 | |
The upper section demonstrates cases that had mRNA down-regulation of CHFR in absence of promoter DNA methylation (≤20%). The lower section shows a combination of DNA methylation (≥20%) and loss of DNA copy numbers.
Immunohistochemistry of CHFR protein expression in EACs
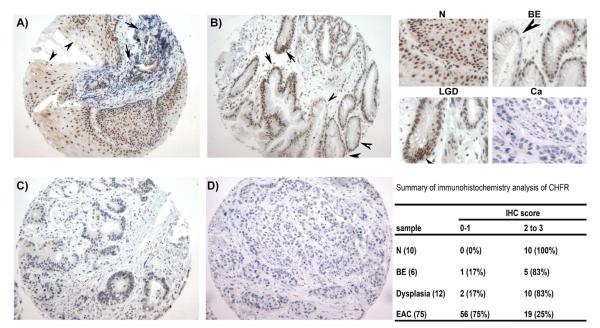
Because protein expression is the ultimate mediator of the biological processes, we followed on our findings by using immunohistochemistry in primary tumor samples. The immunohistochemical analysis of CHFR protein expression in a tumor tissue microarray demonstrated absence or weak nuclear immunostaining (score 0 and 1) for the CHFR protein in 75% (56 of 75) of the EACs (Figure 5). In contrast, normal esophageal and gastric tissue samples demonstrated moderate to strong nuclear immunostaining (Score 2 and 3) in all tested samples (Table 1, and Figure 5). Although the number of cases with premalignant lesions was small, we observed a moderate to strong immunostaining of CHFR in BE, whereas adjacent dysplasia and adenocarcinoma tissues displayed weak to absent immunostaining (Table 1 and Figure 5).
Figure 5. Immunohistochemistry analysis of CHFR expression.
A) Histologically normal esophageal squamous epithelium (arrowheads) demonstrates strong (3+ score) nuclear immunostaining for CHFR. Adjacent poorly differentiated esophageal adenocarcinoma, indicated by arrows, demonstrates weak or absent nuclear immunostaining. B) Barrett's esophagus epithelium (arrowhead) and Barrett's low grade dysplasia (arrows) show moderate nuclear immunostaining (2+). C-D) moderately differentiated (C) and poorly differentiated (D) esophageal adenocarcinoma demonstrate weak (+1) or absent (0) nuclear immunostaining for CHFR, respectively. The upper right corner demonstrates insets of A-D panel at ×100 magnification of normal (N), Barrett's esophagus (BE), low grade dysplasia (LGD) and poorly differentiated adenocarcinoma (Ca). The IHC data are summarized on the lower right corner.
Due to the relatively small number, we did not have sufficient statistical power to analyze correlation with histopathological parameters.
Discussion
In this study, we have shown that silencing of CHFR expression in esophageal adenocarcinomas is mediated by both epigenetic and genetic mechanisms. These findings indicate that cancer cells develop the mechanisms that ensure loss of function of CHFR. In this context, our findings that CHFR expression is frequently lost in EACs further confirm the reports showing that loss of CHFR is critical for cellular transformation and carcinogenesis 14. Previous studies have demonstrated variable results for CHFR methylation (10 −40%) in carcinomas of the colon, head and neck, lung, and stomach 19-23. None of these studies has investigated changes in copy numbers of CHFR.
While promoter DNA hypermethylation has been described for several genes in EAC such as APC, GPX3, GPX7, CDKN2A, DAPK, ID4, MGMT, RBP1, RUNX3, SFRP1, TIMP3, and TMEFF2 24-27, CHFR has not been investigated in EAC. We have applied state-of-the-art pyrosequencing technology for quantitative analysis of promoter DNA methylation of CHFR. We demonstrated a strong inverse correlation between the DNA methylation and gene expression levels. We also showed that promoter DNA methylation of CHFR was significantly higher in EACs as compared to adjacent normal tissue from the same patient. Using in vitro model for validation of the impact of DNA methylation on CHFR expression, we showed that treatments with 5-Aza alone can restore the expression of CHFR with a reversal in the DNA methylation level. Taken together, our data confirm that aberrant hypermethylation of the 5′CpG island of the CHFR gene is closely associated with the transcriptional inactivation and might be involved in tumor development.
A striking finding was the presence of significant loss of expression of CHFR without a notable increase in promoter DNA methylation in several cases. Interestingly, CHFR is located in chromosome 12q24, a genomic area that often displays loss of DNA copy numbers in EAC 28, 29. As shown in this report, our analysis demonstrated a reduction in DNA copy numbers of CHFR in 59% of analyzed tumors; 33% of these cases (9/27) had mRNA down-regulation of CHFR in absence of promoter DNA hypermethylation (<20%). However, 26% of the analyzed cases showed a combination of DNA hypermethylation and loss of DNA copy numbers, suggesting that the loss of CHFR expression could be due to a combined effect of the loss of one allele and methylation of the second allele. Alternatively, this could be explained by the existence of heterogeneous clones where in some cells the loss of CHFR expression was driven by promoter hypermethylation whereas in other cells, this phenomenon was mediated by loss of gene copy numbers. The first explanation is more plausible, although single cell based approaches are needed to confirm this hypothesis. An additional layer of gene expression control includes regulation by miRNAs. Although we have not investigated the role of miRNA in regulating CHFR this paper, there are recent reports suggesting that miRNAs are involved in gene regulation and progression of EACs 30. Of note, the IHC results in our study have confirmed the significant reduction in CHFR expression in primary EACs.
Cell cycle progression is monitored by checkpoint mechanisms to ensure the integrity of the genome and the fidelity of sister chromatid separation. CHFR (checkpoint with forkhead-associated and ring finger) is an E3 ubiquitin ligase and an early mitotic checkpoint protein implicated in many cancers and in the maintenance of genomic stability 13, 31-33. Failure of such checkpoint functions results in genomic instability, a condition that predisposes cells to neoplastic transformation and tumor progression 21. Recent studies have shown that CHFR can interact with polo-like kinase 1 (PLK1), a key regulator of G2/M check point 34, 35. Moreover, there are several studies indicating that CHFR is a negative regulator of the expression levels of Aurora kinase A (AURKA). AURKA encodes a centrosome associated cell cycle regulated serine/threonine kinase 36 that functions to establish mitotic spindles by regulating centrosome duplication and separation, as well as microtubule-kinetochore attachment, spindle checkpoint and cytokinesis. Cytological analysis revealed that over-expression of AURKA results in centrosome amplification, cytokinesis failure and aneuploidy 37. The tumor development and progression cascades are often characterized by a high incidence of DNA copy number variations and aneuploidy 29, 38-41. Indeed, we and others have shown that chromosomal instability and DNA copy number variations are common features of esophageal adenocarcinomas 28, 42-44, pointing out the disruption of key mechanisms that regulate mitosis and chromosomal segregation. In this context, CHFR, acting as a mitotic stress checkpoint gene, could be critical in regulating mitosis and protecting against stress induced mitotic errors.
Taken together, our data indicate interplay between epigenetic and genetic mechanisms for inactivation of CHFR, suggesting its involvement in development and progression of esophageal adenocarcinomas.
Acknowledgments
This work was supported by the National Cancer Institute Grant CA106176 and R01CA131225 (WER). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or Vanderbilt University.
References
- 1.Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83(10):2049–53. [PubMed] [Google Scholar]
- 2.Jankowski JA, Wright NA, Meltzer SJ, Triadafilopoulos G, Geboes K, Casson AG, et al. Molecular evolution of the metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Pathol. 1999;154(4):965–73. doi: 10.1016/S0002-9440(10)65346-1. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10233832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veugelers PJ, Porter GA, Guernsey DL, Casson AG. Obesity and lifestyle risk factors for gastroesophageal reflux disease, Barrett esophagus and esophageal adenocarcinoma. Dis Esophagus. 2006;19(5):321–8. doi: 10.1111/j.1442-2050.2006.00602.x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16984526. [DOI] [PubMed] [Google Scholar]
- 4.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22(1):106–9. doi: 10.1038/8816. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10319873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang Z, Vortmeyer AO, Mark EJ, Odze R, Emmert-Buck MR, Merino MJ, et al. Barrett's esophagus: metaplastic cells with loss of heterozygosity at the APC gene locus are clonal precursors to invasive adenocarcinoma. Cancer Res. 1996;56(9):1961–4. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8616831. [PubMed] [Google Scholar]
- 6.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60(18):5021–6. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11016622. [PubMed] [Google Scholar]
- 7.El-Rifai W, Frierson HJ, Moskaluk C, Harper J, Petroni G, Bissonette E, et al. Genetic Differences between adenocarcinomas arising in Barrett's esophagus and gastric mucosa. Gastroenterology. 2001;121:592–98. doi: 10.1053/gast.2001.27215. [DOI] [PubMed] [Google Scholar]
- 8.Metzger R, Schneider PM, Warnecke-Eberz U, Brabender J, Holscher AH. Molecular biology of esophageal cancer. Onkologie. 2004;27(2):200–6. doi: 10.1159/000076913. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15138356. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–28. doi: 10.1038/nrg816. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12042769. [DOI] [PubMed] [Google Scholar]
- 10.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–16. doi: 10.1038/nrc1799. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16491070. [DOI] [PubMed] [Google Scholar]
- 11.Chan AO, Rashid A. CpG island methylation in precursors of gastrointestinal malignancies. Curr Mol Med. 2006;6(4):401–8. doi: 10.2174/156652406777435417. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16900663. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–54. doi: 10.1056/NEJMra023075. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14627790. [DOI] [PubMed] [Google Scholar]
- 13.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406(6794):430–5. doi: 10.1038/35019108. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10935642. [DOI] [PubMed] [Google Scholar]
- 14.Privette LM, Petty EM. CHFR: A Novel Mitotic Checkpoint Protein and Regulator of Tumorigenesis. Transl Oncol. 2008;1(2):57–64. doi: 10.1593/tlo.08109. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18633460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlemper RJ, Kato Y, Stolte M. Diagnostic criteria for gastrointestinal carcinomas in Japan and Western countries: proposal for a new classification system of gastrointestinal epithelial neoplasia. J Gastroenterol Hepatol. 2000;15(Suppl):G49–57. doi: 10.1046/j.1440-1746.2000.02266.x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11100994. [DOI] [PubMed] [Google Scholar]
- 16.El-Rifai W, Moskaluk CA, Abdrabbo MK, Harper J, Yoshida C, Riggins GJ, et al. Gastric cancers overexpress S100A calcium-binding proteins. Cancer Res. 2002;62(23):6823–6. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12460893. [PubMed] [Google Scholar]
- 17.El-Rifai W, Moskaluk CA, Abdrabbo M, Harper JC, Yoshida C, Riggins G, et al. Gastric Cancers Overexpress S100A calcium binding proteins. Cancer Res. 2002;62:6823–26. [PubMed] [Google Scholar]
- 18.Siewert JR, Feith M, Stein HJ. Biologic and clinical variations of adenocarcinoma at the esophago-gastric junction: relevance of a topographic-anatomic subclassification. J Surg Oncol. 2005;90(3):139–46. doi: 10.1002/jso.20218. discussion 46. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15895452. [DOI] [PubMed] [Google Scholar]
- 19.Toyota MSY, Satoh A, Ogi K, Kikuchi T, Suzuki H, Mita HTN, Itoh F, Issa JPJ, Jair KW, Schuebel KE, T IKaT. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci USA. 2003;100:7818–23. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuno K, Osada H, Konishi H, Tatematsu Y, Yatabe Y, Mitsudomi T, et al. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene. 2002;21(15):2328–33. doi: 10.1038/sj.onc.1205402. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11948416. [DOI] [PubMed] [Google Scholar]
- 21.Shibata Y, Haruki N, Kuwabara Y, Ishiguro H, Shinoda N, Sato A, et al. Chfr expression is downregulated by CpG island hypermethylation in esophageal cancer. Carcinogenesis. 2002;23(10):1695–9. doi: 10.1093/carcin/23.10.1695. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12376479. [DOI] [PubMed] [Google Scholar]
- 22.Corn PG, Summers MK, Fogt F, Virmani AK, Gazdar AF, Halazonetis TD, et al. Frequent hypermethylation of the 5′ CpG island of the mitotic stress checkpoint gene Chfr in colorectal and non-small cell lung cancer. Carcinogenesis. 2003;24(1):47–51. doi: 10.1093/carcin/24.1.47. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12538348. [DOI] [PubMed] [Google Scholar]
- 23.Kang HC, Kim IJ, Park JH, Shin Y, Park HW, Ku JL, et al. Promoter hypermethylation and silencing of CHFR mitotic stress checkpoint gene in human gastric cancers. Oncol Rep. 2004;12(1):129–33. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15201973. [PubMed] [Google Scholar]
- 24.Kuester D, Dar AA, Moskaluk CC, Krueger S, Meyer F, Hartig R, et al. Early involvement of death-associated protein kinase promoter hypermethylation in the carcinogenesis of Barrett's esophageal adenocarcinoma and its association with clinical progression. Neoplasia. 2007;9(3):236–45. doi: 10.1593/neo.06802. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17401463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng DF, Razvi M, Chen H, Washington K, Roessner A, Schneider-Stock R, et al. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett's adenocarcinoma. Gut. 2009;58(1):5–15. doi: 10.1136/gut.2007.146290. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18664505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee OJ, Schneider-Stock R, McChesney PA, Kuester D, Roessner A, Vieth M, et al. Hypermethylation and loss of expression of glutathione peroxidase-3 in Barrett's tumorigenesis. Neoplasia. 2005;7(9):854–61. doi: 10.1593/neo.05328. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16229808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith E, De Young NJ, Pavey SJ, Hayward NK, Nancarrow DJ, Whiteman DC, et al. Similarity of aberrant DNA methylation in Barrett's esophagus and esophageal adenocarcinoma. Mol Cancer. 2008;7:75. doi: 10.1186/1476-4598-7-75. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18831746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Rifai W, Frierson HF, Jr., Moskaluk CA, Harper JC, Petroni GR, Bissonette EA, et al. Genetic differences between adenocarcinomas arising in Barrett's esophagus and gastric mucosa. Gastroenterology. 2001;121(3):592–8. doi: 10.1053/gast.2001.27215. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11522743. [DOI] [PubMed] [Google Scholar]
- 29.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, et al. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155(3):683–94. doi: 10.1016/S0002-9440(10)65166-8. Available from http://www.ncbi.nlm.nih.gov/cgibin/Entrez/referer? http://www.amjpathol.org/cgi/content/full/155/3/683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136(5):1689–700. doi: 10.1053/j.gastro.2009.02.002. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19422085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortez D, Elledge SJ. Conducting the mitotic symphony. Nature. 2000;406(6794):354–6. doi: 10.1038/35019227. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10935617. [DOI] [PubMed] [Google Scholar]
- 32.Kang D, Chen J, Wong J, Fang G. The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J Cell Biol. 2002;156(2):249–59. doi: 10.1083/jcb.200108016. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11807090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Privette LM, Weier JF, Nguyen HN, Yu X, Petty EM. Loss of CHFR in human mammary epithelial cells causes genomic instability by disrupting the mitotic spindle assembly checkpoint. Neoplasia. 2008;10(7):643–52. doi: 10.1593/neo.08176. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18592005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erson AE, Petty EM. CHFR-associated early G2/M checkpoint defects in breast cancer cells. Mol Carcinog. 2004;39(1):26–33. doi: 10.1002/mc.10161. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14694445. [DOI] [PubMed] [Google Scholar]
- 35.Shtivelman E. Promotion of mitosis by activated protein kinase B after DNA damage involves polo-like kinase 1 and checkpoint protein CHFR. Mol Cancer Res. 2003;1(13):959–69. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14638868. [PubMed] [Google Scholar]
- 36.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, et al. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156(3):437–51. doi: 10.1083/jcb.200108135. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11827981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–93. doi: 10.1038/2496. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9771714. [DOI] [PubMed] [Google Scholar]
- 38.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36(3):125–31. doi: 10.1016/S1673-8527(08)60099-5. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19302968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39(7 Suppl):S16–21. doi: 10.1038/ng2028. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17597776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knuutila S, Björkqvist A-M, Autio K, Tarkkanen M, Wolf M, Monni O, et al. DNA copy number amplifications in human neoplasms. Review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–23. [PMC free article] [PubMed] [Google Scholar]
- 41.Langer C, Gunawan B, Schuler P, Huber W, Fuzesi L, Becker H. Prognostic factors influencing surgical management and outcome of gastrointestinal stromal tumours. Br J Surg. 2003;90(3):332–9. doi: 10.1002/bjs.4046. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12594669. [DOI] [PubMed] [Google Scholar]
- 42.Walch AK, Zitzelsberger HF, Bruch J, Keller G, Angermeier D, Aubele MM, et al. Chromosomal imbalances in Barrett's adenocarcinoma and the metaplasia-dysplasia-carcinoma sequence. Am J Pathol. 2000;156(2):555–66. doi: 10.1016/S0002-9440(10)64760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller J, Werner M, Siewert JR. Malignant progression in Barrett's esophagus: pathology and molecular biology. Recent Results Cancer Res. 2000;155:29–41. doi: 10.1007/978-3-642-59600-1_3. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10693236. [DOI] [PubMed] [Google Scholar]
- 44.van Dekken H, Vissers CJ, Tilanus HW, Tanke HJ, Rosenberg C. Clonal analysis of a case of multifocal oesophageal (Barrett's) adenocarcinoma by comparative genomic hybridization. J Pathol. 1999;188(3):263–6. doi: 10.1002/(SICI)1096-9896(199907)188:3<263::AID-PATH374>3.0.CO;2-Y. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10419593. [DOI] [PubMed] [Google Scholar]