Abstract
There is a complex relationship between the evolution of segmental duplications and rearrangements associated with human disease. We performed a detailed analysis of one region on chromosome 16p12.1 associated with neurocognitive disease and identified one of the largest structural inconsistencies with the human reference assembly. Various genomic analyses show that all examined humans are homozygously inverted relative to the reference genome for a 1.1-Mbp region on 16p12.1. We determined that this assembly discrepancy stems from two common structural configurations with worldwide frequencies of 17.6% (S1) and 82.4% (S2). This polymorphism arose from the rapid integration of segmental duplications, precipitating two local inversions within the human lineage over the last 10 million years. The two human haplotypes differ by 333 kbp of additional duplicated sequence present in S2 but not in S1. Importantly, we show that the S2 configuration harbors directly oriented duplications specifically predisposing this chromosome to disease rearrangement.
INTRODUCTION
Numerous studies have shown that segmental duplications and the flanking unique regions are sites of both rare and common copy-number polymorphism (CNP) 1–3. Segmental duplications are blocks of DNA >1 kb in size that occur at more than one site within the genome and typically share a high level (>90%) of sequence identity 4–6. Duplicated blocks may be substrates for non-allelic homologous recombination (NAHR) resulting in large structural polymorphisms and chromosomal rearrangements that directly lead to genomic disorders 5,7–13. NAHR between directly oriented segmental duplications results in deletions or reciprocal duplications of the genomic segment between them, whereas NAHR between inverted segmental duplications leads to an inversion of the intervening sequence.
Recently, a recurrent microdeletion on chromosome 16p12.1 was reported as a risk factor for childhood intellectual disability and developmental delay 14. The microdeletion was found to be inherited in 95.6% of the cases and 24% of the probands carried an additional large duplication or deletion elsewhere in the genome. The data suggested a two-hit copy-number variation (CNV) model in which the 16p12.1 microdeletion results in severe neurodevelopmental phenotypes when coupled to an additional genetic, epigenetic, or environmental abnormality.
Using high density and targeted array-based comparative genomic hybridization (CGH) experiments, we mapped the 16p12.1 microdeletion breakpoints to large blocks of segmental duplications, which we posited might mediate the recurrent rearrangement associated with disease 15. The extensive copy-number variation and inconsistencies between the reference genome and various genomic analyses, however, complicated breakpoint assessment suggesting that large alternative structural configurations might exist within the human population 16,17. We therefore investigated this region by conducting a detailed analysis by fluorescence in situ hybridization (FISH), arrayCGH, optical mapping, and sequencing of large-insert BAC clones in order to understand the extent of human genetic variation, its origin, and the impact on disease.
RESULTS
Resolution of a reference genome assembly error
We initially began our investigation of the region by testing whether the gene order within this ~1-Mbp region was consistent with published reference genome assemblies (GRCb37 and build 36). We performed a series of cohybridization FISH experiments on 10 HapMap cell lines using probes corresponding to unique sequences flanking the duplication blocks (Supplementary Note). FISH results showed that 20/20 chromosomes tested were inverted relative to build 36 and GRCb37 suggesting a potential error in the orientation of the reference genome assembly involving 18 genes (Supplementary Note). To confirm this surprisingly large-scale difference, we used optical mapping 18,19 to generate single-molecule restriction maps from the genomes of GM18994 and GM10860 cell lines. We compared the consensus maps to a restriction map generated in silico from the build 36 human genome reference sequence. Maps from both genomes confirm a large inversion spanning from the duplication blocks defined as breakpoint regions BP1 and BP3 (build 36, chr16:21421324-22464053) (Supplementary Note; Supplementary Figure S1).
As a final test, we generated a map of contiguous clones of the region from the CHORI-17 BAC library from a hydatidiform (haploid) mole derived human cell line (CHM1hTERT) 20 (http://bacpac.chori.org/library.php?id=231). Complete hydatidiform moles arise from the fertilization of an enucleated egg from a single sperm and, therefore, carry a haploid complement of the human genome eliminating allelic variation that may confound mapping and assembly. We constructed a contiguous set of 10 BAC clones corresponding to this 1.6-Mbp region on 16p12.1 and then sequenced the inserts using Illumina technology. We generated 406 Mbp of sequence (270-fold coverage) from these clones and aligned it to both the human reference genome assembly and our reconstructed inverted version of the region (see below). The mapped sequence data from these clones were consistent with the entire region being inverted within the hydatidiform mole (Supplementary Note). Thus, all three analyses indicate that orientation of the sequence between BP1 and BP3 should be flipped with respect to published versions of the human genome (Figure 1).
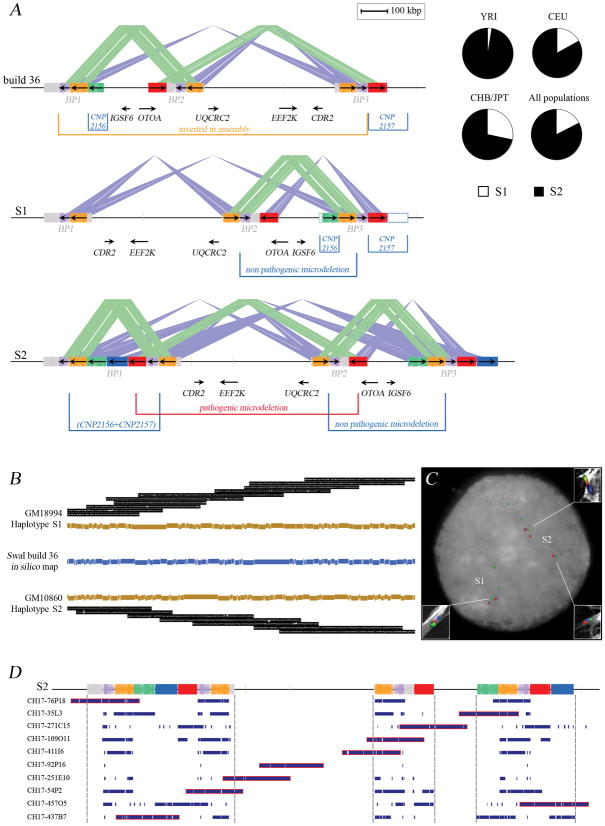
Figure 1.
Alternate structural configurations of the 16p12.1 region. (A) The organization in the reference genome (build 36, top schematic) is compared against two experimentally validated structural configurations (S1 and S2). The locations of the inversion, copy-number polymorphisms 2 (CNP2156 and CNP2157), a rare (20/6712) non-pathogenic deletion variant 1 and segmental duplications (colored rectangles) are indicated. Dashed empty boxes at the S1 structure correspond to regions duplicated in S2 but present in single copy in the S1 haplotype. The S1 and S2 structures differ because of the presence of the distal duplication segment (CNP2156 and CNP2157 at BP1) on the S2 haplotype. Based on this structure, the S1 configuration is predicted to be protective against occurrence of the 16p12.1 pathogenic microdeletion. The red block corresponds to the 68-kbp segmental duplication that likely mediates, through NAHR, the recurrent 16p12.1 microdeletion in patients 14. Segments duplicated in a direct orientation are connected by green lines while sequences duplicated in an inverted orientation are connected by blue lines. (B) The organization of the region was experimentally validated by optical mapping. SwaI single-molecule restriction maps are depicted and summarized for both configurations (Supplementary Note). (C) The large-scale orientation of each block was confirmed by FISH experiments on interphase nuclei and stretched chromosomes (white rectangles) using probes mapping at the red, blue and green segmental duplications. (D) A contig of 10 BAC clones from the genome of the complete hydatidiform mole (CHM1hTERT) along the 16p12.1 region was sequenced. All clones mapped against the S2 structure were concordant.
Copy number and structural polymorphism
One of the predicted consequences of this inverted orientation of the human genome is that the location of previously described segmental duplications and copy-number polymorphisms change with respect to disease-associated breakpoints. The deletion breakpoints associated with intellectual disability now map to BP1 and BP2 based on the correct orientation (build 36, chr16:21716331-22464053) (Figure 1A). These variable regions correspond, in part, to two sites of common copy-number polymorphism (CNP2156 and CNP2157) identified in the HapMap sample collection by McCarroll and colleagues 2. Both loci have three reported copy-number (CN) states (diploid copy numbers of 2, 3, and 4), with the highest copy-number state (CN = 4) having a frequency of 73% in Europeans (CEU), 95% in Yorubans (YRI), and 52% in Asians (CHB/JPT) (Supplementary Note). We performed a series of FISH and arrayCGH experiments to determine the absolute copy number, location and extent of copy-number polymorphism within this region (Supplementary Note).
We analyzed 11 DNA control samples (Supplementary Note) using a customized oligonucleotide microarray and found good correspondence between predicted CNP2157 genotypes and expected signal intensity differences between samples (Figure 2). ArrayCGH data for CNP2156 was less clear and the data suggested more extensive copy-number variation than was originally defined, although the location of this variation could not be determined based solely on hybridization data. We therefore designed a series of three-color FISH experiments to investigate copy number and location. FISH analysis showed that the absolute copy number of the 68-kbp segment corresponding to the distal region of CNP2157 differed by a count of two with respect to previous reports (CN = 4, 5 and 6). Similarly, FISH analysis for the CNP2156 region showed an absolute count that is four copies greater than previously reported genotype estimates (Supplementary Note) 2. FISH mapping showed that the variable sequences corresponding to CNP2156 and CNP2157 map adjacent to one another within the BP1 region (Supplementary Note; Figure 1). Thus, the two reported CNP regions actually correspond to a single segment of variable sequence that has been duplicatively transposed from BP3 to BP1. In total, these experiments revealed the presence of two distinct structural configurations for the 16p12.1 region, which we refer to as S1 and S2, with the S2 haplotype showing the greater duplication complexity (Figure 1).
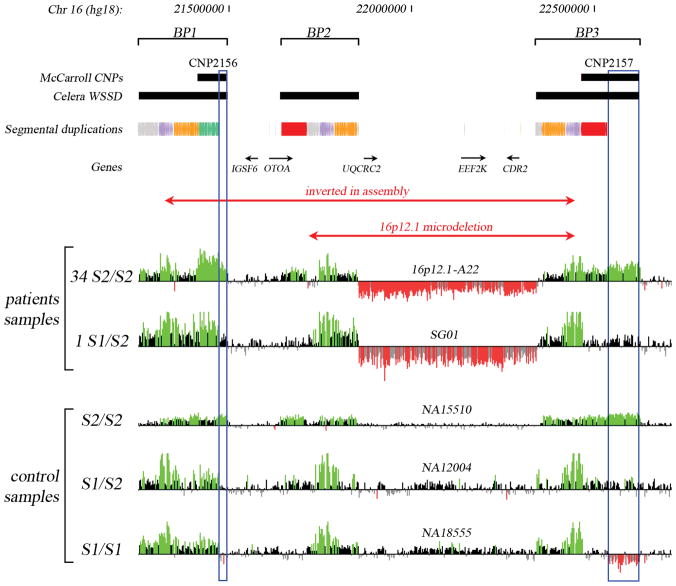
Figure 2.
ArrayCGH data for 16p12.1 microdeletion patient samples and control HapMap samples (NA15510, NA12004 and NA18555). Probes with log2 ratios above or below a threshold of 1.5 standard deviations from the normalized mean log2 ratio are colored green (duplication) or red (deletion), respectively. The positions of copy-number polymorphisms (CNP2156 and CNP2157) and segmental duplications are indicated. Blue empty boxes highlight the S2-specific duplications that have a diploid copy number of 2 in S1/S1 individuals, 3 in S1/S2 heterozygotes, and 4 in S2/S2 homozygotes. HapMap sample NA18956 with S1/S2 genotype was used as reference.
Since our analyses predicted a large, alternate structural polymorphism, we searched GenBank for additional sequenced BACs from this region. We identified clones anchored within the unique region distal to BP1 and constructed an alternate assembly from four BAC clones not included in the human reference genome assembly (Supplementary Note). We assembled a 433-kbp alternate sequence haplotype corresponding to most of the additional duplicated sequence in BP1. Detailed comparisons with FISH, optical mapping and fosmid end-sequence pair data all provide strong support for the orientation and location of the additional duplicated copies on the S2 chromosomal configuration (Supplementary Note).
The combined analysis identifies one of the largest, common copy-number polymorphisms in human euchromatin. We identify a total of 333 kbp of duplicated sequence that is specific to S2 when compared to the BP1 region of S1. Since this additional sequence is homologous to BP1 and BP2, this polymorphism creates additional direct and inverted blocks of high sequence identity making S2 prone to rearrangement events mediated by NAHR 15. Only the S2 configuration has segmental duplications in the direct orientation necessary to drive the formation of microdeletions associated with disease. We note that the S2-specific segmental duplications at BP1 show the highest sequence identity (99.85%) with BP3 when compared to BP2 (99.47%), consistent with a recent duplicative transposition event from BP3 placing a large inverted duplication within BP1.
Disease risk
The large-scale structural polymorphism between S1 and S2 allows us to make some testable predictions regarding differences in susceptibility to microdeletion and disease. Since only the S2 configuration possesses directly oriented duplications, we hypothesized that the breakpoints would map to this 68-kbp segment and that only carriers of the S2 configuration would be predisposed to the 16p12.1 microdeletion. Interestingly, we find that the S2 structure is the most common world-wide haplotype with frequencies of 97.5% in Africans (YRI), 83.1% in Europeans (CEU) and 71.6% in Asian populations (CHB/JPT) 2 (Table 1). This general observation is confirmed by an examination of a larger group of African samples, which show the almost complete absence of the protective S1 haplotype (Supplementary Note). Thus, we hypothesize that African and European populations should be more at risk for the 16p12.1 microdeletion “syndrome” than Asians.
Table 1.
S1 and S2 haplotype frequencies.
| Population | S1 frequency | S2 frequency |
|---|---|---|
| Asians (CHB/JPT) | 0.28 | 0.72 |
| Yorubans (YRI) | 0.03 | 0.98 |
| Europeans (CEU) | 0.17 | 0.83 |
| microdeletion samples | 0.01 | 0.99 |
The frequencies of S1 and S2 haplotypes in 3 HapMap populations are shown. Analysis of 35 patients with the 16p12.1 microdeletion confirmed a non-Hardy-Weinberg equilibrium enrichment of the S2 haplotype (p-value=0.0088) suggesting that this structural polymorphism predisposes to deletion and disease.
One way to test if the S2 haplotype predisposes to microdeletion is to determine on which structure the microdeletion occurs. However, most of the identified cases are inherited and parental DNA for additional genotyping is not available 14. We therefore determined the structural genotype present in each of the cases using array comparative genomic hybridization. The presence of any S1/S1 homozygotes that also have 16p12.1 microdeletion would be inconsistent with the proposed rearrangement structures and mechanism. Since the S2 haplotype has a more extended segmental duplication architecture than S1, differences in the chromosomal configuration can be easily deduced (Figure 2). In particular, the S2-specific duplication block corresponding to the distal segment of CNP2157 (blue empty box in Figure 2) has a diploid copy number of 2 in S1/S1 individuals, 3 in S1/S2 heterozygotes, and 4 in S2/S2 homozygotes.
We examined 35 microdeletion samples by arrayCGH using two reference samples with known genotypes (NA15724 = S2/S2 and NA18956 = S1/S2). Self-identified ethnicity was provided for 27 of these patients (21 European and 6 African descent). Based on the observed mean log2 values for the S2 specific duplication block, the genotype of each sample was determined (Figure 2; Supplementary Figures S2 and S3; Supplementary Note). We found that 97% (34/35) of the cases were homozygous for the S2/S2 haplotype with only a single heterozygous carrier (S1/S2) being identified in the patient population (Table 1). This represents a significant enrichment of the S2 haplotype when matching for ethnicity of the sample collection (p-value = 0.0088, Hardy-Weinberg equilibrium test). Furthermore, arrayCGH data from 15/16 patients were consistent with breakpoints mapping within the 68-kbp S2-specific duplication (Supplementary Note). These combined data strongly suggest that the S2, and not S1, haplotype predisposes to the 16p12.1 microdeletion associated with intellectual disability and neurocognitive disease (Table 1).
Evolutionary origin
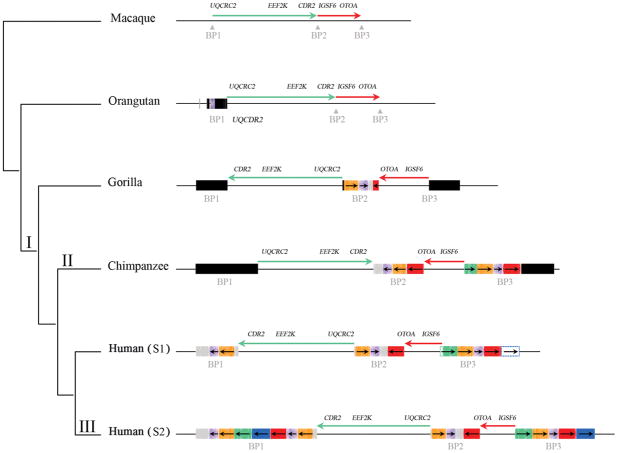
In order to investigate the ancestral configuration of the 16p12.1 region, we compared the orientation of the region in human with other non-human primate species. Notably, sequence comparison of the orangutan (WUGSC 2.0.2/ponAbe2) and human sequence at 16p12.1 revealed an expansion of the region in human due to the integration of segmental duplications accompanied by two local inversions of 481 kbp and 142 kbp (Supplementary Note). We tested for the presence of the larger inversion between BP1 and BP2 (481 kbp) by FISH analysis of cell lines from three chimpanzees (Pan troglodytes), three orangutans (Pongo pygmaeus), two gorillas (Gorilla gorilla) and one macaque (Macaca mulatta) (Supplementary Note). Macaque, orangutan and chimpanzee were found to be inverted when compared to the true human genome orientation suggesting that this represents the likely ancestral state. To resolve the status of the smaller inversion (BP2-BP3) as well as duplications at the boundaries, we identified and sequenced nine large-insert chimpanzee, orangutan and gorilla BAC clones generating 1.8 Mbp of high quality ape sequence from the region (Supplementary Figure S4). Our results indicated that all African great apes are inverted for the smaller BP2-BP3 interval (142 kbp) when compared to orangutan (ponAbe2) and macaque (rheMac2) genome assemblies. We conclude that the two inversions occurred in the human-African great ape ancestor and that the region spanning BP1 to BP2 likely flipped back to the ancestral orientation in the chimpanzee lineage (Figure 3). Alternatively, the chimpanzee configuration may represent incomplete lineage sorting of an ancestral state.
Figure 3.
Expansion and multiple inversions of the 16p12.1 region in humans and the syntenic regions in non-human primates during primate evolution. The genomic organization is compared within a generally accepted phylogeny of macaque, orangutan, gorilla, chimpanzee and human. The region has expanded from 726 kbp (macaque) to 1.6 Mbp (human S2) as a result of segmental duplication accumulation (black and colored rectangles). Sequence and FISH data indicate that the inverted configuration as found in orangutan and macaque is likely the ancestral state in all mammals (I). The expansion of segmental duplications in the African great ape ancestor occurred in conjunction with two inversions between BP1 to BP2 (green arrow) and BP2 to BP3 (red arrow), which may have reverted back to the direct orientation in the chimpanzee lineage (II). The region has become increasingly complex in human leading to the addition of another polymorphic 333 kbp at BP1 specifically in the human lineage (III). Colored boxes indicate segmental duplications as determined by complete sequencing of large-insert BAC clones from primate genomic libraries (Supplementary Note).
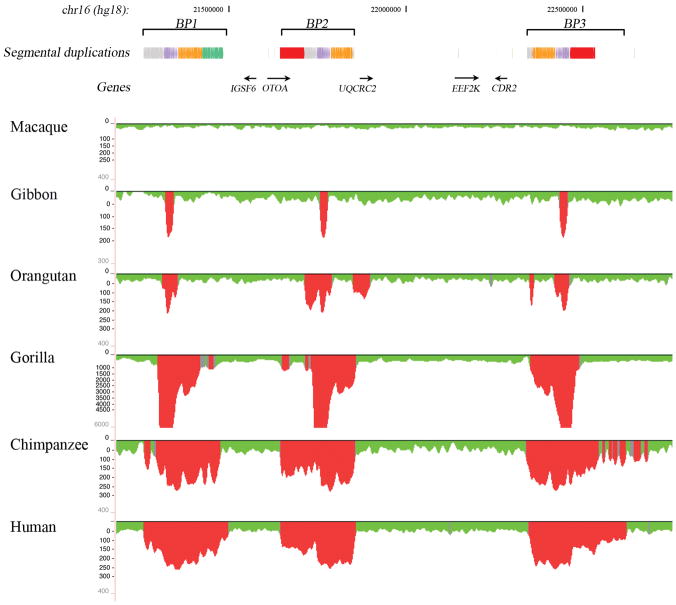
Next, we compared the extent of segmental duplications in the 16p12.1 region among human, chimpanzee, gorilla, orangutan, gibbon and macaque using a whole-genome shotgun sequence (WGS) detection method and interspecies arrayCGH 21,22. These analyses showed an expansion of segmental duplications among African great apes (human, chimpanzee, gorilla) with respect to orangutan, gibbon and macaque (Figure 4; Supplementary Note). Sequencing of orangutan BAC clones suggests that this region was largely devoid of segmental duplications in orangutan with the exception of BP1 where the composition of the duplication block differs radically from that of human (Figure 3). Sequence analysis of the BAC clones reveals the presence of duplicated sequences that are not present at this location in human or chimpanzee with the exception of a 20-kbp segment corresponding to the NPIP gene. Overall, we determined that this particular region of 16p12.1 has increased in size from 726 kbp to 1,259 kbp (S1) or 1,671 kbp (S2) during the last 10 million years primarily as a result of a duplicative transposition of segmental duplications in the region. Our primate analysis suggests that the region has become increasingly complex in the human-African great ape lineage. The euchromatin has expanded 2.3 fold in size. These changes were accompanied by two local inversions of 481 kbp and 142 kbp in length creating the genomic architecture that now predisposes this region to microdeletion and neuropsychiatric disease.
Figure 4.
Regions of segmental duplication based on read-depth mapping of whole-genome shotgun sequences (WGS) against the human genome. The figure shows an expansion of segmental duplications in the African great apes (human, chimpanzee, gorilla) with respect to orangutan, gibbon and macaque. Also shown are the segmental duplications in human annotated using SegDupMasker 44.
DISCUSSION
Our analyses highlight three important properties regarding the organization and evolution of the human genome. First, the data illustrate that the structure and copy number of even very large-scale, euchromatic regions may yet be unresolved in the human reference assembly. We describe a large 333-kbp polymorphism that has changed in copy, orientation and location over a 1-Mbp portion of chromosome 16p12.1. With estimated frequencies of 17.6% and 82.4% for the S1 and S2 configurations respectively, this represents one of the largest copy-number polymorphisms mapping within human euchromatin.
We show that previous analyses of genome structural variation 2,3,16 have failed to adequately decipher the true structure and copy number of this polymorphism. In particular, CNP analysis using Affymetrix 6.0 microarrays 2 did not accurately determine the extent of the CNP (76 kbp at CNP2156 and 146 kbp at CNP2157) due to the insensitivity of probes mapping within the duplicated regions. Moreover, FISH analyses revealed that the absolute copy number was incorrect since a baseline copy number of 2 (diploid) was assumed to represent the population average in previous analyses. This was compounded by the fact that the reference genome (GRC37 and build 36) are missing duplicated copies and present an organization that can not be validated over 1.1 Mbp. We postulate that the presence of the inverted 333-kbp duplication polymorphism led to large-scale misassembly and misorientation of sequence involving 18 genes (Figure 1). It may be somewhat surprising that such a large “error” has been uncovered nearly 10 years after the sequence and assembly of the human genome 23,24; however, it should be pointed out that at least five different types of molecular, optical mapping and cytogenetic analyses were required to resolve the architecture of this region. We anticipate that other regions of comparable complexity and variation will be uncovered and that similar, detailed analyses of large-insert clones will be required to ultimately resolve the true architecture of these regions.
Second, our comparative analyses of human and African great ape genomes reveal the evolutionary rapidity of these complex changes and their intimate association with larger chromosomal rearrangements. The 16p12.1 region has experienced a remarkable “bloating” of euchromatin, doubling the size of this region from 726 kbp to 1.6 Mbp as a result of duplicative transposition of sequences from other portions of chromosome 16. Most of these changes occurred in a ~6 million year window of evolution before the emergence of humans and great apes as distinct lineages (Figure 3) consistent with the burst of duplications in their common ancestor 21. In concert with these changes, there have been multiple local inversions specific to humans and African great apes. These findings reinforce the strong association between evolutionary inversions and segmental duplications 25–28. It is interesting that all of the 16p12.1 changes are associated with the spread of the human-great ape gene family morpheus (NPIP) 29. The core duplicon carrying this gene, LCR16a 30, maps to each of the breakpoint regions, including the boundaries of the complex copy-number polymorphism. Sequencing of large-insert ape clones suggests that these sequences also demarcate the breakpoints of the evolutionary inversions. Interestingly, the segmental duplication associated with the NPIP gene family appears to be at the breakpoints of other recurrent microdeletions on chromosome 1631–36.
Third, our findings emphasize the impact of this genetic variation with respect to human health and genomic susceptibility to neurocognitive disease. The dramatic changes in the S2 chromosome architecture mean that it is the only configuration with homologous segmental duplications in direct orientation flanking the disease-critical region. Accordingly, we find that S1 chromosomes are depleted from microdeletion patients (p-value = 0.0088 rejecting Hardy-Weinberg equilibrium) and that the breakpoints map specifically to the directly oriented duplication on S2. Combined, these results suggest that S2 chromosomes are likely to predispose to 16p12.1 microdeletion while the S1 chromosomes are immune to such rearrangement. Interestingly, Asian HapMap samples are enriched for S1 chromosomes predicting that this particular cause of intellectual disability may be less common among these populations. These results bear striking similarity to another region of the human genome on 17q21.31 where a largely Mediterranean-European-specific duplication arose in direct orientation predisposing H2 chromosomes to microdeletion associated with 17q21.31 syndrome 26,37–40. In both of these cases, changes in disease-causing architecture are also associated with inversions. We posit that this will be the underlying molecular basis for other associations that have been seen with inverted chromosomal haplotypes 41–43. These observations emphasize the importance of correctly defining alternative human genomic configurations in order to assess variable risk of subsequent pathogenic rearrangements. Molecular cytogenetics, genomic approaches, and sequencing of long molecules from single haplotypes remain the only way to correctly resolve these complex architectures of the human genome.
METHODS
FISH analysis
Metaphase spreads were obtained from lymphoblast and fibroblast cell lines from 10 human HapMap individuals (Coriell Cell Repository, Camden, NJ), three chimpanzees (Douglas; Veronica; Cochise), three orangutans (Susie, ISIS #71; PPY9; PPY6), two gorillas (AG20600; AG05251) and one macaque (MMU2). Stretched chromosomes were prepared according to Laan et al. 45. Briefly: cells were resuspended in hypotonic solution (HCM: hepes 100 mM; glycerol 1M; CaCl2 100mM; MgCl2 0.5M) for 15 minutes. The suspension was then centrifuged using a cytospin (800–1200 rpm for 5–15 minutes). FISH experiments were performed using fosmid clones directly labeled by nick-translation with Cy3-dUTP (Perkin-Elmer), Cy5-dUTP (Perkin-Elmer), and fluorescein-dUTP (Enzo) as described by Lichter et al. 46 with minor modifications. Briefly: 300 ng of labeled probe were used for the FISH experiments; hybridization was performed at 37°C in 2xSSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, and 3 μg sonicated salmon sperm DNA, in a volume of 10 μL. Posthybridization washing was at 60°C in 0.1xSSC (three times, high stringency). Nuclei were simultaneously DAPI stained. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, Cy3, Cy5 and fluorescein fluorescence signals, detected with specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software. A minimum of 50 interphase cells were scored for each inversion to statistically determine the orientation of the examined region.
Copy-number variation analysis
Microarray-based comparative genomic hybridization was performed on 35 16p12.1 microdeletion cases with intellectual disability/developmental delay and congenital malformation 14. ArrayCGH experiments on 16p12.1 microdeletion samples and HapMap samples were performed with custom, high-density oligonucleotide arrays (12-plex NimbleGen chip with a density of 1 probe per 40 bp within the 16p12.1 region; 4x180K Agilent chip targeted to copy-number polymorphic regions of the human genome (Campbell et al., unpublished), containing 50 probes in the CNP2157 at chr16:22533636-22618896).
The duplication content of human, chimpanzee, gorilla, orangutan, gibbon and macaque was determined using the whole-genome shotgun sequence detection (WSSD) method 21,47. We also assessed copy-number differences in shared duplications by interspecific array comparative genomic hybridization as previously reported 21 (GEO Accession: GSE13885). We performed cross-species arrayCGH with human, Coriell GM15510 as a reference (GEO accession number: GSE13884) using chimpanzee (Clint, Coriell S006006), gorilla (Bahati), orangutan (Susie, ISIS #71), and macaque (ID17573) samples.
Optical mapping
We examined the 16p12.1 locus in optical mapping data sets for two genomes, those of HapMap panel members GM10860 and GM18994. Briefly, optical mapping 18,19,48,49 is a whole-genome, single-molecule system for the discovery and characterization of structural variation. Individual genomic DNA molecules are restriction mapped using light microscopy, producing large data sets that are assembled into multi-megabase map contigs covering up to 98% of the euchromatic genome. These map contigs provide a global, detailed assessment of genome structure. We recovered consensus restriction maps matching the S1 haplotype from the GM18994 assembly and the S2 haplotype from GM10860; the consensus maps, their alignments back to the build 36 reference sequence (build 36), and a montage of representative single molecule micrographs are depicted in Supplementary Figure S1.
Illumina sequencing
DNA was extracted from 10 BAC clones (CHORI-17) (Supplementary Note) from the genome of a complete hydatidiform mole (CHM1hTERT) using Roche high pure plasmid isolation kit. 3 μg of DNA from each BAC were used for construction of a shotgun sequencing library as described previously 50,51 using adaptors for paired-end sequencing on an Illumina Genome Analyzer IIX (GAIIX). To allow the simultaneous sequencing of multiple BAC clones, we differentially ligated modified adaptors (Supplementary Note) to each sample during library preparation, enabling the in silico separation of samples post-sequencing 52. We obtained a total of 34,206,404 76-bp reads (17,103,202 pairs) and separated into 10 pools using 12-bp barcodes, resulting in 20,316,752 reads of length 64 bp. To control for contamination, we first aligned the reads to the E.coli reference genome (K12 strain) using mrsFAST (http://mrsfast.sourceforge.net) allowing at most 4-bp mismatches. This experiment resulted in removing 2,363,518 reads (1,181,759 pairs) from consideration due to contamination. The remaining reads (a total of 406 Mbp generated sequence) were then mapped to the 16p12 region in build 36 and the S1 and S2 haplotype sequences that we constructed. We tracked all possible map locations for the concordant pairs and discarded the discordant mappings. This resulted in reliably mapping of 6,345,136 reads (3,172,568 pairs; 406,088,704 bp of sequence) to 16p12, S1 and S2 reference sequences, corresponding to 270.7-fold coverage per BAC sequence on the average (min coverage: 132.5X, max coverage: 520.82X). Next, we merged the map locations of the overlapping pairs into contiguous segments and removed any segment <2 kbp from analysis. We reasoned that the smaller segments are mapping artifacts due to short repeats in the sequenced BAC clones and the reference sequences (16p12, S1 and S2). Finally, we visualize the resulting segments using the IGV software (Integrated Genomics Viewer, http://www.broadinstitute.org/igv).
Non-human primate BAC clone sequencing
We selected nine BAC clones from the libraries of chimpanzee (CH251), orangutan (CH276) and gorilla (CH255) genomes mapping to the 16p12.1 segmental duplications in human (Supplementary Note). We generated a clone shotgun sequence library and completely sequenced the insert of each clone. We aligned the sequence to the human genome and to the S1 haplotype that we reconstructed with miropeats 53. Final annotation with common repeats and DupMasker output 44 describing the composition of segmental duplications was also included with customized Perl scripts.
Supplementary Material
Acknowledgments
We thank P. Sudmant for useful discussions, G.M. Cooper and T. Brown for critical review of the manuscript, L. Zhou, Y. Fu, R. Shi, J. Wu, S. Shaull, and B.A. Roe for the sequencing of clone AC120780. This work was supported by a National Science Foundation Graduate Research Fellowship [to J.M.K.], a Marie Curie fellowship [to T.M.-B.], and by the National Institutes of Health [T32 GM07215 and 5T15 LM007359 to B.T.; HG000225 to D.C.S.; HG002385 to E.E.E.]. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
FINANCIAL INTEREST
E.E.E. is a member of the Scientific Advisory Board Member of Pacific Biosciences. J.A.R. is employee of Signature Genomic Laboratories, LLC. L.G.S. is an employee of, owns shares in and sits on the Members’ Board of Signature Genomic Laboratories, LLC.
AUTHOR CONTRIBUTIONS
This study was designed by F.A. and E.E.E. F.A. performed FISH experiments and shotgun sequencing libraries construction. J.M.K. performed sequence analysis and haplotypes reconstruction. B.T. and D.C.S. performed optical mapping analysis. T.M.-B., T.A.G. and R.K.W. performed non-human primate BAC clones sequencing and analysis. M.V. performed FISH experiments on stretched chromosomes. C.A. performed Illumina sequencing data analysis. S.G., C.D.C. and L.V. performed high-density arrayCGH experiments. M.M. performed PCR experiments. J.A.R., B.C.B. and L.G.S. contributed to 16p12.1 microdeletion data collection. F.A., J.M.K. and E.E.E. contributed to data interpretation. F.A. and E.E.E. wrote the manuscript.
References
- 1.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 3.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung VG, et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–8. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey JA, et al. Recent segmental duplications in the human genome. Science (New York, N Y. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 6.Cheung J, et al. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome biology. 2003;4:R25. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome research. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- 8.Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annual review of genomics and human genetics. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- 9.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends in genetics. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 10.Scherer SW, et al. Human chromosome 7: DNA sequence and biology. Science (New York, N Y. 2003;300:767–72. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichler EE, Clark RA, She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nature reviews. 2004;5:345–54. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 12.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Human molecular genetics. 2004;13(Spec No 1):R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 13.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS genetics. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girirajan S, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 16.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 18.Zhou S, et al. A single molecule scaffold for the maize genome. PLoS Genet. 2009;5:e1000711. doi: 10.1371/journal.pgen.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teague B, et al. High-resolution human genome structure by single-molecule analysis. Proc Natl Acad Sci U S A. 107:10848–53. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan JB, et al. Paternal origins of complete hydatidiform moles proven by whole genome single-nucleotide polymorphism haplotyping. Genomics. 2002;79:58–62. doi: 10.1006/geno.2001.6676. [DOI] [PubMed] [Google Scholar]
- 21.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–81. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 23.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 24.Martin J, et al. The sequence and analysis of duplication-rich human chromosome 16. Nature. 2004;432:988–94. doi: 10.1038/nature03187. [DOI] [PubMed] [Google Scholar]
- 25.Caceres M, Sullivan RT, Thomas JW. A recurrent inversion on the eutherian X chromosome. Proc Natl Acad Sci U S A. 2007;104:18571–6. doi: 10.1073/pnas.0706604104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–83. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehrer-Sawatzki H, Cooper DN. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res. 2008;16:41–56. doi: 10.1007/s10577-007-1207-1. [DOI] [PubMed] [Google Scholar]
- 28.Murphy WJ, et al. A rhesus macaque radiation hybrid map and comparative analysis with the human genome. Genomics. 2005;86:383–95. doi: 10.1016/j.ygeno.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Johnson ME, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–9. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–8. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 31.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 32.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 33.Ullmann R, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–82. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 34.Hannes FD, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–32. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballif BC, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–3. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 36.Bochukova EG, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 38.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 39.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–37. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 40.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–7. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 41.Osborne LR, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–5. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giglio S, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–83. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antonacci F, et al. Characterization of six human disease-associated inversion polymorphisms. Hum Mol Genet. 2009;18:2555–66. doi: 10.1093/hmg/ddp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Z, Hubley R, Smit A, Eichler EE. DupMasker: a tool for annotating primate segmental duplications. Genome Res. 2008;18:1362–8. doi: 10.1101/gr.078477.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laan M, et al. Mechanically stretched chromosomes as targets for high-resolution FISH mapping. Genome Res. 1995;5:13–20. doi: 10.1101/gr.5.1.13. [DOI] [PubMed] [Google Scholar]
- 46.Lichter P, et al. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–9. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 47.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–17. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou S, et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8:278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quail MA, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–10. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craig DW, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–93. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parsons JD. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–9. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.