Abstract
The hepatitis delta virus (HDV) ribozyme is a small self-cleaving RNA with a compact tertiary structure and buried active site that is important in the life cycle of the virus. The ribozyme’s function in nature is to cleave an internal phosphodiester bond and linearize concatemers during rolling circle replication. Crystal structures of the ribozyme have been solved in both pre-cleaved and post-cleaved (product) forms and reveal an intricate network of interactions that conspire to catalyze bond cleavage. In addition, extensive biochemical studies have been performed to work out a mechanism for bond cleavage in which C75 and a magnesium ion catalyze the reaction by general acid-base chemistry. One question that has remained unclear in this and other ribozymes is the nature of long-distance communication between peripheral regions of the RNA and the buried active site. We performed molecular dynamics simulations on the HDV ribozyme in the product form and assessed communication between a distal structural portion of the ribozyme–the protonated C41 base quartet–and the active site containing the critical C75. We varied the ionization state of C41 in both the wild-type and a C41-double mutant variant and determined the impact on the active site. In all four cases, effects at the active site observed in the simulations agree with experimental studies on ribozyme activity. Overall, these studies indicate that small functional RNAs have the potential to communicate interactions over long distances, and that wild-type RNAs may have evolved ways to prevent such interactions from interfering with catalysis.
Keywords: molecular dynamics, HDV, ribozyme, RNA folding, active site
Introduction
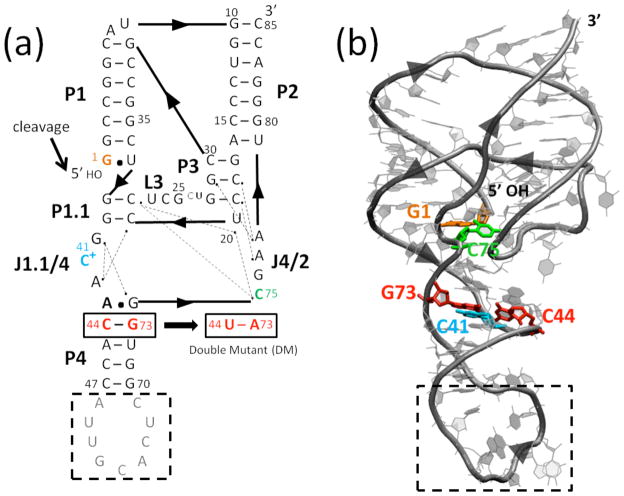
The hepatitis delta virus (HDV) ribozyme is a small, self-cleaving RNA that participates in the double rolling cycle replication of the HDV virus.1 The HDV virus has an RNA genome and contains closely related genomic and antigenomic ribozymes that self-cleave to process the genomic and antigenomic concatemers into linear monomers. The HDV ribozymes are ~85 nt in length and have closely related complex, double-pseudoknotted structures comprised of five pairing regions, P1–P4 and P1.1 (Fig. 1a). The overall shape of the ribozyme is dictated by two stacks: P1, P1.1, and P4 coaxially stack, as do P2 and P3 (Fig. 1b). The active site is found at the cleft where the stacking networks intersect. The short P1.1 pairing was discovered only after the first crystal structure of the ribozyme was solved;2 its catalytic importance was confirmed by compensatory base pair changes and activity studies.3 More recently, HDV-like ribozymes have been found in the human transcriptome,4 and they are prevalent in other forms of life as well.5
Fig. 1.
Structure of the genomic HDV ribozyme. (a) Secondary and (b) tertiary structures of the cleaved (product) form of the genomic HDV ribozyme. Arrowheads denote 5′ to 3′ directionality. (a) Ribozyme is comprised of five pairing regions, P1–P4 and P1.1, and two joining regions, J1.1/4 and J4/2. Nucleotides are numbered from the site of cleavage (black arrow). Dotted lines indicate hydrogen bonding interactions inferred from crystal structures.2,6–8 In the crystallography construct, the binding site for the U1A protein (gray nucleotides) was inserted into the P4 pairing. Black dashed box represents the part of the P4 stem truncated in MD simulations. The site of the C44-G73 to U44-A73 double mutant (DM) is indicated. (b) Tertiary structure of the self-cleaved form of the ribozyme from PDB ID 1CX0,2 generated using VMD.43 Key residues are colored as in panel a.
Crystal structures have been solved for various forms of the wild-type (WT) genomic HDV ribozyme, including both cleaved (product)2,6 and pre-cleaved forms wherein self-cleavage was inhibited by a C75U change or omission of Mg2+ ions.7,8 The product structure is consistent with a wealth of biochemical data, while the C75U pre-cleaved structure disagrees with certain key mechanistic experiments.9,10 As such, our study is performed on the product form of the ribozyme. Analysis of naturally occurring isolates of the virus reveal that most of the HDV ribozyme sequence is invariant in nature; in agreement, experiments show that mutating much of it leads to loss of function, although the long P4 stem can be truncated with little effect on activity.11–13 Indeed, for crystallization purposes, the binding sequence for the U1A protein has been inserted into P4,6 and we truncate most of it in our calculations.
Of particular importance to the function of the HDV ribozyme are two protonated cytosines, C75 and C41. At the active site is C75, which has been implicated as the general acid in the reaction, donating a proton from its N3 to the 5′-bridging oxygen of G1.2,9,14 C75 also serves a structural role in the ribozyme, as inferred from both its overall contribution to catalysis and its intramolecular interactions in the crystal structure.2,15
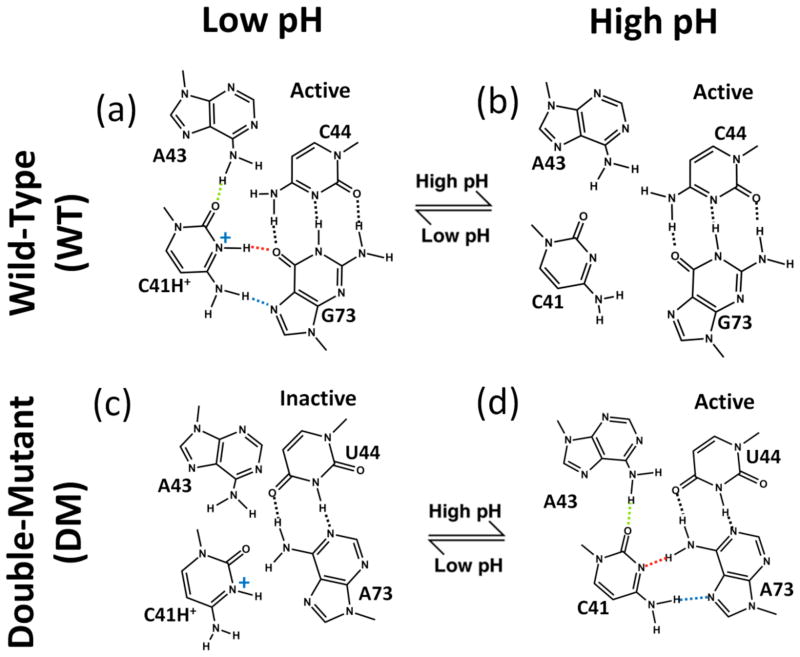
At a structural site buttressing the active site is C41, which participates in a base triple that is important for ribozyme function (Fig. 2a).6,16,17 This motif is found in several other viral RNAs as well,18 making its structural and dynamical roles of general interest. Crystal structures of the genomic HDV ribozyme revealed that C41 of the product and pre-cleaved structures forms a reverse Hoogsteen interaction with G73 that relies on protonation of the N3 of C41, and that C41 also hydrogen bonds to the N6 of A43 through its carbonyl O2.2,6,7 In addition, G73 interacts with a fourth base, C44, to form a Watson-Crick base pair, which provides a base quartet motif overall. There is another base triple near this region involving G40 interacting A42 and G74, which interacts with a fourth base, A43 (Fig. 1a), using the Watson-Crick base pairing faces of G74 and A43 to give an imino GA base pair.
Fig. 2.
Relationship between C41 ionization and HDV ribozyme structure and self-cleavage activity. ‘WT’ is wild-type and ‘DM’ is the C44-G73 to U44-A73 double mutant. DM allows for WT-like C41 base triple interactions with a neutral C41 (see panel d). For WT (top panels), upon moving from low pH (panel a) to high pH (panel b), kinetic experiments reveal that the ribozyme is still active17 denoted with “active.” For DM (lower panels), upon moving from high pH (panel d) to low pH (panel c), experiments reveal that ribozyme activity is lost17 denoted with “inactive.” In panel a, dotted lines indicate hydrogen bonding interactions from crystal structures, with protonation of C41(N3) inferred from positioning of heteroatoms.2 Dotted line coloring for C41 base triple hydrogen bonding is used in subsequent figures.
To complement the structural biology are extensive kinetic, mutagenesis, and Raman spectroscopic experimental studies.12,19–23 Of particular importance to protonation and coupling between C41 and C75 are kinetics and mutagenesis experiments from the Been lab16 and our lab. 10,17 In these studies, a double mutant (DM) of the ribozyme was examined in which the 44:73 Watson-Crick base pair was mutated from CG to UA. Notably, this change leads to formation of the C41 base triple with neutral rather than protonated C41 (Fig. 2d). Detailed kinetics studies of DM in the presence and absence of Mg2+ allowed the pH dependence of C41 quartet formation and C75 general acid-base catalysis to be untangled.10,17 Of particular importance to the present study, for WT, deprotonation of C41 did not affect the rate, but for DM, protonation of C41 caused loss of self-cleavage.17
A major question regarding the mechanism of RNA enzymes is how interactions distal from the active site impact catalysis. Since the distance between C41 and C75 is ~15 Å, interaction between these residues is relatively long-range. Moreover, the body of well-defined structural biology and extensive experimental data make this ribozyme ideal for investigating long-distance interactions. In an effort to elucidate the molecular and dynamical basis of the aforementioned experimental observations, we conducted molecular dynamics (MD) studies on cleaved WT and DM HDV ribozyme structures with neutral and protonated C41. Our data are in full agreement with outcomes of experimental studies and provide insight into how interactions can or cannot communicate over long distances in RNA.
Results and Discussion
Overview
The goal of the present study is to understand if protonation of C41 at a structural base triple communicates to the active site involving protonated C75, over 15 Å away. Our approach was to perform MD simulations of the HDV ribozyme with a WT or DM base quartet, using both C41+ and C41° states. In all studies, catalytically non-essential residues were deleted from P4 (see Methods). Analyses at several key positions in WT and DM C41-protonated and -deprotonated ribozymes are presented in this order: C41 base triple, P1.1 pairing, G40 base triple, active site near C75, and reverse GU wobble. Conformational changes among these variants are depicted by overlaying snapshots of equilibrated structures, and fluctuations observed in the MD simulations are depicted by plotting specified distances as a function of time along the trajectories. Relevant thermally averaged distances for various states, as well as distances from the crystal structure, are summarized in Table 1. Root-mean-square deviations (RMSD) between thermally averaged structures, as defined below in Methods, of protonated and deprotonated states for WT and DM are provided in Table 2.
Table 1.
Average distances in the C41 base triple and active sitea
| Heteroatoms | Wild-type (Å) | Double-mutant (Å) | Crystal (Å) | ||
|---|---|---|---|---|---|
| C41+ | C41° | C41+ | C41° | ||
| C41 Base Triple | |||||
| C41(O2) – A43(N6) | 3.24 (0.52) | 8.11 (1.03) | 2.97 (0.27) | 3.06 (0.25) | 2.83 |
| C41(N3) – G73(O6) | 2.84 (0.11) | 10.35 (1.02) | 4.55 (0.48) | 3.03 (0.16) | 2.88 |
| C41(N4) – G73(N7) | 2.94 (0.12) | 11.65 (1.27) | 6.01 (0.51) | 3.06 (0.22) | 3.03 |
| Active Site | |||||
| C75(N4) – C22(O2P) | 2.96 (0.42) | 6.27 (0.56) | 2.85 (0.12) | 2.95 (0.38) | 2.83 |
| C75(N4) – U20(O2′) | 3.88 (0.50) | 4.41 (0.43) | 4.96 (0.21) | 4.25 (0.72) | 2.95 |
| C75(N3) – G1(O5′) | 3.65 (0.50) | 4.67 (0.42) | 2.92 (0.23) | 3.27 (0.62) | 2.83 |
Distances provided are between heavy atoms averaged over the 20 ns MD trajectory.
Numbers in parenthesis are standard deviations of the averaged distances.
Table 2.
RMSD between C41+ and C41° in WT and in DMa
| Region | Residues | RMSD (Å) | |
|---|---|---|---|
| WT | DM | ||
| P1.1 | C21, C22, G38, G39 | 1.61 | 0.64 |
| P2-P3 | C18, G29, A78 | 0.93 | 0.81 |
| J4/2-P3 | C18, C19, A77, A78 | 1.17 | 1.44 |
| P3 | C18, C19, G28, G29 | 0.97 | 0.69 |
| C41 base triple | C41, A43, C/U44, G/A73 | 2.66 | 1.42 |
| Active site | U20, C22, C24, G25, C75, A77 | 1.71 | 1.39 |
| Active site – no C24, A77 | U20, C22, G25, C75 | 1.69 | 0.84 |
RMSD is between protonated and deprotonated thermally averaged structures. The RMSD for a specific region (e.g. P1.1) was calculated by aligning only the residues stated in column 2.
MD of C41-protonated WT product
Overview
We begin analysis of HDV ribozyme motions by presenting results from MD trajectories of the C41+ WT product structure. Overall this structure is fairly stable throughout the trajectories, supporting the inferred protonation of C41 in the crystal structure and the force field developed herein for protonate cytosine. Some minor conformational changes relative to the WT product crystal structure were observed at the active site.
In the C41+ WT product structure, C41 is protonated at the N3 and forms a base triple with A43 and G73, making the key hydrogen bonding interactions: C41(N4)-G73(N7), C41(N3+)-G73(O6) and C41(O2)-A43(N6) (Fig. 2a). Note that the crystal structure of the product WT HDV ribozyme was solved at 2.3 Å resolution, so hydrogen atoms could not be directly observed in the electron density map;6 thus our present study also probes experimentally inferred protonation of C41.
Since no parameters are present in the AMBER99 forcefield for protonated bases, we first parameterized the partial charges for protonated cytosine using quantum mechanical calculations according to the prescription for the RESP method (see Methods). These partial charges are provided in the Supplementary Data (Fig. S1). The remaining parameters were assumed to be the same as those for the standard cytosine base. The RESP calculations indicate positive partial charges localized on H3, H4s, C4, and N1. The positive charge on H3 of C41 should strengthen hydrogen bonding to G73. The positive partial charge on N1 can be understood in terms of electron donation from N1 into the ring system to delocalize positive charge from N3.
The C41 base triple
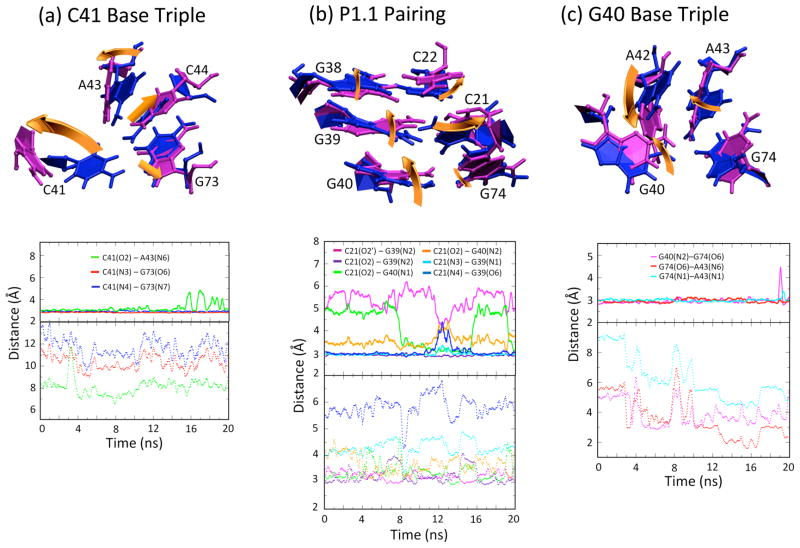
Figures 3 and 4 depict results from MD trajectories of the WT product structure, both in the protonated (solid lines) and deprotonated (dotted lines) states (deprotonated results are presented in the next section). The upper panel in Figure 3a depicts the position of the C41 base triple for an equilibrated structure, and the lower panel shows specified distances along the trajectories. The time evolutions of the three key hydrogen-bonding contacts in the C41+ base triple reveal that the base quartet is largely maintained in the orientation found in the product crystal structure (Fig. 3a, solid lines). Hydrogen bonding distances are all approximately 3.0 Å, and the hydrogen bonds remain at this approximate distance throughout the trajectory, with minor excursions of the hydrogen bond between C41(O2)-A43(N6) near the end of the trajectory. Similar behavior was observed in an independent trajectory (Fig. S4 (ii) solid lines). The less stable hydrogen bond is the only hydrogen bond to A43.
Fig. 3.
C41 base triple, P1.1 pairing region, and G40 base triple of WT product undergo major rearrangements upon C41 deprotonation. (a, b, c) Overlays of protonated C41 (blue) and deprotonated C41 (magenta) structural elements (upper panels), along with time evolution of distances between heteroatoms of specified residues (lower panels). Equilibrated structures in the upper panels were obtained from a snapshot after 5 ns of data collection following an extensive equilibration procedure. Overlays were obtained by minimizing RMSD of residues shown (values given in Table 2). Solid lines (upper panels) and dotted lines (lower panels) in the distance plot represent protonated and deprotonated C41 states, respectively. Distances (in Å) are between heteroatoms and colored according to the legend. Gold arrows depict key structural rearrangements upon C41 deprotonation. Independent trajectories are provided in Fig. S4.
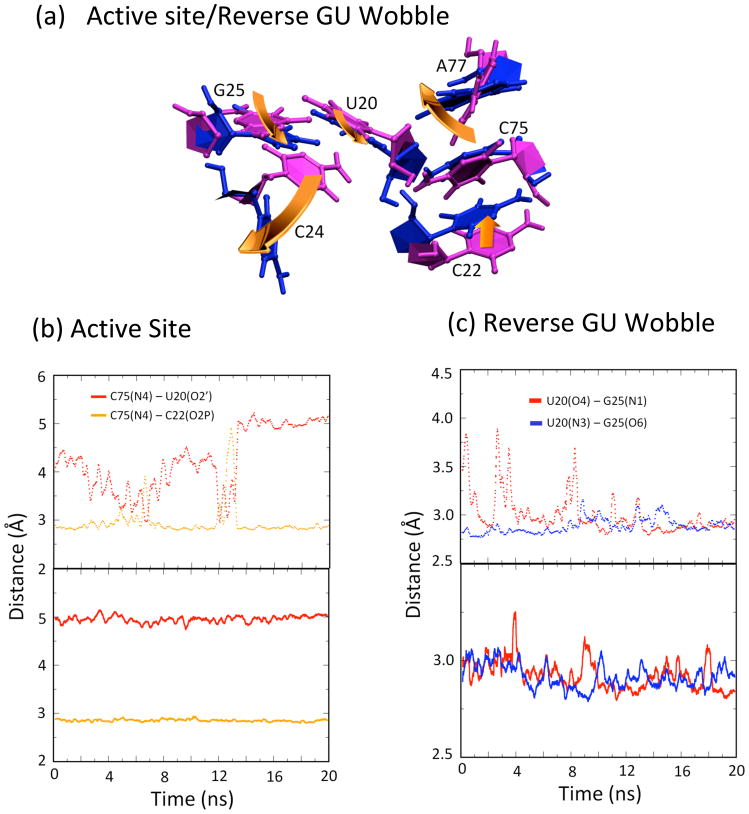
Fig. 4.
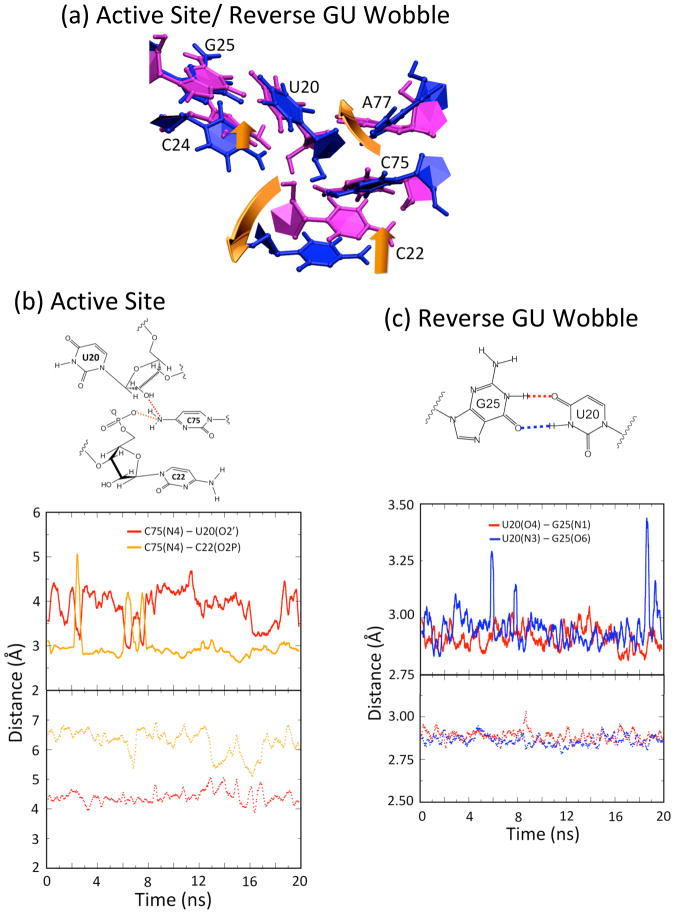
Active sites and GU reverse wobble of WT product show only minor rearrangements upon C41 deprotonation. (a) Overlays of protonated C41 (blue) and deprotonated C41 (magenta) structural elements. Equilibrated structures were obtained from a snapshot after 5 ns of data collection following an extensive equilibration procedure. Overlays were obtained by minimizing RMSD of residues shown (values given in Table 2). Chosen are key residues important for self-cleavage. Overall, the geometry of the active site is maintained in both the protonated and deprotonated structures. (b) Time evolution of distances of key residues at the active site. (c) Time evolution of distances of the GU reverse wobble. Solid lines (upper panels) and dotted lines (lower panels) in the distance plot represent protonated and deprotonated C41 states, respectively. Distances (in Å) are between heteroatoms and colored according to the legend. Structures of the interactions in panels (b) and (c) are provided to aid in interpretation of the plots. Gold arrows depict key structural rearrangements upon C41 deprotonation. Independent trajectories are provided in Fig. S5.
To summarize, C41 does not exhibit significant fluctuations or conformational changes during the trajectory. The RMSD1 at the C41 base triple between the thermally averaged structure for this trajectory and the crystal structure is just 0.65 Å. Even though excursions of the amine of C41 are observed in some parts of the trajectories, these excursions are small and return to the native state. We also note that these trajectories are consistent with protonation of C41 providing an anchor to the structural site, thereby providing validation of the force field for protonated cytosine used herein.
The P1.1 pairing
Above the C41 base triple are the two GC base pairs that comprise P1.1: C21-G39 and C22-G38 (Fig. 1). The position of P1.1 for an equilibrated structure and fluctuations along the trajectories are presented in Figure 3b (solid lines). Hydrogen bonds between both base pairs are maintained throughout this trajectory. In addition to Watson-Crick base pairing with G38, C22 intermittently forms hydrogen bonding interactions with the N4 and O2′ of C75 via its phosphate (Fig. 4b, gold solid line) and N4, respectively, which are described below.
The G40 base triple
In close proximity to the C41 base triple is the G40 base triple, which is comprised of G40, A42, and G74. The position of the G40 triple for an equilibrated structure, as well as fluctuations of specified distances along the trajectories, are depicted in Figure 3c (solid lines). In the G40 triple, G40(N2) forms a hydrogen bond with G74(O6), which is stable throughout the trajectory (Fig. 3c, solid magenta line). The interaction connecting the G40 base triple to the remainder of the ribozyme, the A43-G74 imino base pair, is also maintained throughout the trajectory (Fig. 3c, cyan solid line), providing structural support to the G40 base triple motif. Both G40 and A42 are mobile, forming non-native hydrogen bonds (not shown). In particular, at around 8 ns, A42 pulls out of the base triple such that its N7 is now accessible to the exocyclic amine of G40. Also, A42 is oriented such that its exocyclic amine is within hydrogen bonding distance of C21(O2′) and C21(O2). The complex hydrogen bonding network in the G40 base triple is able to absorb the structural rearrangements observed at this region, perhaps preventing its transmission to the active site.
The active site near C75
The position of the active site for an equilibrated structure and fluctuations along the trajectories are presented in Figure 4 (solid lines). Two key rearrangements relative to the WT product crystal structure occur near the active site: C75 is shifted 1.3 Å away from the active site (Fig. S3, black arrow a), and a reverse GU wobble forms between G25 and U20 (Fig. S3, black arrow b), which is described in the next subsection. This shift in C75 relative to the crystal structure is maintained throughout the MD trajectories. The exocyclic amine (N4) of C75 makes one of two hydrogen-bonding interactions throughout the trajectory, which appear to be mutually exclusive: either to the 2′OH of U20 or to a non-bridging phosphate oxygen (O2P) of C22 (Fig. 4b, red and gold solid lines, respectively). This alternating hydrogen bonding is consistent with the crystal structure, which was interpreted in terms of one or the other hydrogen bond forming.2 In addition, proximity of C75(N3) and G1(O5′) (Table 1), which is the proposed site of general acid catalysis, was found to be maintained throughout the trajectory at a value of 3.65±0.50 Å, which is close enough for hydrogen bonding with the shorter distances. Also, the scissile phosphate is absent in these trajectories, which may be important for hydrogen bonding in this region.
The reverse GU wobble
The last feature of the C41+ WT product MD trajectory to note is formation of a reverse GU wobble between G25 and U20. The positions of U20 and G25 in an equilibrated C41+ WT structure and fluctuations of specified distances along the trajectories are depicted in Figure 4c (solid lines). Note that the hydrogen-bonded reverse wobble is not observed in either product or pre-cleaved crystal structures of the ribozyme (e.g. Fig. S3, gold structure).2,7 During equilibration, the base of U20 shifted by one hydrogen bond register such that strong hydrogen bonding interactions formed between U20(O4)-G25(N1) and U20(N3)-G25(O6) (Fig. S3, blue structure). These interactions are maintained at values near, or just below, 3.0 Å and at angles of ~165° throughout all MD trajectories (Fig. 4c, solid lines), and experience only occasional excursions, consistent with strong hydrogen bonding. Notably, in this reverse wobble, the G of the GU is in the rare syn conformation wherein the base resides over the ribose sugar, which has adopted the rare C2′-endo conformation. This feature was also observed previously in MD simulations of the product form of the ribozyme.24 The recent solution of a pre-cleaved crystal structure exhibiting this GU reverse wobble with U20(O4)-G25(N1) and U20(N3)-G25(O6) hydrogen-bonding distances of 2.85–3.0 Å, suggests that it may be catalytically relevant.25
Other regions
In addition to the structural regions discussed above, we monitored several other regions of the ribozyme, including P2 and P3 (see Table 2). The RMSD values between C41° and C41+ for these regions are quite small (~1 Å), indicating that protonation of C41 does not perturb the structural helices, as expected. Also, we noticed a solvent Na+ ion move into and remain in a negative pocket formed by U20(O2), G25(O6), G25(N7), and the phosphates of C22 and U23 for both WT and DM protonated and deprotonated trajectories.
MD of C41-deprotonated WT product
Overview
Next, MD trajectories of the deprotonated and neutral C41 (C41°) WT product structure are discussed. In contrast to similarity of the base triple domain in the equilibrated C41+ WT and crystal structures, substantial rearrangements occur in the base triple of C41° WT upon equilibration. Nonetheless, despite these rearrangements, the active site remains fairly undisturbed. Similar to C41+ WT, a small rearrangement of C75 occurs at the active site, a reverse GU wobble pair forms between G25 and U20, and some conformational changes are observed at the active site involving the C22 backbone upon equilibration.
The C41 base triple
Results of MD trajectories for WT product structure in the deprotonated state are presented in Figures 3 and 4, dotted lines. Inspection of the time evolutions of the three key hydrogen-bonding contacts in the C41 base triple reveal that the native C41 base triple interactions are lost in the deprotonated state (Table 1, Fig. 3a, dotted lines). Structural changes are anticipated because hydrogen bonding within the triple is interrupted upon deprotonation of C41. Heteroatom distances C41(O2)-A43(N6), C41(N3)-G73(O6) and C41(N4)-G73(N7) all move into the 6 to 14 Å range, much greater than hydrogen bonding distances. Both C41 and A42 move away from their native position in the crystal structure of the C41+ state such that they are almost perpendicular to the plane of the C44:G73 WC base pair, while this Watson-Crick base pair itself remains intact through the simulations.
In sum, C41 and other bases in the region of C41° undergo significant conformational changes relative to the WT product crystal structure. The RMSD between the thermally averaged structure for this trajectory and the crystal structure at the C41 base quartet is ~2.8 Å versus just ~0.65 Å for the C41+ thermally averaged structure. In addition, the fluctuations within this region are greater for the deprotonated than the protonated state.
The P1.1 pairing
The position of P1.1 in an equilibrated C41° structure, as well as fluctuations of specified distances along the trajectories, are presented in Figure 3b (dotted lines). It can be first noted that many interactions in P1.1 formed in the C41+ state are lost, while others are gained (Fig 3b, compare solid and dotted lines). Several movements specific to the C41° state of WT can be noted. First, the C21-G39 base pair has been altered relative to the crystal structure. Specifically, G39 has moved toward the sugar of C21, forming a G39(N2)-C21(O2′) hydrogen bond (Figure 3b, magenta dotted line). Despite its new interactions with G39, C21 does maintain some native interactions with G39; in particular, C21(O2) appears to interact with G39(N1) (not shown) despite the N3 and N4 of C21 having completely lost its native Watson-Crick interaction with G39(N1) and G39(O6) respectively (Fig. 3b, cyan and dark blue dotted lines). The RMSD between the thermally averaged structure for this trajectory and the crystal structure at the P1.1 is ~1.7 Å, and the movement of C21 is the dominant rearrangement in this region. Overall, the hydrogen bonding network in the P1.1 area seems to provide for the anchoring of G39, despite loss of certain native interactions with C21, which buffers the rearrangements seen at the G40 base triple (next subsection) and thus prevents significant disruption of the active site.
The G40 base triple
The position of the G40 triple for an equilibrated C41° WT product structure and specified distances along the trajectories are depicted in Figure 3c (dotted lines). In the G40 triple, the G40(N2)-G74(O6) hydrogen bond is not consistently observed (Fig. 3c, magenta dotted line); this contrasts with C41+ WT, wherein this interaction is maintained throughout the trajectory. Instead, G40 moves upwards and towards C21 (as depicted in Fig. 3b, gold arrow) occasionally forming a hydrogen bonding interaction with C21(O2′) through its N2 (not shown). Rearrangement of A43 causes a twist in the planarity of the A43-G74 interaction and loss of their imino base pairing interaction (Fig. 3c, red and cyan dotted lines); this also contrasts with C41+ WT, wherein this interaction is stable throughout the trajectory (Fig. 3c, red and cyan solid lines). Also, A42 undergoes substantial rearrangement through a ~70° anti-clockwise rotation (Fig. 3c, gold arrow). Based on the fluctuations of these distances along the trajectories, the G40 triple is significantly more mobile in C41° WT than in C41+ WT.
The active site near C75
Next, we turn our attention to the active site for C41° WT. The position of the active site for an equilibrated structure and distances along the trajectories are provided in Fig. 4. Three key rearrangements relative to the WT product crystal structure are observed in C41° WT involving C75, C22 and U20. First, there is formation of the reverse GU wobble between G25 and U20. Second, C75 moves towards the U20 sugar in the C41°-WT, as compared to a ~1.3 Å retraction of C75 observed in the C41+-WT. Third, C22 undergoes a twist of its sugar phosphate backbone (Fig. 4a and S3, left gold arrow), which causes loss of the C75(N4)–C22(O2P) hydrogen bonding interaction throughout the MD trajectories (Fig. 4b, gold dotted line). Thus, the alternating hydrogen bonding of C75(N4) observed in C41+ WT is not present in C41° WT (Fig. 4b, compare solid and dotted lines). We also note that the average distance between C75(N3) and G1(O5′), which is the proposed site of general acid catalysis, is increased to ~4.7±0.4 Å in the C41° WT (Table 1). In C41°, the carbonyl oxygen of C75 seems to have taken the place of the imino N3 with a C75(O2)-G1(O5′) distance of ~2.95 Å, which leaves C75 near the site of catalysis. Again, in the absence of the scissile phosphate, it is difficult to assign any catalytic relevance to this movement. In the WT crystal structure, the C22 phosphate served to anchor the C75 such that the donor and acceptor groups are aligned. The movement of C75 observed in the C41°-WT is understandable in light of the rotation of the C22 phosphate observed in the simulations. Overall, these contacts within the active site are largely consistent with maintenance of catalytic activity in the WT-C41° state.
The reverse GU wobble
The last feature we note is formation of the reverse GU wobble between G25 and U20. Similar to the C41+ WT simulations, these interactions occur and are maintained at values near or just below 3.0 Å throughout all MD trajectories (Fig. 4c, dotted lines), suggesting that they are especially stable and could potentially be related to catalysis since both states are catalytically active.
Other regions
We monitored the P2 and P3 pairings, and the RMSD values between the thermally averaged protonated and deprotonated WT product structures are provided in Table 2. The RMSD values are quite small (~1 Å), indicating that deprotonation of C41 does not significantly impact these regions.
MD of C41-deprotonated DM product
Overview
Next, we consider the C41° DM product state. As mentioned, DM ribozyme is active when C41 is deprotonated but loses activity when protonated (Fig. 2, lower right hand corner);17 thus we begin our discussion of DM with C41°. Overall the structure of C41° DM is fairly stable throughout the trajectory (Fig. 5a, dotted lines), which supports the predicted hydrogen bonding pattern at the C41 base quartet. In C41° DM, C41 is deprotonated at the N3 and forms a base triple with A43 and A73, making the following key hydrogen bonding interactions: C41(N4)-A73(N7), C41(N3)-A73(N6) and C41(O2)-A43(N6) (Fig. 2d).
Fig. 5.
C41 base triple and G40 base triple of DM product undergo minor rearrangements upon C41 protonation. (a, b) Overlays of protonated C41 (blue) and deprotonated C41 (magenta) structural elements (upper panels), along with time evolution of distances between heteroatoms of specified residues (lower panels). Equilibrated structures were obtained from a snapshot after 5 ns of data collection following an extensive equilibration procedure. Overlays were obtained by minimizing RMSD of residues shown (values given in Table 2). Dotted lines (upper panels) and solid lines (lower panels) in the distance plot represent deprotonated and protonated C41 states, respectively. (Note that the trajectories in DM figures are displayed such that the natively folded C41° triple is kept as the upper panel.) Distances (in Å) are between heteroatoms and colored according to the legend. Gold arrows depict key structural rearrangements upon C41 protonation. Independent trajectories are provided in Fig. S6.
The C41 base triple
Figures 5 and 6 present results of MD trajectories for DM product, both in the deprotonated (dotted lines) and protonated (solid lines) states (protonated results are discussed in the next section). The position of the C41 triple for an equilibrated structure, as well as selected distances along the trajectories, are provided in Figure 5a. Time evolutions of the three key hydrogen bonding contacts in the C41° base triple reveal that the base triple is largely maintained in the orientation found in the WT product crystal structure (Fig. 5a, dotted lines). Hydrogen bonding distances are all approximately 3.0 Å (Table 1) and remain relatively constant throughout the trajectory. The RMSD between the thermally averaged structure for this trajectory and the crystal structure (with the DM created in silico) at the C41 base triple is low at just 0.91 Å. Thus, the C41° state does not exhibit significant fluctuations or conformational changes at the C41 base triple during the trajectory.
Fig. 6.
Active site of DM product undergoes major rearrangements upon C41 protonation. (a) Overlays of protonated C41 (blue) and deprotonated C41 (magenta) structural elements. Equilibrated structures were obtained from a snapshot after 5 ns of data collection following an extensive equilibration procedure. Overlays were obtained by minimizing RMSD of residues shown (values given in Table 2). C24 and A77 are not included in the alignment shown here (last row of Table 2). Chosen are key residues important for self-cleavage. Overall, the geometry of the active site is maintained in the deprotonated structure but not in the protonated structure. (b) Time evolution of distances of key residues at the active site. (c) Time evolution of distances of the reverse GU wobble. Dotted lines (upper panels) and solid lines (lower panels) in the distance plot represent deprotonated and protonated C41 states, respectively. Distances (in Å) are between heteroatoms and colored according to the legend. Gold arrows depict key structural rearrangements upon C41 protonation. Independent trajectories are provided in Fig. S7.
The P1.1 pairing
Both of the P1.1 base pairs, C21-G39 and C22-G38, are maintained throughout the trajectory (data not shown). Absence of significant fluctuations or conformational changes in P1.1, which is connected to the active site, is consistent with the active site remaining largely intact (see below).
The G40 base triple
The position of the G40 triple for an equilibrated structure and distances along the trajectories are presented in Figure 5b (dotted lines). The hydrogen bond between G40(N2) and G74(O6) is stable throughout the trajectory (Fig. 5b, magenta dotted line). The two other interactions comprising the G40 triple in the WT product crystal structure–between G40(N3) and A42(N6), and between G40(O2′) and A42(N7)–are not observed during the trajectories (data not shown). However, A42 is held in place by a strong hydrogen bonding interaction between G40(N2) and A42(N7) (data not shown). Also, the A43-G74 imino base pair is maintained throughout the trajectories (Fig. 5b, red and cyan dotted lines), providing structural support to the G40 triple motif.
The active site near C75 and the reverse GU wobble
The active site in an equilibrated structure, as well as selected distances along the trajectories, are depicted in Figure 6a and b (dotted lines). Only one key rearrangement relative to the WT product crystal structure occurs near the active site: the reverse GU wobble between G25 and U20 forms (Fig. S3, black arrow b), as described for C41 WT. Similar to C41+ WT, the exocyclic amine (N4) of C75 exhibits the hydrogen-bonding interaction with the non-bridging phosphate oxygen of C22 through most of the trajectory, with occasional alternating interaction with the 2′OH of U20, (Fig. 6b, gold and red dotted lines). Overall, these observations are consistent with maintenance of catalytic activity in the DM-C41° state.
MD of C41-protonated DM product
Overview
Next, MD trajectories of the C41+ DM product structure are considered. As mentioned above, DM ribozyme is inactive when C41 protonates (Fig. 2, lower left hand corner).17 Large rearrangements relative to the WT product crystal structure occur in the C41+ base triple domain. Somewhat surprisingly, despite these changes, the P1.1 and G40 regions remain fairly intact. Nonetheless, there are substantial rearrangements of C75 at the active site, which are consistent with loss of activity in this state.17
The C41 base triple
When C41 is protonated in the product DM, distinct and large rearrangements occur in the base triple (Fig. 5a, solid lines), as expected on the basis of interrupted hydrogen bonding. Initially, in the first 2 ns, C41+ residue twists in order to avoid the unfavorable C41+(N3)-A73(N6) interaction (Fig. 5a, gold arrow), which in turn positions the O2′ of C41+ ~3.1 Å from A43(N6), leading to a stable non-native interaction. The C41+ is then seen to move upwards such that it is out of plane with A73. The two native base hydrogen bonding triple interactions between C41 and A73 are not observed (Fig. 5a, red and blue solid lines).
The P1.1 pairing
Somewhat surprisingly, the P1.1 region undergoes only small rearrangements, which allow the majority of the interactions to be maintained. The P1.1 interactions remain strong (not shown).
The G40 base triple
As mentioned previously, the protonation of C41 causes it to shift upwards. This is transmitted to G40, which loses its planarity with G74, however, still maintaining its G40(N2)-G74(O6) interaction (Figure 5b, magenta dotted line). A42 appears to be pulled out of the base triple but still contacts G40 through a new A42(N7)-G40(N2) hydrogen bonding interaction. Also, the exocyclic amine of A42 is found to form two new interactions with the 2′OH and O2 of C21 (not shown). In spite of this, C21 maintains its native Watson Crick base pairing with G39. The A43-G74 interaction through their Watson Crick faces are maintained, but A43 undergoes a twist. Again, this could be attributed to the motion of C41+.
The active site near C75
A number of significant changes due to C41 protonation in the DM are observed at the active site (Fig. 6a, gold arrows). As observed in previous cases, U20 has moved by one hydrogen-bonding registry, toward C75, to form the reverse GU wobble (Fig. 6a, c). Also, C24, which is found to be ~4.5 Å from U20(O2′) in the WT product crystal structure, undergoes a very large movement of ~5 Å downward. Notably, C24 is known to be important for self-cleavage activity, as changing it to U abrogates self-cleavage activity.26
Despite the above changes, some of the active site structure remains intact in the trajectories. In particular, C75(N4) maintains its hydrogen bonding interaction with C22(O2P) (Fig. 6b, solid gold line). Also, positionings between the G1(O5′) and the N3 and O2 of C75 are largely maintained at ~3.3 Å and ~2.7 Å, respectively. Perhaps longer trajectories would show opening of this interaction that drives proton transfer; nonetheless, losses of interactions involving U20 and C24 are consistent with loss of function observed in experiments for C41° DM.17
In summary, the extent of rearrangements in DM C41+ but not C41°, supports loss of key active site interactions consistent with experiments.17
Possible contributors to ribozyme activity
MD trajectories of WT and DM C41+ and C41° product structures indicated rearrangements of active site atoms that largely agreed with experiments in which only DM C41+ was inactivated.17 This effect appears to originate within the C41 base triple itself, wherein the DM C41+(N3H+) has a repulsion with the formerly cognate A73(N6). This causes an upward shift of the plane of the C41+ base. Somewhat surprisingly, C41+ itself is rather stationary, possibly owing to favorable alignment of its partially positive N3 and N1 (Fig. S1) and negative atoms in G40. Notably these rearrangements are absent in WT C41° since a proton is lost rather than gained in the WT upon moving from the native triple.
Protonation of C41 in the DM causes A42 and G40 to pull further out of the G40 base triple (Fig. 5b, gold arrows), making it accessible to the dangling U23. This pulling out is exploited to form U23(N3)-G40(O2′) and U23(O2)-A42(N6) interactions (not shown) perhaps causing C24 to pull out of the active site (Fig. 6a, gold arrows). We notice in the simulations of WT C41+,° that C24 lends support to C75 by interacting with the other non-bridging phosphate oxygen of C22. Drop of C24 out of the active site may interfere with the positioning of the phosphate of C22, and hence destabilize C75(N4) and perhaps contribute to loss of activity. Also, movement of C24 could be the cause of the loss of C75(N4) interaction with the 2′OH of U20 by causing a displacement of the G25.
Further, we hypothesize that the degree of rigidity of P1.1 and the G40 base triple plays an important role in regulating activity of the ribozyme. The less rigid P1.1-G40 base triple region in WT C41° appears able to absorb motion and thus prevent communication of disruptions at the C41 triple to the active site. In contrast, in the inactive DM C41+, the P1.1-G40 region is more rigid, which appears to allow communication of the C41 triple disruptions to the active site.
Conclusions
Long distance communication has been implicated as contributing to catalysis in the mechanism of numerous protein enzymes.27 Herein, we demonstrate that in a small ribozyme interactions distal from the active site can affect active site geometry and thereby potentially impact ribozyme activity. Importantly, outcomes from MD are consistent with activity studies from experiments. The main behavior observed in the present study enhances understanding of how distal non-native interactions communicate to the active site and interfere with RNA catalysis. Future studies are needed to determine whether distal native interactions communicate to the active site and facilitate RNA catalysis. These studies will require careful considerations of the scissile phosphate and upstream nucleotide in catalytic conformations.
Methods
Crystal structure of the product2,6 form of the HDV ribozyme was used as the starting structure (PDB: 1CX0, 2.3 Å resolution). The catalytically dispensable P4 stem was truncated by removing residues 48–69 in the product structure, which reduced the system to 62 nt. Double mutants were generated by replacing C44-G73 with U44-A73 using Accelrys Discover Studio Visualizer 2.0. Only the functional groups were modified. Hydrogen atoms were added to the entire ribozyme using the GROMACS simulation package version 3.3.1.28
The MD simulations were performed using both the DLProtein29 and Desmond30,31 MD packages with the Amber99 force field.32,33 Since this force field does not include parameters for cationic residues, we obtained partial atomic charges for base atoms of a protonated cytosine using the RED-II program.34 First, the Gaussian03 program35 was used to perform a geometry optimization of protonated cytosine at the HF/6-31G* level of theory, and the RESP method36,37 was subsequently used to fit the atom-centered charges to the molecular electrostatic potential. Resulting partial atomic charges for protonated cytosine are provided in Supplementary Data (Fig. S1). The remaining parameters were assumed to be the same as those for the standard cytosine base.
The ribozyme was solvated with ~8300 rigid TIP3P waters38 in a periodically replicated parallelepiped box extending at least 10 Å beyond the ribozyme on all sides. Nine Mg2+ ions resolved in the RNA portion of the crystal structure were included, but no additional divalent ions were added. The structure was neutralized with 42 Na+ ions for the C41° structures and 41 Na+ ions for the C41+ structures, and physiological concentration of free salt (0.1 M NaCl) was added to the solvent. The concentration of free salt was calculated from the box volume. Long-range electrostatic interactions were calculated with the Smooth Particle Mesh Ewald method39 using a direct space cutoff of 15 Å with DLProtein and 12 Å with Desmond, and SHAKE40 constraints were applied to bonds involving hydrogen. For the double mutant, the mutated portion was initially optimized by minimizing the energy with the remaining system fixed. For WT and DM ribozymes, an initial energy minimization of the solvent and ions was conducted with the ribozyme fixed, followed by an energy minimization of the entire system.
For each system studied, a simulated annealing equilibration procedure was performed by propagating 150 ps of MD for an isobaric, isothermal ensemble (NPT) at each temperature sequentially for 0, 100, 200, and 300 K, followed by 5 ns of MD at 300 K for a canonical ensemble (NVT). For each system studied, this equilibration procedure was performed with the DLProtein package for two independent trajectories. These data, which consist of two independent sets of 5 ns trajectories, are provided in Supporting Information. For each system studied, one trajectory was further extended for 20 ns with the Desmond MD package. First an additional equilibration of 100 ps with the NPT ensemble and 1 ns with the NVT ensemble at 300 K was performed with Desmond for each trajectory. Subsequently, data were collected at 300 K with the NVT ensemble for 20 ns per trajectory. A Nosé-Hoover thermostat41,42 was used to maintain temperature and pressure, and the time step was 1 fs for all MD trajectories.
The equilibrated structures shown in Figures 3 through 6 were obtained after 5 ns of data collection following an extensive equilibration procedure that also includes 5 ns of MD. During the data collection, snapshots were output every 5 ps, and distance plots were generated with a running average over 50 snapshots. Root mean square deviations (RMSD) were calculated by selecting specific residues using the GROMACS simulation package, version 3.3.1.28 Thermally averaged structures were obtained by minimizing the RMSD of the heavy atoms with respect to the crystal structure for ~4000 configurations sampled over the 20 ns MD trajectory using the g_confrms utility in GROMACS and calculating the average coordinates of the atoms. The heavy-atom RMSD for the entire 20 ns trajectory for the four variants studied are given in Supporting Information (Figure S8).
Supplementary Material
Acknowledgments
Supported by National Institutes of Health Grants GM58709 (PCB) and GM56207 (SHS).
Abbreviations
- BT
base triple
- C41+
structure of ribozyme when C41 is protonated
- C41°
structure of ribozyme when C41 is deprotonated
- DM
double-mutant
- HDV
hepatitis delta virus
- MD
molecular dynamics
- RESP
restrained electrostatic potential
- RMSD
root mean square deviation
- WT
wild-type
Footnotes
Only residues C41, A43, C44, and G73 are considered for this RMSD calculation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Philip C. Bevilacqua, Email: pcb@chem.psu.edu.
Sharon Hammes-Schiffer, Email: shs@chem.psu.edu.
References
- 1.Lai MM. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 2.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 3.Wadkins TS, Perrotta AT, Ferre-D’Amare AR, Doudna JA, Been MD. A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA. 1999;5:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 5.Webb CHT, Riccitelli NJ, Ruminski DJ, Lupták A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferre-D’Amare AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: Use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 7.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 8.Ke A, Ding F, Batchelor JD, Doudna JA. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 10.Cerrone-Szakal AL, Siegfried NA, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: solvent isotope effects and proton inventories in the absence of divalent metal ions support C75 as the general acid. J Am Chem Soc. 2008;130:14504–14520. doi: 10.1021/ja801816k. [DOI] [PubMed] [Google Scholar]
- 11.Suh YA, Kumar PK, Nishikawa F, Kayano E, Nakai S, Odai O, Uesugi S, Taira K, Nishikawa S. Deletion of internal sequence on the HDV-ribozyme: elucidation of functionally important single-stranded loop regions. Nucleic Acids Res. 1992;20:747–753. doi: 10.1093/nar/20.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur J Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 13.Chadalavada DM, Cerrone-Szakal AL, Bevilacqua PC. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA. 2007;13:2189–2201. doi: 10.1261/rna.778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 15.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 16.Wadkins TS, Shih I, Perrotta AT, Been MD. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J Mol Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 17.Nakano S, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: a mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- 18.Nixon PL, Rangan A, Kim YG, Rich A, Hoffman DW, Hennig M, Giedroc DP. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J Mol Biol. 2002;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- 19.Been MD. Molecular biology. Versatility of self-cleaving ribozymes. Science. 2006;313:1745–1747. doi: 10.1126/science.1133259. [DOI] [PubMed] [Google Scholar]
- 20.Been MD. HDV ribozymes. Curr Top Microbiol Immunol. 2006;307:47–65. doi: 10.1007/3-540-29802-9_3. [DOI] [PubMed] [Google Scholar]
- 21.Bevilacqua PC, Brown TS, Nakano S, Yajima R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- 22.Bevilacqua PC, Yajima R. Nucleobase catalysis in ribozyme mechanism. Curr Opin Chem Biol. 2006;10:455–464. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Koo S, Novak T, Piccirilli JA. Catalytic mechanism of the HDV ribozyme. In: Lilley DM, Eckstein F, editors. Ribozymes and RNA Catalysis. RSC Publishing; Cambridge, UK: 2008. pp. 92–122. [Google Scholar]
- 24.Krasovska MV, Sefcikova J, Spackova N, Sponer J, Walter NG. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J Mol Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Yajima R, Chadalavada D, Chase E, Bevilacqua PC, Golden BL. A 1.9 Å crystal structure of the HDV ribozyme pre-cleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester bond cleavage. Biochemistry. 2010 doi: 10.1021/bi100670p. in press. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami J, Kumar PK, Suh YA, Nishikawa F, Kawakami K, Taira K, Ohtsuka E, Nishikawa S. Identification of important bases in a single-stranded region (SSrC) of the hepatitis delta (delta) virus ribozyme. Eur J Biochem. 1993;217:29–36. doi: 10.1111/j.1432-1033.1993.tb18214.x. [DOI] [PubMed] [Google Scholar]
- 27.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 29.Melchionna S, Luise A, Venturoli M, Cozzini S. DLPROTEIN: A molecular dynamics package to simulate biomolecules. In: Voli M, editor. Science and Supercomputing at CINECA-1997 report. Supercomputing Group; CINECA: 1998. pp. 496–505. [Google Scholar]
- 30.Desmond Molecular Dynamics System. D. E. Shaw Research; New York: 2008. [Google Scholar]
- 31.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary IK, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (SC06); Tampa, Florida. 2006. [Google Scholar]
- 32.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KMJ, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 33.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comp Chem. 2000;21:1049–1074. [Google Scholar]
- 34.Pigache A, Cieplak P, Dupradeau F-Y. Automatic and highly reproducible RESP and ESP charge derivation: Application to the development of programs RED and X RED. 227th ACS National Meeting; Anaheim, CA. March 28-April 1, 2004. [Google Scholar]
- 35.Frisch MJ, et al. Gaussian03 (Revision C.02) Gaussian, Inc; Wallingford, CT: 2003. [Google Scholar]
- 36.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: The RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 37.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the multimolecule and multiconformational RESP methodology to biopolymers: Charge derivation for DNA, RNA, and proteins. J Comput Chem. 1995;16:1357–1377. [Google Scholar]
- 38.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 39.Darden T, York D, Pedersen L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 40.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 41.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol Phys. 1984;52:255–268. [Google Scholar]
- 42.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.