Abstract
Single molecule force spectroscopy is a powerful method that uses the mechanical properties of DNA to explore DNA interactions. Here we describe how DNA stretching experiments quantitatively characterize the DNA binding of small molecules and proteins. Small molecules exhibit diverse DNA binding modes, including binding into the major and minor grooves and intercalation between base pairs of double-stranded DNA (dsDNA). Histones bind and package dsDNA, while other nuclear proteins such as high mobility group proteins bind to the backbone and bend dsDNA. Single-stranded DNA (ssDNA) binding proteins slide along dsDNA to locate and stabilize ssDNA during replication. Other proteins exhibit binding to both dsDNA and ssDNA. Nucleic acid chaperone proteins can switch rapidly between dsDNA and ssDNA binding modes, while DNA polymerases bind both forms of DNA with high affinity at distinct binding sites at the replication fork. Single molecule force measurements quantitatively characterize these DNA binding mechanisms, elucidating small molecule interactions and protein function.
Keywords: force spectroscopy, DNA binding, DNA melting, DNA replication, nucleic acid chaperones
1 INTRODUCTION
Single molecule methods have provided a clearer understanding of a wide range of fundamental biological processes, including DNA replication, transcription, and repair. Single molecule force spectroscopy began with the capture and manipulation of single DNA molecules. Techniques such as optical tweezers, magnetic tweezers, and atomic force microscopy (AFM) apply forces to single molecules, probing conformational changes and structural dynamics in a variety of conditions. Such measurements explore the interactions of DNA with molecules ranging from small ligands to complex proteins. Quantifying the thermodynamics and kinetics of these interactions leads to substantial insights into DNA binding mechanisms in important biological systems.
1.1 Single molecule force spectroscopy techniques
Optical tweezers, magnetic tweezers, and AFM are the predominant force spectroscopy techniques used to trap and manipulate single DNA molecules. Single-beam optical tweezers instruments focus a high power laser through a high numerical aperture microscope objective to form an optical trap. Dual-beam optical tweezers instruments use microscope objectives to bring two counter-propagating laser beams to an overlapping focus to form an optical trap. The trap captures one typically streptavidin-coated polystyrene bead, while a second bead is attached to a micropipette tip fixed to a flow cell or is held in another optical trap. A single biotin-labeled DNA molecule is tethered to the beads through a biotin-streptavidin linkage or some other attachment method that can withstand the forces to be applied. Translation of the flow cell or optical trap pulls the bead affixed to the pipette tip or held in the trap, resulting in extension of the captured DNA molecule. This displaces the bead in the optical trap, which provides a measurement of the force on the DNA molecule with piconewton (pN) accuracy [1].
Magnetic tweezers use a glass slide and magnetic bead, both coated with streptavidin or another attachment ligand, in order to capture a biotin-labeled DNA molecule. Translation of the glass slide through a magnetic field gradient results in a force on the DNA molecule, measured as three-dimensional motion of the magnetic bead in video acquisition. Advantages of this technique include single molecule manipulation in three dimensions and detection of forces as low as 0.05 pN [2].
Although AFM is predominantly used in imaging applications, the technique may be used for single molecule force spectroscopy. A single DNA molecule is immobilized between the surface and the AFM tip, and force is measured as a function of extension and relaxation. A typical DNA attachment technique functionalizes opposite ends of the molecule with thiol and biotin. The thiolated end binds covalently to a gold surface, while the streptavidin-coated AFM tip captures the biotin-labeled end of a single DNA molecule [3]. Resolution of the DNA stretching curves is on the order of 5-10 pN [4-5].
1.2 Stretching single DNA molecules
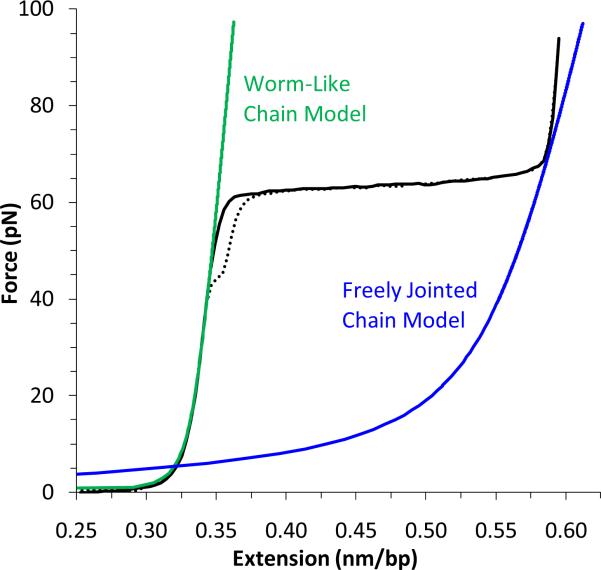
Single molecule DNA stretching experiments determine force as a function of extension (Figure 1). At low extensions, the measured tension increases gradually as the duplex uncoils in what is known as the entropic regime. As extension approaches the dsDNA contour length, the backbone resists further extension and the force increases dramatically in an elastic response. At ~65 pN, dsDNA undergoes an overstretching transition, increasing to ~1.7 times its contour length at nearly constant force. There is a second transition at the end of this overstretching plateau, near the contour length of ssDNA. If the extension is reduced at this point, the relaxation curve will match the stretching curve. Some hysteresis, where the relaxation curve does not match the stretching curve may occur, depending upon solution conditions. DNA stretching and relaxation cycles exhibit similar force-extension curves on the timescale of the experiment in typical solution conditions, indicating that the process is reversible.
Figure 1.
DNA stretching experiments measure the force on a dsDNA molecule as a function of extension. The extension data for a typical λ-DNA molecule is shown as a solid black line, and a dotted line represents the relaxation data. The Worm-Like Chain (WLC) model (green line) describes dsDNA. Near the dsDNA contour length, the molecule undergoes a force-induced melting transition, from dsDNA to ssDNA. The Freely-Jointed Chain (FJC) model describes ssDNA (blue line). Minimal hysteresis is evident in these solution conditions (100 mM Na+, 10 mM Hepes, pH = 7.5, T = 20 °C).
1.2.1 Models of polymer elasticity
Polymer models of dsDNA and ssDNA effectively characterize DNA force-extension curves. The Worm-Like Chain (WLC) model assumes a smooth distribution of bending angles, and describes dsDNA in terms of observed length bds of an elastic polymer under the influence of tension F [6-10]. Though no exact solutions to this model are known, an approximate solution is appropriate for high forces:
| (1) |
where Pds is the persistence length, Bds is the end-to-end or contour length, and a stretch modulus Sds is added to account for backbone extensibility. Here kB is Boltzmann's constant and T is temperature. The Freely Jointed Chain (FJC) model describes the polymer elasticity of ssDNA as a collection of independent monomers with varying bond angles [11]:
| (2) |
Figure 1 shows the WLC (green) and FJC (blue) polymer models with typical values for the parameters B, P, and S [1, 12].
1.3 Force-induced structural transitions
A thermodynamic model quantitatively describes the overstretching transition at 65 pN in terms of force-induced melting. The force exerted on the dsDNA molecule does work to increase the length of the DNA, converting dsDNA to ssDNA and disrupting both base pairing and base stacking interactions. In this model, the second transition at the end of the melting plateau is a non-equilibrium process involving the remaining base pairs of dsDNA which must break for strand separation. Force-induced melting is analogous to thermal melting, and the model predicts that solution conditions which influence thermal melting, such as salt, pH, and temperature, also affect the force-induced melting transition. DNA stretching experiments quantitatively confirmed these predictions [10, 13-14], and recent modeling studies also support a force-induced melting model [15-17]. Furthermore, experiments demonstrated that solution conditions [18] and DNA binding ligands [19-26] known to inhibit DNA reannealing induce strong hysteresis in the force-extension curves, providing additional evidence for melting of the DNA strands.
In an alternate model of the overstretching plateau, B-form duplex DNA lengthens in response to the applied force, undergoing a structural transition to a new form of DNA referred to as “S-DNA” [4, 27-29]. This form of DNA is predicted to preserve base pairing but not base stacking, a distinction based upon the observation that strand separation occurs at high forces [30-32]. An early modeling study predicted a transition to this form of DNA at a significantly larger force than experiments observed. Recent studies use the proposed existence of S-DNA as a means to generate new parameters to fit stretching curves and other experimental results. However, it is not clear that additional fitting parameters are needed to explain DNA stretching experiments. In addition, these models do not make predictions that can be tested with other experiments, making it difficult to test proposed S-DNA models [33-35]. Magnetic tweezers experiments with both strands of a dsDNA molecule tethered to beads did not observe the transition at 65 pN, but instead measured a transition at 110 pN over a similar extension attributed to a combination of S-DNA and P-DNA, a form of melted DNA which is overwound [36]. Although this particular transition of torsionally constrained DNA is consistent with force-induced melting, it was suggested that some features of DNA stretching curves were incompatible with force-induced melting [37-38] and that the structure of DNA in the overstretching transition remains unclear [39-40]. It is essential to establish the nature of the overstretching transition in order to use single molecule force spectroscopy techniques to characterize DNA binding. Recent experiments use glyoxal, intercalating dyes, and single-stranded binding proteins (SSBs) to establish that this conformational transition involves base pair disruption, and therefore DNA overstretching is force-induced melting of dsDNA into ssDNA.
1.3.1 Glyoxal binds ssDNA bases exposed in the force-induced melting transition
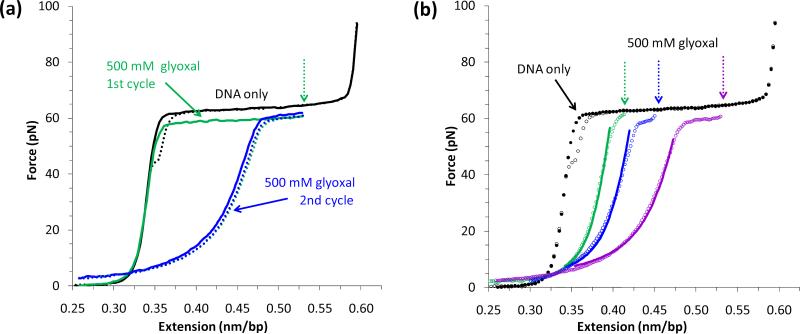
Glyoxal (C2H2O2) is a small molecule which binds irreversibly to exposed guanine bases of DNA with slow kinetics [41]. The modified guanine bases have three rings instead of two, introducing steric constraints that hinder base pair reannealing [42]. λ-DNA molecules were held at fixed extensions for ~30 min in the presence of glyoxal, which is the timescale required for DNA binding [41]. The DNA stretching curve exhibits a decrease in melting force and strong hysteresis (Figure 2a), indicating that guanine bases exposed to solution are subject to glyoxal modification and subsequently prevent DNA reannealing [41]. Therefore extension into the overstretching plateau exposes ssDNA bases to solution, reflecting force-induced melting of the dsDNA molecule.
Figure 2.
Glyoxal binds ssDNA bases exposed in the force-induced melting transition. (a) Extension (solid line) and relaxation (dotted line) data of a λ-DNA molecule alone is shown in black. After the addition of 500 mM glyoxal, the molecule is extended (solid green line) and held fixed (green dotted arrow) for ~30 min. The significant hysteresis upon relaxation (dotted green line) reflects that the two DNA strands do not reanneal, indicating glyoxal binding to exposed nucleotides. The second stretch (solid blue line) follows the previous relaxation curve, which suggests that modification is permanent. (b) Relaxation data (open circles) for a series of fixed extensions (dotted arrows), in which the DNA molecule is stretched in the presence of 500 nM glyoxal. Fits to a linear combination of the WLC and FJC models (Equation (3)) are shown as solid lines. Figures reproduced with permission from [1].
As the DNA molecule is held at larger fixed extensions, the corresponding relaxation curves exhibit additional hysteresis (Figure 2b). These results demonstrate that glyoxal binding increases as the DNA molecules are held further into the overstretching plateau, despite constant solution conditions. This indicates that greater extensions into the stretching transition result in exposure of additional bases, and the relaxation curves in the presence of glyoxal are a combination of dsDNA and ssDNA. The experimental data fits well to a linear combination of the FJC and WLC polymer models (fits shown in Figure 2b), where the measured contour length b is a function of the ssDNA fraction γss [41]:
| (3) |
and bds and bss are force-dependent DNA extensions from the WLC (Equation (1)) and the FJC model (Equation (2)), respectively. The fractional extension along the transition plateau agrees well with the fraction of glyoxal-stabilized ssDNA obtained from fits to Equation (3), which provides structural evidence that DNA overstretching is indeed the force-induced melting of dsDNA into ssDNA.
1.3.2 Visualizing force-induced melting with intercalators and SSBs
The significant presence of ssDNA exposed to glyoxal modification in the overstretching transition is unlikely to arise from nicks in the DNA backbone [43], and further experiments with small molecules and SSBs confirm the force-induced melting model. Recent single-molecule studies have directly visualized the nature of the structural transition in a combination of optical tweezers and fluorescence imaging techniques [44]. A DNA molecule stretched to a fixed extension in the absence of ligand is briefly transferred into the presence of YOYO, a fluorescent dye which intercalates into the paired bases of dsDNA [45]. Subsequent imaging reveals only regions of dsDNA, to which the intercalator can bind [44]. The fraction of dsDNA present at each fixed extension corresponds directly to fractional extension along transition plateau, illustrating a structural conversion from dsDNA into a form of DNA to which YOYO is unable to bind [44].
Experiments with fluorescent dye-labeled SSBs demonstrate that the form to which dsDNA is converted upon overstretching is ssDNA. Human mitochondrial SSB (mtSSB) binds and wraps relaxed ssDNA [46], but does not affect the overstretching transition or bind ssDNA which is under tension greater than ~40 pN. When DNA is extended into the stretching transition and briefly placed in the presence of mtSSB, the images show fluorescent spots at both ends of the DNA molecule, indicating the presence of protein-wrapped ssDNA [44]. These spots increase in brightness and move toward the center of the molecule as a function of extension, illustrating the relative increase of ssDNA with progressive movement into the stretching transition. This method also visualizes nicks in the DNA backbone, since the mtSSB wraps the relaxed ssDNA in the middle of the molecule. Molecules without nicks do not exhibit these binding events, and mtSSB fluorescence is confined to the ends of the DNA. Two-color fluorescent measurements with both YOYO and mtSSB confirm that mtSSB-wrapped ssDNA forms at an interface with YOYO-labeled dsDNA [44].
In contrast with mtSSB, the SSB Replication Protein A (RPA) binds ssDNA under tension of at least 70 pN. RPA is able to bind both ssDNA under tension and relaxed ssDNA without wrapping it [47]. Two-color fluorescence measurements with eGFP-labeled RPA and bis-intercalator POPO-3 show three fluorescent regions [44]. The dsDNA segment has two bright spots of relaxed ssDNA on either side, followed by two ssDNA strands extending out to their respective attachment sites on each bead. Application of flow perpendicular to the axis of the molecule stretches out the relaxed ssDNA, clearly illustrating both strands of ssDNA created upon dsDNA overstretching [44].
Similar experiments with a DNA molecule attached to beads on both strands reveal the torsionally-constrained transition at 110 pN, with sites of POPO-3-labeled dsDNA and RPA-labeled ssDNA throughout the molecule [44]. The negative correlation of dsDNA and ssDNA areas on the same DNA molecule implies spatial separation of melted regions, with no evidence to support an interpretation of separate S-DNA and P-DNA phases [48]. The data also indicate that short regions of dsDNA remain when DNA is stretched to forces beyond the overstretching transition (in the second transition at the end of the overstretching plateau, near contour length of ssDNA). Thus, complete separation of the strands require application of unexpectedly high forces, but most of the DNA has been melted by force during overstretching [44]. Although this pulling-rate dependent transition [4, 49] is not well-described, it exists even in the presence of ssDNA binding ligands, which is unexpected in the S-DNA model [48].
The results of these single-molecule fluorescence imaging experiments are consistent with formation of ssDNA during both structural transitions, an observation which is incompatible with the prediction of unexposed individual bases of the S-DNA model. Thus the overstretching transition is a force-induced melting transition, in which the applied force does work to melt dsDNA into ssDNA. Therefore DNA stretching experiments involve melting of the two strands. This result can be used as a basis for investigation of the biophysical mechanisms of DNA-ligand interactions with single molecule force spectroscopy techniques.
2 DNA BINDING LIGANDS: SMALL MOLECULES
DNA-ligand interactions are relevant to fundamental intracellular processes such as DNA replication, transcription, and the regulation of gene expression. Small molecules that bind DNA can interfere with these processes, and thus play a key role in rational drug design for complex diseases such as cancer and AIDS. Furthermore, a detailed understanding small molecule binding to DNA may provide insight into the DNA binding properties of larger, more complex molecules such as proteins. Small molecules may bind DNA covalently, which is an essentially irreversible interaction, or non-covalently in a reversible process. Although covalent DNA binding has been examined with single molecule force spectroscopy, the focus has been on reversible binding of small molecules. Intercalation and groove binding are reversible binding modes which may be distinguished with optical tweezers experiments. When DNA is stretched in the presence of small ligands, their influence on the mechanical properties of the DNA molecule may be measured in order to determine their binding mechanisms. Single molecule stretching methods may also resolve multiple binding modes such as bis-intercalation and threading intercalation.
2.1 Cross-linkers
Force spectroscopy studies of irreversible DNA binding has been limited to cross-linkers, which are small molecules that bind DNA covalently, forming inter-strand or intra-strand cross-links between specific dsDNA bases. Cisplatin irreversibly binds dsDNA and triggers apoptosis, or programmed cell death, and has been used in cancer therapy. This ligand creates intra-strand cross-links between guanine and neighboring guanine or adenine bases. It also creates inter-strand cross-links between guanine bases [50]. AFM stretching studies of λ-DNA [50-51] and synthetic dsDNA [50] with sequences p(dGdC)-p(dGdC), p(dAdC)-p(dGdT), and p(dG)-p(dC) reveal that high concentrations of cisplatin decrease the cooperativity of the melting transition and demonstrate slow binding kinetics. The absence of hysteresis in the presence of cisplatin suggests that the two single strands are in close proximity after the cross-linking, allowing them to re-anneal on the timescale of the experiment. Stretching curves of synthetic p(dAdT)-p(dAdT) dsDNA [50] did not exhibit any of these effects, confirming that cross-links only form with guanine bases. The drug psoralen is also a cross-linker, but it intercalates within the dsDNA base pairs as well, and its dual binding modes are discussed below, with other small molecules which exhibit multiple and complex binding modes.
2.2 Intercalators
Intercalators have flat aromatic rings that slide between adjacent dsDNA base pairs for π-electron system interactions, lengthening and unwinding dsDNA [52]. Intercalators stabilize dsDNA, and the well-studied dye ethidium is known as a classical intercalator. Initial force spectroscopy studies with ethidium [27] explored the nature of melting transition observed in DNA stretching experiments using optical fiber force sensors. In these experiments, saturated concentrations of ethidium (~25 μM) clearly demonstrate lengthening of the DNA molecule, but the force-extension curves do not display a melting transition or hysteresis.
AFM stretching experiments with λ-phage BstE II-digested DNA [51] and duplex poly(dG-dC) [53] in the presence of high concentrations of ethidium (~5-10 μM) reflect similar results. At lower concentrations (~1 μM), however, force-extension curves of λ-phage BstE II-digested DNA exhibit a melting transition at a higher force relative to that of DNA without drug, indicating dsDNA stabilization, and hysteresis between stretching and relaxation cycles [51]. Force spectroscopy studies with intercalators such as ethidium [54-59], YO [57, 59], psoralen [56], the psoriasis and herpes virus drug proflavin [56, 60], the chemotherapy drug daunomycin [57, 59], and high concentrations of berenil [56, 58] also illustrate increases in DNA extension and melting transition force at low concentrations, but no melting transition or hysteresis at high concentrations. Furthermore, proflavin studies show that these effects are independent of solution pH [60].
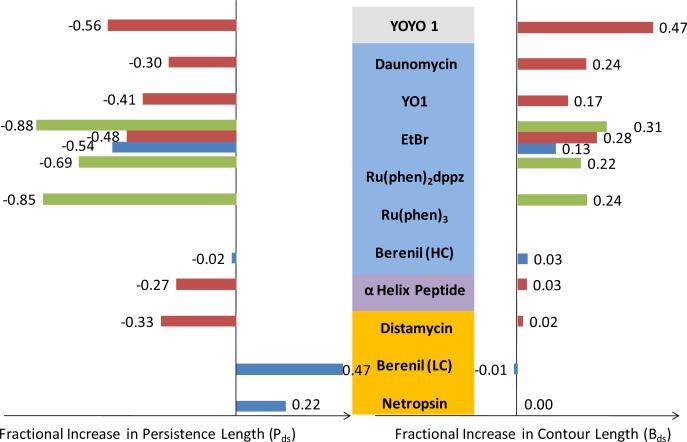
The first quantitative approach was to fit the force extension curves obtained in the presence of intercalators to the worm like chain (WLC) model [61], Equation (1). Fits of high concentration ethidium data at both low [58] and high force [59] limits indicate an increase in contour length and decrease in persistence length (Figure 3, blue and brown bars) relative to dsDNA in the absence of drug. The first experiments to observe non-equilibrium binding kinetics of mononuclear intercalators stretched DNA at various pulling rates in the presence of daunomycin [59]. DNA stretched to a maximum retention force was held at that extension to measure the force decay. Time constants obtained from these measurements are linearly dependent on the maximum retention force up to 45pN.
Figure 3.
Fractional increase in contour length (right) and persistence length (left) of the DNA-ligand complex for different drugs that exhibit a variety of binding modes, obtained from three types of fits to the WLC model (Equation (1)). Low force limit fits are shown in blue bars [58], high force limit fits are shown in brown bars [59, 66], and fits at drug saturation are shown in green bars [55]. Concentrations are relatively high (~1 μM) with the exception of saturated concentrations, which depends on the drug, and beneril, for which high concentration studies (HC) are at 3 μM and low concentrations studies (LC) are at 0.3 μM. The minor grove binders (drug names shaded yellow) show minor change in contour length, increase in persistence length in the low force limit and decrease in persistence length in the high force limit. Major groove binders (names shaded purple) show no change in contour length and decrease in persistence length. Intercalators (names shaded light blue) and bis-intercalator (names shaded gray) exhibits an increase in contour length and decrease in persistence length.
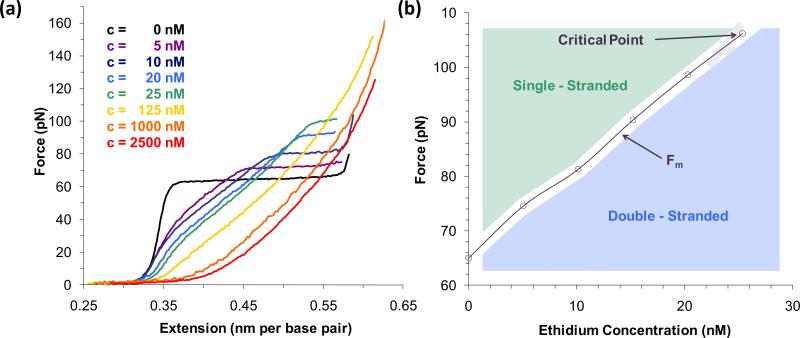
Rigorous concentration-dependent studies of ethidium with optical tweezers clearly illustrate that the DNA contour length increases with ethidium concentration, saturating at ~2.5 μM ethidium [54]. Although the melting force increases as a function of concentration, the transition plateau becomes progressively shorter as result of the DNA lengthening (Figure 4a). The phase diagram [54] in the force-extension-ethidium concentration space, which is analogous to PVT space for gas, showed a critical concentration for ethidium (~25nM) where phase separation between dsDNA and ssDNA becomes impossible (Figure 4b), which explains the disappearance of the melting transition at high concentration. Integrating the area under the dsDNA force extension curve in the presence and absence of ethidium provides the change in melting free energy due to the binding of ethidium, which is a function of concentration [54]. The results agree well with those from thermal melting experiments, demonstrating that increase in melting free energy corresponds to the increase in ethidium concentration.
Figure 4.
(a) DNA stretching curves in the presence of different ethidium bromide concentrations shows the increase in melting force, shortening of the melting transition with increasing concentration, and vanishing of the melting transition at a critical concentration around 25 nM. (b) The dependence of melting force on ethidium bromide concentration separates the two phases of the DNA (dsDNA shaded area in blue and ssDNA shaded area in green). The phase diagram shows that beyond 25 nM these phases cannot be distinguished by stretching experiments. Figures adopted from [12].
The site exclusion binding isotherm of McGhee and von Hippel relates the occupancy γ to the protein concentration c, for a protein with an equilibrium association binding constant K and binding site size n [62-63]:
| (4) |
Generally, the occupancy γ may refer to dsDNA (γds), ssDNA (γss), or intercalation (γint). For very large n, it may not be possible to obtain a DNA molecule fully saturated with ligand. However, the impact of this caveat decreases with ligand mobility rate, and the effect is unlikely to change experimental results within error.
Fractional occupancy γint, determined from change in DNA extension upon intercalation, fit to the McGhee von Hippel isotherm (Equation (4)) as a function of force yields K = 107 M-1 and n = 2 at F ≤ 10 pN and K = 1.5 × 107 M-1 and n = 1 at F ≥ 20 pN for ethidium [54]. The low force values agree particularly well with those obtained in bulk experiments. The high force value of n = 1 suggests that ethidium stacks between every base pair, which contradicts the traditional view that ethidium stacks only with every other base pair. It is possible that the conformation of the DNA backbone which excludes binding sites at low force may change as the DNA is held under higher tensions, allowing intercalation between every base pair.
Optical tweezers studies examining the concentration dependence of ethidium and daunomycin at low force (F ≤ 2pN) suggest that the contour length and persistence length of the drug-DNA complex increases with concentration until saturated binding, upon which the contour length remains constant and the persistence length decreases sharply to nearly that of drug-free DNA [64].
Optical tweezers experiments also examined the DNA binding properties of ruthenium(II) polypyridyl complexes Ru(phen)2dppz2+, Ru(phen)32+ and Ru(bpy)32+ [65]. The first two complexes clearly demonstrate the increase in melting force and contour length with high concentration that is characteristic of intercalation. The third complex, Ru(bpy)32+, does not intercalate at zero force. Fits of γint to the McGhee von Hippel binding isotherm (Equation (4)) suggest a binding site size of n = 3 for both intercalators, with binding constants of K = 3.2 (± 0.1) × 106 M-1 and K = 8.8 (± 0.3) × 103 M-1 for Ru(bby)2dppz2+ and Ru(phen)32+, respectively.
Single molecule studies propose a method of quantifying force-dependent intercalation which extrapolates the results to zero force, characterizing ligand binding to relaxed DNA [55]. This method is now widely used to analyze intercalation of small molecules. The force-dependent fractional occupancy of the DNA lattice ν is defined in terms of the binding site size n:
| (5) |
Thus ligand binding may be described in terms of fractional occupancy γ, which runs from 0 to 1, or fractional occupancy per base pair ν, which accounts for binding site size and therefore runs from 0 to 1/n. The fractional occupancy per base pair for intercalators, νint, at a given force F is:
| (6) |
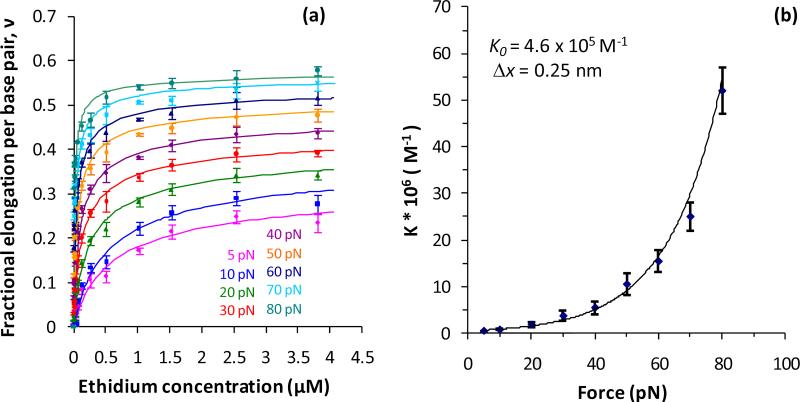
where bds(F,c) is the extension in the presence of the intercalator at concentration c, bds(F,0) is the extension in the absence of intercalator, and Bds is the DNA contour length in the absence of intercalator. This factional binding per base pair ν is fit to the McGhee von Hippel isotherm (Equations (4) and (5) combined) to obtain the equilibrium association constant KF and binding site size nF at a given force F (Figure 5a).
Figure 5.
(a) Fractional elongation per base pair (ν) as a function of ethidium bromide concentration c, fit to the McGhee von Hippel model at different forces F. (b) Force-dependent binding constants KF obtained from the fits shown in (a) yield the zero force binding constant K0 and the DNA lengthening upon a single intercalation event Δx.
The force F applied to stretch the DNA molecule reduces the free energy of intercalation ΔG0 by FΔx, where Δx is the dsDNA elongation upon a single intercalation event, leading to an exponential force dependence of the binding constant [55]:
| (7) |
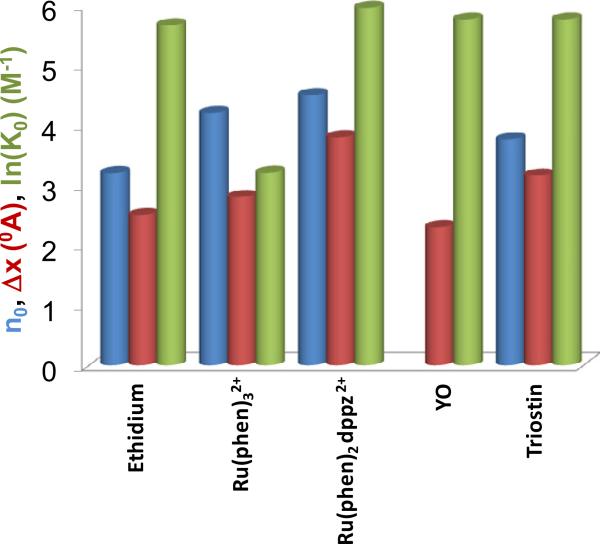
where K0 is the binding constant in the absence of the force, kB is the Boltzmann constant and T is the absolute temperature (Figure 5b). Figure 6 presents K0, Δx, and n for ethidium [55], Ru(phen)2dppz2+ [55], Ru(phen)32+ [55] and YO [66].
Figure 6.
Binding site size n in base pairs (blue), lengthening upon a single intercalation event Δx in 0A (brown) and log of the zero force binding constant K0 in M-1 (green) estimated for ethidium, Ru(phen)32+, Ru(phen)2dppz2+, YO, and triostin using the method of Vladescu et al. [55] for molecules reported in [55, 66, 73].
Fits to the WLC model at saturated intercalator concentrations yield the persistence length, contour length, and elastic modulus of the drug-DNA complex at saturation. These results show that intercalator binding increases DNA contour length and decreases DNA persistence length (Figure 3, green bars). The saturated stretch modulus of the drug-DNA complex is reduced nearly five-fold relative to that of dsDNA in the absence of drug [55, 66].
Recent experiments with intercalator YO using the combined techniques of optical tweezers and fluorescence microscopy measure fluorescence intensity during DNA stretching [66]. Since YO molecules fluoresce only upon binding to dsDNA, the fluorescence intensity may be used to quantify intercalation. Fluorescence intensity and fractional elongation have the expected linear relationship, but with two regions of different slopes. This suggests a force-induced structural transition of the YO-DNA complex at ~0.14 fractional elongation at 100 nM YO. Additionally, fits to the WLC model (Equation (1)) provide the persistence length and contour length as a function of concentration for the YO-DNA complex. The results agree with those from the high force limit, demonstrating that the contour length increases with concentration, while the persistence length decreases.
2.3 Groove binders
The majority of molecules that bind in dsDNA grooves are positively charged, and thus binding is dominated by electrostatic interaction and assisted by hydrogen bonds and van der Waals interactions [51, 57]. Initial AFM stretching experiments explored the minor groove binding activity of berenil [51]. High concentrations of berenil results in dsDNA intercalation, and further investigations of groove binding used netropsin [56], an antibiotic that also exhibits antitumor and antiviral activity. Force-extension curves in the presence of netropsin, which is known to bind only in the minor groove of dsDNA, exhibit an increase in melting force similar to that observed in the presence of berenil at low concentrations. However, there was no broadening of the melting plateau, an effect which is only evident with berenil. To clarify this difference, AFM experiments examined the minor groove binder Hoechst 33258 [56]. These DNA stretching curves are similar to those in the presence of netropsin, without melting plateau broadening. Force extension curves in the presence of Hoechst 33258 do not depend on pH, and the slightly lower melting force at high pH (~10.5) relative to that at neutral pH (7.5) [60] may be attributed to the pH dependence of the DNA melting force [14].
Initial AFM stretching experiments with poly(dG-dC) dsDNA in the presence of the peptide distamycin-A found that the minor groove binder lowered the melting force to 50 pN [57]. Although this result contrasts with an expected increase in melting force, the discrepancy was attributed to the distamycin-A preference for AT-rich regions [57], although this observation remains unclear. Optical tweezers experiments stretching λ-DNA in the presence of distamycin-A measured an increase in the melting force [59], an observation consistent with results from other minor groove binding experiments. Therefore, molecules which bind in the minor groove are believed to increase the melting force and preserve melting transition cooperativity.
AFM experiments stretching poly(dG-dC) dsDNA examined two synthetic amphipathic peptides which bind to the major groove of dsDNA, an α-helix Ac-(Leu-Ala-Arg-Leu)3-NH-linker (linker:1,8-diamino-3,6-dioxaoctane) and 310-helix Ac-(Aib-Leu-Arg)4-NH-linker containing β-loop builder α-aminoisobutyric acid (Aib) [57]. Although the stretching curves of both major groove-binding peptides are similar to those of minor groove binders, both exhibit a decrease in melting transition cooperativity. DNA stretching experiments using optical tweezers in the presence of the α-helical peptide studied with AFM [59] and the major groove binder SYBR-Green I [67] confirm these characteristics of dsDNA major groove binding.
Fits to the WLC model (Equation (1)) for dsDNA-minor groove binding complexes with netropsin [58], berenil at low concentrations [58], and distamycin-A [59] did not indicate a change in contour length or persistence length in either the low or high force limits (Figure 3). In contrast, WLC model fits for the major groove binder α-helix Ac-(Leu-Ala-Arg-Leu)3-NH-linker (linker:1,8-diamino-3,6-dioxaoctane) did demonstrate an increase in contour length and decrease in persistence length (Figure 3).
2.4 Multiple and complex binding modes
Small molecules such as berenil and psoralen bind DNA with multiple binding modes, while molecules such as binuclear ruthenium complexes, YOYO, and triostin have complex modes such as threading intercalation and bis-intercalation. Berenil binds into the minor groove, favoring AT-rich regions, at low concentrations, but intercalates within the base pairs at high concentrations. AFM stretching experiments in the presence of berenil show minimal DNA lengthening, but demonstrate an increase in melting force at low concentration which resembles minor groove binding [51, 56]. At an order of magnitude higher concentration of berenil, the stretching curves exhibit loss of cooperativity and no hysteresis, resembling intercalators at high concentrations. A quantitative study fit force extension curves at varying berenil concentration to the WLC model (Equation (1)) in the low force limit [58]. The results reveal a significant increase in persistence length Pds at low berenil concentrations relative to that of dsDNA in the absence of drug. As berenil concentration increases, Pds decreases until it is less than the persistence length of dsDNA without ligands. This relationship indicates that the binding mode of berenil changes from minor groove binding, which is characterized by higher persistence lengths in the low force limit, to intercalation, which is characterized by low persistence lengths in the low force limit. The contour length Bds increases with berenil concentration, which is consistent with a change in DNA interaction from minor groove binding to intercalation.
Psoralen is a drug used in Psoralen Ultra Violet A (PUVA) therapy, which involves exposure of administered psoralen to Ultra Violet (UV) A light as treatment of specific skin diseases. The drug intercalates with dsDNA, then forms a covalent bond with pyrimidine in one DNA strand to form a mono-adduct. Exposure to UV A light results in a covalent bond on the other DNA strand for formation of a cross-link. Although exposure to UV B light may break cross-links, it does not affect the mono-adducts. DNA stretching experiments explore the binding mechanisms of psoralen, measuring the persistence length as a function of time in the presence of the drug when exposed to UV A light [68]. Fits of the DNA-drug stretching curves to the WLC model (Equation (1)) in the low force limit provide the persistence length Pds. Pds initially increases with exposure time as the drug begins to intercalate, then decreases in value as intercalation saturates. After this saturation point, Pds increases again to its maximum value after ~35 minutes, indicating formation of cross-links which make the complex more rigid. Exposing the drug-DNA complex to UV B results in a dramatic drop in Pds, indicating the breakage of cross-links, and the mono-adducts which remain are significantly less rigid.
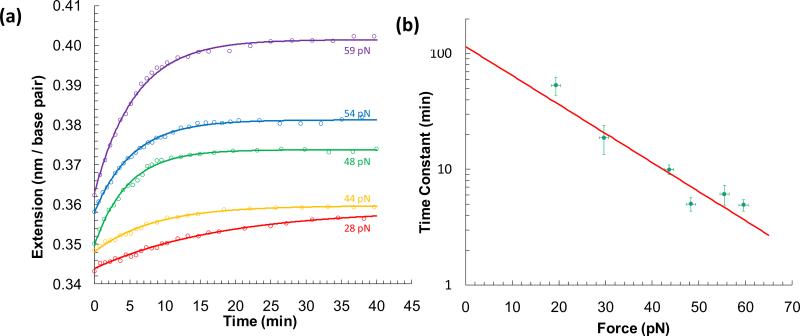
Binuclear ruthenium complexes are two covalently linked Ru(phen)2dppz2+ moieties, and they initially bind in the major grooves of dsDNA. These complexes then thread through the DNA bases, and the bidppz bridge intercalates between the dsDNA base pairs [69]. In order for this dumb-bell shaped binuclear ruthenium complex to thread, the dsDNA must melt so that the bulky end can slide through the unpaired bases and adopt the final threaded conformation. Although this requires hours to occur in traditional bulk experiments, single molecule DNA stretching in the presence of these complexes facilitates threading. Extrapolating the force-dependent kinetics measured then quantifies DNA binding kinetics in the absence of force. DNA that is stretched and held at a constant force in the presence of these complexes (Figure 7a) reveals a characteristic force-dependent time constant [70]. Applying force F favors the melted state by decreasing the melting free energy by FnΔx, and therefore increasing the probability of melting by [18]:
| (8) |
where n is the number of melted base pairs and Δx is the length increase due to conversion of dsDNA to ssDNA, a value which is expected to be 0.22nm in the linear approximation region [20]. This leads to an exponential dependence of the time constant on applied force:
| (9) |
where kBT is ~4.1pN-nm. Fits of the force dependence of these time constants (Figure 7b) yields that only one base pair must melt to thread the binuclear ruthenium complex. The extrapolation to zero force provided a time constant of 120 (± 30) min, which is the time constant associated with threading in the absence of force [70].
Figure 7.
(a) Extension measurements (open circles) as a function of time obtained at constant forces of 28 pN (red), 44 pN (yellow), 48 pN (green), 54 pN (blue) and 59 pN (purple) in the presence of threading intercalator ΔΔ-[μ-bidppz(phen)4Ru2]4+ and single exponent fits (solid lines) to these measurements. (b) Characteristic time constants (green circles) obtained from the fits in (a) for corresponding constant force measurements. Exponential dependence on force (fit, red line) yields the key result that only one base pair must melt in order for this molecule to thread through the DNA bases. Figures adopted from [70].
YOYO-1 is a bridged oxazole yellow (YO) dimer which stacks two aromatic ring systems into two intercalating sites, causing a clamp-like binding known as bis-intercalation. It can also bind in the DNA major groove at high concentrations. This dye is particularly useful to study the properties of DNA, since it is non-fluorescent in solution and highly fluorescent upon binding to dsDNA [71]. DNA stretching experiments with YOYO-1 show lengthening of dsDNA upon YOYO-1 binding, which is similar to other intercalators [57, 59, 66, 72]. In contrast to many intercalators, however, DNA stretching curves exhibit hysteresis during relaxation, and the dsDNA lengthening observed is strongly dependent on the velocity of stretching, which indicates slow binding kinetics [59, 66, 72]. When the DNA molecule is stretched to a maximum retention force, in an experiment designed to reach different fixed extensions, and allowed to relax in the presence of YOYO-1, the exponential decay of the retention force may be used to calculate the association time constant of YOYO binding [59, 66]. The results show that time constants are linearly dependent on the maximum retention force up to 60pN [59].
In experiments which combine optical tweezers with fluorescence microscopy, fluorescence intensity is used as a measure of the number of YOYO molecules bound [66]. The fluorescence intensity has a linear relationship with the fractional elongation, indicating that both quantities increase with the number of YOYO molecules bound. The equilibrium DNA lengthening may be obtained from this linear relationship. Furthermore, fits to the WLC model (Equation (1)) may be used to calculate the pulling rate-dependent contour length, persistence length, and stretch modulus. The results suggest that slow pulling rates, which are closer to equilibrium, yield the lowest persistence length and stretch modulus, but the highest contour length. The method of quantifying force-dependent intercalation kinetics developed by Vladescu et al. [55] was used to obtain the zero force binding constant K0 = 38.75 (± 1.28) × 105 M-1 and DNA lengthening upon single YOYO intercalation event, Δx = 0.095 (± 0.002) nm, although both quantities were inconsistent with results from bulk experiments.
A recent study shows that bis-intercalator triostin [73] exhibits properties similar to YOYO, with very slow kinetics. Thus the force extension curves obtained are non-equilibrium even at very slow pulling rates. Constant force measurements similar to those employed for threading intercalators [70] were used to obtain the equilibrium lengthening due to triostin binding dsDNA. Fractional elongation was calculated according to Equation (5), with a factor of 0.5 used to incorporate bis-intercalators. The method introduced by Vladescu et al. [55] for intercalators was used to analyze force-dependent kinetics in order to determine the binding site size n and binding constant K. The zero force binding constant of K0 = (5.8 ± 0.3) × 105 M-1 and DNA lengthening per intercalation event Δx = 0.316 nm were determined from the fits.
2.5 Classification of different binding modes
Although several studies discussed qualitative discrimination between intercalation, minor and major groove binding [51, 56-57], the first quantitative approach was with low force (F≤15pN) extension measurements obtained using optical tweezers [58], followed by fits of the data to the WLC model (Equation (1)) in the near-full extension limit [61]. Several more recent studies use the WLC in both the and low high force limits to obtain the contour length Bds and persistence length Pds of different small molecules (Figure 3).
The contour length obtained from fits to the WLC model in both low and high force limits provides the same insights. Minor groove binders [58-59] and major groove binders do not change the contour length, while intercalators [58-59, 66] and the bis-intercalator YOYO [59, 66] exhibit an increase in contour length for DNA-ligand complexes. Concentration-dependent studies on intercalators and bis-intercalators show that the contour length increases with the increase in concentration until saturation at a specific concentration [54-55, 65, 73].
Measurements of minor groove binders at low force (less than 15 pN) generally show an increase in DNA persistence length [58], whereas a measurement of minor groove binder Distamycin-A at higher force showed an increase in DNA persistence length [59]. Major groove binders cause a decrease in persistence length [58]. Concentration-dependent studies of intercalators show that the persistence length of the DNA-intercalator complex initially increases with concentration, dropping to the value of dsDNA [58, 64], and then dropping even lower at very high or saturated concentrations [55, 59, 66].
Other features that distinguish different binding modes qualitatively are the melting force, cooperativity of the transition and the hysteresis observed during relaxation. All of the dsDNA binding modes increase the melting force, as expected thermodynamically. The significant hysteresis observed in the presence of bis-intercalators and threading intercalators reflects slow dissociation kinetics [59, 66, 73]. Major and minor groove binders have similar effects on DNA stretching curves, but major groove binders decrease the cooperativity of the melting transition [57]. These results show that single molecule force spectroscopy is a useful method for quantitatively characterizing the thermodynamics and kinetics of small molecule interactions with DNA.
3 DOUBLE-STRANDED DNA BINDING PROTEINS
Force spectroscopy experiments on small molecules generally serve to assay length and force changes as ligands bind to double- or single-stranded DNA. Changes in the observed DNA force-extension curves indicate the mode and binding affinity. Proteins that preferentially bind to dsDNA also induce changes in DNA force-extension curves. In many instances, protein binding must be disrupted observe melting or extend the DNA-protein complex, as indicated by an increase in the observed melting force or by discrete increases in the measured DNA length. Protein binding may also alter the elasticity of the DNA backbone. dsDNA binding proteins often alter the overall organization of DNA in the cell. Examples of such DNA packaging proteins include histones that package DNA into chromatin in eukaryotic cells, as well as HU and H-NS proteins in Escherichia coli (E. coli). High mobility group (HMG) proteins facilitate the reorganization of packaged DNA in eukaryotic cells by altering DNA elasticity, which in turn allow other proteins access to the DNA. We discuss below single molecule force spectroscopy experiments that elucidate the mechanisms by which these proteins alter the biophysical properties of DNA to facilitate important cellular interactions.
3.1 Packaging DNA: Histones and chromatin
DNA is highly organized and compacted in the nucleus, interacting with histone proteins to form the basic higher order structure of the genome, known as the nucleosome core particle. First discovered in 1974 [74-76], the core particle is composed of pairs of the histones H2A, H2B, H3 and H4 [77-78]. Each histone is a small protein (~120 amino acids) that contains three D-helices connected by two loops, a motif known as the histone fold [79]. These highly conserved proteins, rich in basic residues, assemble to form an octamer in the shape of a rough disk. A length of 146 dsDNA base pairs wraps around this structure 1.65 times, forming a somewhat larger cylinder 11 nm in diameter and 5-6 nm in height [77]. The contacts between the histone core and the DNA include over 120 direct DNA-protein interactions [80], though the wrapped structure may include significant kinks and transient fluctuations [81]. This assembly of DNA and the octamer hub is known as the nucleosome core particle. Adjacent core particles may interact, stacking together to form a higher order structure known as chromatin [82-84]. The structure and stability of chromatin is driven by nucleosome stacking interactions, solution conditions and auxiliary histones that stabilize linker strands of DNA between adjacent core particles. The length of DNA between the start of adjacent core particles, known as the nucleosome repeat length, ranges from 165 to 212 base pairs, and determines the length of the linker DNA [82, 85]. Assembled chromatin fibers, roughly 30 nm in diameter, are further compacted in the cell into euchromatin and ultimately heterochromatin [78].
3.1.1 Fundamental interactions: Histones and nucleosomes
Early experiments on chromatin were not realized on the fully assembled structure, but on single strands of DNA wrapped around successive nucleosome core particles. The core particles were typically unstacked and separated by additional linker segments of DNA, the length of which depend upon the nucleosome repeat length. These experiments thus probed the stability of individual core particles, in a conformation sometimes referred to as beads on a string [86-92]. The ends of the assembly are tethered to polystyrene beads which are then manipulated in optical tweezers experiments. The results show small deformations in the extension curves at low forces that may be the result of nucleosomenucleosome interactions [89], while others have shown distinct unbinding ‘rips’ that begin at a threshold of ~ 20 pN, depending somewhat upon the solution conditions [86-88]. These rips, corresponding to DNA unwrapping from the nucleosome, show characteristic lengths of multiples of ~60 nm, corresponding to complete unwrapping and release of core particles [86]. Others have seen additional opening events with a smaller length scale, indicating stepwise breaking of histone contacts, opening of the core particle and ultimately the release of the octamer [87-88, 91].
3.1.2 Higher order interactions: Unraveling chromatin
Recent studies probe the higher order structure of assembled chromatin [93]. In these studies, 25 tandem repeats of 601 DNA, which localizes the chromatin to specific sequence locations [94-95], are used to facilitate the formation of a uniformly repeating structure. Two repeat lengths of 167 and 197 base pairs are used, which vary length of the linker segments between adjacent nucleosomes. In the presence of Mg2+, the assembled nucleosomes then reconstitute a 30 nm diameter chromatin fiber. The fiber, approximately 50 nm in length, is tethered to a 1 μm diameter magnetic bead and a microscope slide via additional linker DNA, raising the total contour length to 150 nm. Magnetic tweezers may pull this structure over several pN, with a sub-pN resolution. Here the instrument is configured with a force feedback mechanism, so that the tweezers act as a force clamp. When the chromatin fiber is unfolded to a string of DNA and nucleosomes, increases in extension are observed out to a length of ~750 nm. The applied tension is kept below the threshold force required for core particle disruption (~15 pN).
A key result reveals that chromatin, when constructed with a 197 nucleosome repeat length, displays a Hooke-like spring constant of 0.02 pN/nm (48,500 base pair phage λ DNA exhibits a spring constant of ~0.07 pN/nm). This spring constant is independent of the presence of linker histones H1 and H5, though there is some variability in the fitted parameters. However, a key difference in chromatin without linker histones is the observation of a plateau above 3 pN, where nucleosome–nucleosome interactions are reversibly broken. The length of the structure at the beginning of this transition suggests that the nucleosomes are stacked directly above each other, known as a one-start topology. When chromatin is constructed with a 167 nucleosome repeat length, the linker segments are correspondingly shorter. This causes an increase in the stiffness and length of these chromatin fibers. Furthermore, the structure observed just before the rupture of the nucleosome–nucleosome contacts consists of two twisted stacks, known as a two start topology. Thus the width of the assembled fibers is altered as well, an observation which AFM studies confirm, though the width may also be altered by the presence of linker histones [96]. However, it has been noted that the structure of the 30 nm fiber may be influenced by the techniques used to assemble the nucleosomes and there is some controversy about the exact structure of chromatin in vivo [97]. Given that the stacking interaction energy of the nucleosomes was estimated to be 13.8 kBT, molecular motors such as polymerases, known to typically stall at forces of 10-25 pN [87], can successfully force structural changes in chromatin, though the nucleosome presents strong forces that may stall such motors [98]. However, individual core particles, have been observed to partially and fully unwrap at much lower forces of several picoNewtons [99], suggesting that thermal fluctuations may play a substantial role in chromatin organization that remains poorly characterized. Nonetheless, these ongoing force spectroscopy experiments have provided insight into the protein-DNA contacts that stabilize the nucleosome core particle as well as the interactions between adjacent nucleosomes that favor the formation of chromatin.
3.2 Bending DNA and distorting chromatin: High mobility group proteins
High mobility group proteins are an abundant family of eukaryotic chromosomal proteins characterized by an array of DNA binding modes, including sequence-specific AT hooks and a variety of non-specific motifs [100-103]. The HMGB subgroup is distinguished by a single or double 80 amino acid domain that forms a box structure composed of three D-helices and a disordered tail. This simple structure is present during a variety of cellular processes which it may facilitate, including transcription [104-111], DNA repair [112] and immune response signaling [113-114]. These proteins are also known to facilitate DNA looping, a function also seen in HU [115-117]. A primary function of HMGB proteins is believed to be the disruption of chromatin through strong binding to dsDNA (possibly as a prelude to transcription) [100, 102, 104, 116, 118-121].
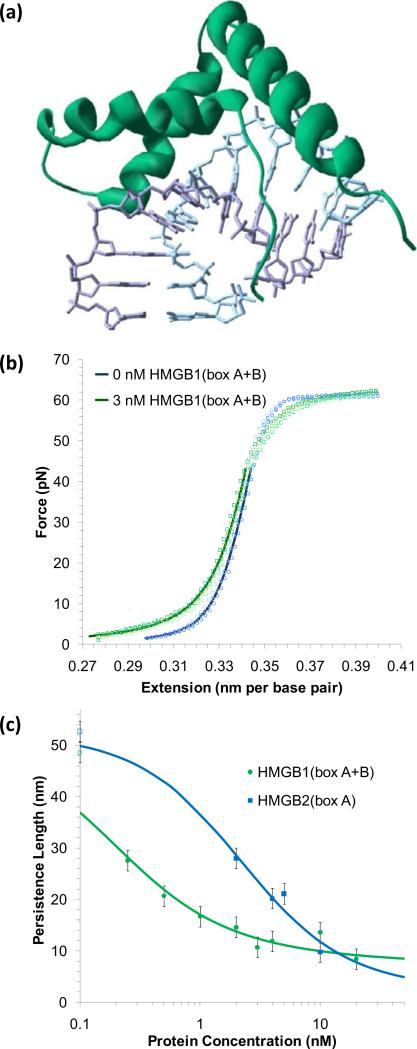
As the protein binds into the minor groove, interactions with the backbone occur through van der Waals contacts, direct and water-mediated hydrogen bonds and partial intercalation. A strong, continuous bend is induced in the backbone, which has been measured to be ~111.1° for structures determined for the single box (shown in Figure 8a) [122-123], and 101.5 ± 9.1° for the double box motifs of HMGB like proteins [124]. In addition to measuring the bending angles induced by HMGB protein binding, optical tweezers experiments may determine the equilibrium association binding constant, distinguishing the binding strength from the bending ability.
Figure 8.
Distortions in the structure of DNA induced by HMGB family proteins. (a) The crystal structure of a single HMGB box motif is shown here for HMG-D from D. melanogaster [123]. Three helices and positively charged C-terminus become more ordered upon binding to DNA. The protein binds along the minor groove, intercalates into and induces a bend of ~ 111° over a region of ~ 7 base pairs (PDB code: 1QRV). Double box motifs are consecutively joined. (b) Extension data for a single molecule of phage-λ DNA (blue circles) and the same molecule in the presence of a solution containing 3 nM human HMGB1(box A+B) (green squares). Four separate extension cycles are shown, as are fits (solid lines) to the average, using the WLC model of Equation (1). Only forces below 45 pN are included for the fitted data, to minimize the effect of melting transition. Fits reveal a decrease in the persistence length of dsDNA from 46 ± 2 nm to 22 ± 2 nm as the protein is added. (c) The persistence length decreases continuously with the addition of single box HMGB2(box A) (blue) or HMGB1(box A+B) (green), and may be fit to the model of Equations (10) and (4) as described in the text. The equilibrium association binding constant per ligand is determined to be 5.9 (± 1.6) × 108 M-1 for HMGB1(box A+B) and 0.15 (± 0.06) × 108 M-1 for HMGB2(box A). Open symbols are persistence lengths from control fits to the same DNA molecules in the absence of protein [1, 12]
3.2.1 HMGB alters the elasticity of dsDNA
During a fully reversible cycle of extension and relaxation, dsDNA exhibits an entropic and elastic response until increasing force favors the stabilization of ssDNA. Upon extension, the entropic elasticity is characterized measuring its persistence length, Pds. Extension data for dsDNA have been fit to Equation (1) under a variety of temperature, pH and salt concentration conditions. Under typical conditions (10 mM Hepes, 100 mM Na+, pH = 7.5, T = 20 °C), as shown in Figure 8b, fits determine Bds = 0.340 ± 0.001 nm/bp, Pds = 48 ± 2 nm, and Sds = 1200 ± 100 pN [11, 125] Interpolated solutions to this model have given more accurate results over a wide range of forces. [126] Furthermore, the WLC model is limited to long sequences (n > 100 base pairs). [9, 127-129] Finally, the bends introduced by discrete protein binding events may distort the continuum of bending angles. However, long molecules (such as the 48 kbp λ DNA) with multiple bound proteins can be described by Equation (1), and changes to the fitted parameters characterize the effect of protein binding on dsDNA.
In the presence of just 3 nM of the two binding motif HMG protein HMGB1(box A+B), the extension data for dsDNA shows a clear change, in Figure 8b. Fits to Equation (1) give Bds = 0.339 ± 0.003 nm/bp, Pds = 22 ± 2 nm, and Sds = 939 ± 100 pN. While the contour length and stiffness show little definite change, the persistence length evinces a clear decrease. As protein binds to dsDNA, the persistence length decreases continuously, from a value for bare dsDNA PDNA to a value when dsDNA is fully covered PPR, as shown in Figure 8c. The observed persistence length Pds will vary according to the factional occupancy γds [130]:
| (10) |
The site exclusion binding isotherm of McGhee and von Hippel provides the occupancy γ (Equation (4)). To limit the number of free parameters, the binding site sizes are held fixed, using sizes determined in previous work [n = 7 for HMGB2(box A) and n = 18 for HMGB1(box A+B)] [122-124, 131]. Figure 8c shows the results of these fits, which yields an equilibrium association binding constant per ligand of 5.9 ± 1.6 × 108 M-1 for HMGB1(box A+B) and 0.15 ± 0.06 × 108 M-1 for HMGB2(box A).
The saturated dsDNA persistence length PPR is also determined from the fits. Assuming that the protein bending angles are induced at discrete sites, an average induced bending angle β may be estimated by [130, 132]:
| (11) |
The contour length at saturation BPR is determined from fits to Equation (1). A particular strength of this assay is that the affinity of protein binding and the degree of the protein-induced bend into the backbone may be deduced separately. Average induced bending angles for protein-DNA binding are β = 99 ± 9° for the single box protein HMGB2 and β = 77 ± 7° for the double box of HMGB1. Thee angles are in good agreement with measurements from other techniques [116, 124, 133-135].
3.2.2 Distributions of bending angles and enhanced DNA flexibility
The bending angles determined in optical tweezers experiments suggest an enhancement of the flexibility of dsDNA. Yet there remains a fundamental question about the nature of that flexibility. While the HU proteins described below appear to create a flexible hinge about the binding site [117], inducing a change in the local flexibility of DNA. The deduced structures for HMG proteins seem to indicate a fixed angle, or a static kink in the DNA generated by protein binding [123-124], which would enhance DNA flexibility through a mechanism of rapid binding and unbinding. Thus the observed distribution of bending angles will differ according to the binding mechanism. A protein that induces a flexible hinge will show a flat distribution of bending angles, as there is no single favored bending angle at the binding site. A distribution composed of a single peak about a well-defined fixed angle will characterize a static kink. Atomic force microscopy (AFM) measurements are particularly well suited to measure the distributions of protein bending angles [136-139].
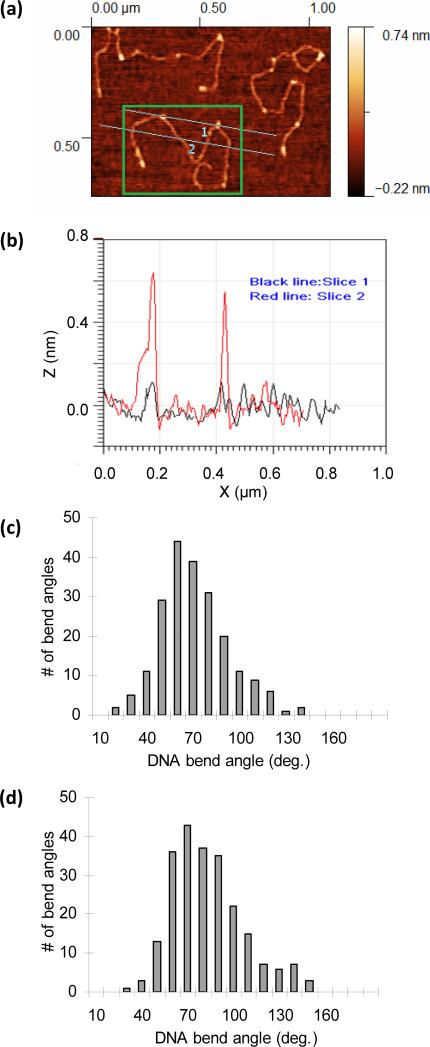
A caveat to AFM experiments is that interactions with the surface may influence the results. DNA–protein interactions are inherently three-dimensional, and the resulting complexes must be deposited upon a two-dimensional surface. If the surface interaction is strong, and the interaction with the substrate is relatively rapid, then the molecules are kinetically trapped and the observed images will have been significantly affected by the surface. If the molecules may approach the surface more slowly, an equilibrium may be established that has been shown to be remarkably unaffected by deposition [136, 140]. Simulations have also proven helpful, allowing the quality of deposition to be estimated for certain proteins [141]. Typical AFM images for pBR322 (4361 base pairs) DNA are shown in Figure 9a [142]. Bound HMG appears as individual bright spots associated with the DNA. The lighter color indicates a higher profile, as shown for two slices drawn through the image. The height profile along these images shown in Figure 9b indicates two protein binding events along the first slice, and that these correspond to the locations of the bright spots in Figure 9a. Although the second slice intercepts the DNA in several places, it crosses no spots and shows no assessable change in the height profile. Thus the spots indicate protein binding and the bend in the DNA backbone may be measured directly at these individual protein binding sites.
Figure 9.
Force microscopy reveals the distributions of bending angles. (a) Gradient surface scans of three DNA molecules in an HMG solution fixed to a mica surface and observed with an oscillating cantilever AFM. The green box frames a single DNA molecule and includes two cross slices. (b) Measured height profiles along cross slice one (black) and slice two (red). Along slice one, bound HMGB proteins are clear as bright spots in the surface image, which correlate to 0.6 nm changes in height. No proteins appear bound along slice two. The angle subtended by the backbone across these bound proteins may be measured directly. (c) Distribution of measured bending angles for HMGB1(box A+B) shows an average induced bending angle of 67 ± 1° (standard error). d) A larger average induced bending angle of 78 ± 1° may be deduced from the distribution for DNA in the presence of HMGB2(box A). The widths of the distributions for both proteins indicate a wide range of angles induced by protein binding [142].
Distributions for the single and double box motifs appear in Figure 9c and 9d. The average bending angles for HMGB1(box A+B) of 67 ± 1° for HMGB2(box A) of 78 ± 1° compare fairly well with the averages found from optical tweezers experiments. Both experiments show that the single box motif induces a greater bending angle into DNA versus the double box, and that the double box is not more effective at increasing the flexibility of dsDNA versus the single box. The standard deviations of these distributions (23° for the double box and 21° for the single box), however, signify that the binding mechanism created is neither a static kink nor a purely flexible hinge. In contrast to the results for HU discussed below, the results for both HMG motifs are peaked, though somewhat weakly. Thus HMG appears to alter the flexibility of DNA by a combination of two mechanisms. In the first, protein binding introduces local flexibility directly at the binding site. In the second, HMG induces local bends, and by rapidly binding and unbinding, alters the flexibility of the overall molecule.
3.2.3 Stabilizing dsDNA
While optical tweezers experiments may characterize DNA binding with changes in elasticity, binding to dsDNA is also evident in the force-induced melting plateau. HMG binds and stabilizes dsDNA, as higher forces are required to melt DNA as shown in Figure 10a and 10b. Protein binding must be at least partially disrupted for melting to occur. In this instance, binding to dsDNA is characterized by the measured melting force, averaged over the extension range from 0.42 to 0.48 nm per base pair. As more protein is added, the average melting force increases, according to Figure 10c. The occupancy γds may be found by comparing the observed melting force Fm with the force measured in the absence of (Fm0) and fully saturated with protein (Fms) [143-144]:
| (12) |
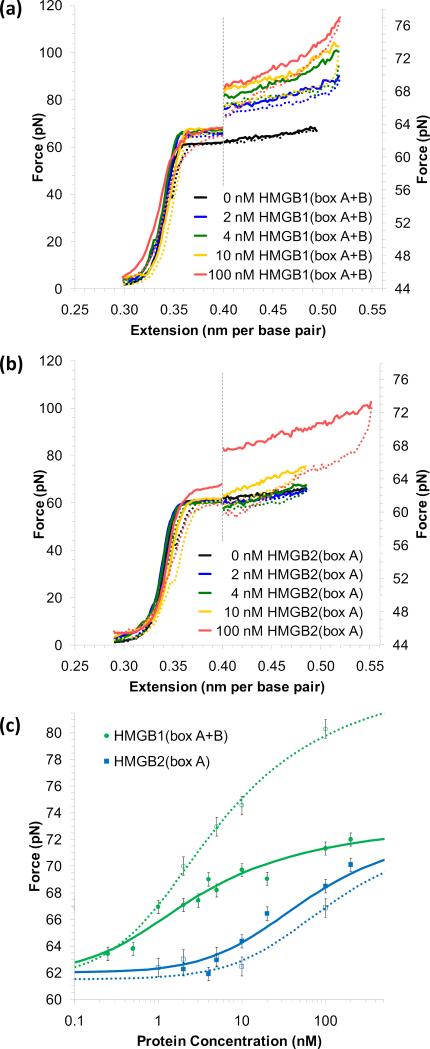
Combining Equation (12) with the site exclusion model of Equation (4) gives the fits shown in Figure 10c and determines the equilibrium association binding constant per ligand to be 7.2 ± 1.7 × 108 M-1 for the double box HMGB1 and 0.28 ± 0.10 × 108 M-1 for the single box HMGB2.
Figure 10.
HMG proteins stabilize dsDNA. (a) Cycles of extension (solid lines) and relaxation (dotted lines) for DNA in the presence of no protein (black) and 2 nM (blue), 4 nM (green), 10 nM (yellow) and 100 nM (red) of human HMGB1(box A+B). The graph is split along the dotted line; data to the right is expanded along the scale shown. The observed melting force increases with escalating protein binding. Moderate hysteresis indicates some protein unbinding during the melting transition. (b) Stretching and relaxation data for DNA in the presence of no protein (black) and 2 nM (blue), 4 nM (green), 10 nM (yellow) and 100 nM (red) of rat HMGB2(box A). Stabilization of dsDNA requires greater amounts of the single box protein. Significant hysteresis is observed for high protein concentration, as protein–protein contacts must be dislocated for melting to occur. (c) The increase in the observed melting force as a binding assay for HMGB proteins. The averaged midpoint of the melting force plotted versus protein concentration for HMGB1(box A+B) (green) and HMGB2(box A) (blue). Error bars reflect instrumental uncertainty and standard deviations from a minimum of four individual extension curves. Fits are to the binding model of Equations (10) and (4), as described in the text. Fits determine an equilibrium association binding constant per ligand of 7.2 ± 1.7 × 108 M-1 for HMGB1(box A+B) and 0.28 ± 0.10 × 108 M-1 for HMGB2(box A).
The fact that the proteins exhibit such dissimilar binding characteristics is initially surprising, since the essential difference between the two proteins is simply the repetition of the box motif. The double box does exhibit a much stronger binding affinity, and the affinity should scale quadratically with the number of binding sites [145-147]. The discrepancy for the overall induced bending angles is particularly unexpected, as a structure containing two identical boxes might reasonably be expected to induce a much greater angle than the single box motif. Thus it is not correct to assume that the double box protein will simply exhibit the characteristics of two consecutive sites. The presence of the flexible linker may explain part of the difference, hindering the effective binding of box boxes simultaneously. Furthermore, at least one of the boxes must be disrupted to melt DNA, while DNA in the presence of the single box motif may be melted without protein dissociation until higher concentrations are reached. The fact that the single box motif must be dissociated in order to melt DNA only at high concentrations suggests that protein–protein interactions must be disrupted at high concentrations, in addition to protein-DNA interactions. Still higher concentrations have even been shown to induce filament formation in HMG [115, 148] and other proteins as well [117]. Thus, though both proteins stabilize dsDNA while enhancing local flexibility, HMGB1(box A+B) appears better suited toward the former while the single box motif of HMGB2(box A) appears to induce stronger bending.
3.3 The bacterial nucleoid: Comparisons to eukaryotes
In comparison to the strong organization of chromatin found in eukaryotes, in E. coli DNA is compacted into the nucleoid, a loose structure that is still poorly understood. Proteins that participate in DNA organization include a factor for inversion stimulation (FIS), histone-like nucleoid structuring protein (H-NS), integration host factor (IHF), and histone-like HU. While the roles of these proteins in bacteria appear to be analogous to those played by histones and HMGB proteins in eukaryotes, the structures of the bacterial nucleoid proteins share no similarities with the nuclear proteins of eukaryotes. Though these proteins (~100 amino acids) involved in DNA condensation have been identified, their exact function and relationships to each other are not well-understood. Additionally, many of the same proteins responsible for bacterial DNA condensation also function as transcription factors and possibly as part of other nucleoid processes [149-150]. Furthermore, the binding properties and functions of these E. coli proteins appear to overlap significantly, though subtle differences are known [149, 151-152]. FIS, IHF and H-NS appear to favor AT-rich segments, while IHF and FIS show some sequence specificity. HU, in contrast, appears to bind sequence-nonspecifically, but favors bent and damaged DNA [153]. It seems likely that experiments involving combinations of these proteins will be needed to fully elucidate their function. Yet single molecule experiments have given interesting insights into the behavior of these proteins, which present interesting comparisons with their eukaryotic counterparts.
3.3.1 IHF binds with sequence specificity
The sequence-specific IHF protein is a heterodimer of α-helices and β-sheets that bend dsDNA in two distinct kinks, producing an overall bend angle of nearly 180° [152, 154]. Though the protein has some non-specific DNA binding affinity, certain sequences are favored, possibly due to greater DNA flexibility in these regions [152]. Magnetic tweezers experiments have shown that IHF binding does serve to compact DNA, and that binding is quickly disrupted by even small increases in the tension along the DNA molecule [155]. While the structure of IHF appears similar to the histone-like HU protein, the proteins are not interchangeable, as both must be present to permit transcription [156]. Yet the specificity of IHF binding biases sites to non-coding regions of the bacterial genome, suggesting a regulatory function [149]. While the role of IHF in transcription is not certain, these results suggest that IHF serves a particular function, possibly facilitating loop formation necessary to the assembly of several structures during initiation [152].
3.3.2 HU and non-specific binding
The E. coli protein HU is another nuclear protein associated with binding to both double- and single-stranded DNA, and is often considered the bacterial analogue to histone-like proteins found in eukaryotes. HU resembles IHF in the appearance of an β-helical body capped by E-ribbon arms that bind into the minor groove of dsDNA [152-153]. HU binding produces a sharp bend in dsDNA, similarly to IHF, inducing bend angles between 105° and 140° [153-154]. However, unlike IHF, HU appears to show no sequence specificity. HU also binds strongly to bent or damaged (nicked) DNA, though binding to undistorted DNA has also been observed [117]. Much of the experimental data collected so far is on specifically damaged structures, which facilitates binding. This technique, commonly used to overcome the weak binding associated with non-specific binding proteins (including many X-ray and NMR structures), imposes important qualifiers on the known properties of HU.
Force spectroscopy experiments on HU bound undamaged DNA have combined magnetic tweezers studies and AFM imaging [117]. Both experimental approaches observe a change in the curvature of dsDNA at low protein concentrations below 100 nM. This is consistent with the role of enhancing DNA flexibility, also seen with the eukaryotic homologue HMG, as described above. While the measured persistence decreases, the distribution of measured angles is isotropic, in contrast to the peaked distributions for HMGB proteins. Thus HU appears to bind almost purely as a flexible hinge, inducing a highly variable bending angle when singly bound to DNA. At higher concentrations, optical tweezers experiments show an increase in the persistence length to over 100 pN, while AFM images show that the protein forms a fully rigid filament around DNA, an effect also seen with HMGB proteins as described above [148, 157]. Cooperative binding has been implicated as a way that HU may stabilize DNA. In this mode DNA is compacted in a histone-like manner [158-159]. Increases in the concentration of HU with respect to the concentrations of IHF have also been shown to inhibit the binding of the latter, as the cooperative binding mechanism apparently excludes IHF binding [156]. Thus an increase in concentration triggers a dramatic change in the interaction of HU with DNA, from a protein that may promote the flexibility of DNA in conjunction with other binding factors, to a protein that stabilizes and condenses dsDNA, excluding other proteins. Strikingly, the function of HU is significantly different from the function of IHF, though the structures bear strong similarities.
3.3.3 Non-specific H-NS binding dynamically bridges DNA
While HU influences higher order bacterial nucleoid structure by inducing changes in the flexibility of DNA, H-NS has been observed to directly bind two distinct DNA molecules together, forming a DNA-H-NS-DNA bridge. A novel assay tethers a pair of DNA molecules between four individually trapped beads, allowing the ends of each molecule to be separately manipulated. H-NS binds in multiple places between the two strands. Pulling on one of the free ends disrupts the bridges along the DNA, revealing the binding strength and the typical lengths between adjacent binding sites [160]. The results suggest an array of H-NS bridges that must serve to stabilize DNA and inhibit access by transcription factors. This regulatory role has been suggested for IHF and FIS as well [149]. However, the relatively low forces required to unbind these proteins suggest that that these structures may easily be overcome by other proteins or motors [151, 160]. Dynamic loop formation has also been observed in single DNA stretching experiments and visualized in AFM images, suggesting a model of flexible linkage that dynamically condenses the nucleoid, but does not deform bacterial DNA [161].
4 SINGLE-STRANDED DNA BINDING PROTEINS
The double-stranded, helical nature of duplex DNA protects and stabilizes the individual nucleic acid strands. However, these duplexes must melt into single-stranded DNA for cellular processes such as DNA replication, repair, and transcription, leaving exposed segments of ssDNA vulnerable to damage and degradation. Single-stranded binding proteins (SSBs) bind ssDNA to shield it from nucleases, prevent chemical degradation, and inhibit stem-loop and other secondary structure formation [162-163]. Furthermore, SSBs associate with cellular genome maintenance proteins such as polymerases, primases, helicases, and exonucleases, and may actively stimulate DNA replication, recombination, and repair through these interactions [164].
Organisms as diverse as viruses, bacteria, and mammals require SSBs for DNA replication [165]. SSBs therefore vary greatly in size, complexity, and binding mechanism. Bacteriophage SSBs bind DNA as monomers [166], bacterial SSBs are tetramers or dimers [167] with one known exception [168], and the eukaryotic SSB Replication Protein A (RPA) is a heterotrimer [169]. Despite this variation, the oligonucleotide/oligosaccharide binding (OB) fold [170] that binds ssDNA is conserved across SSBs in all domains of life [171]. Bacteriophages have among the simplest SSBs, with only one OB fold per monomer [163]. However, characterization of their DNA binding mechanisms has been relatively recent.
4.1 Bacteriophage SSB proteins
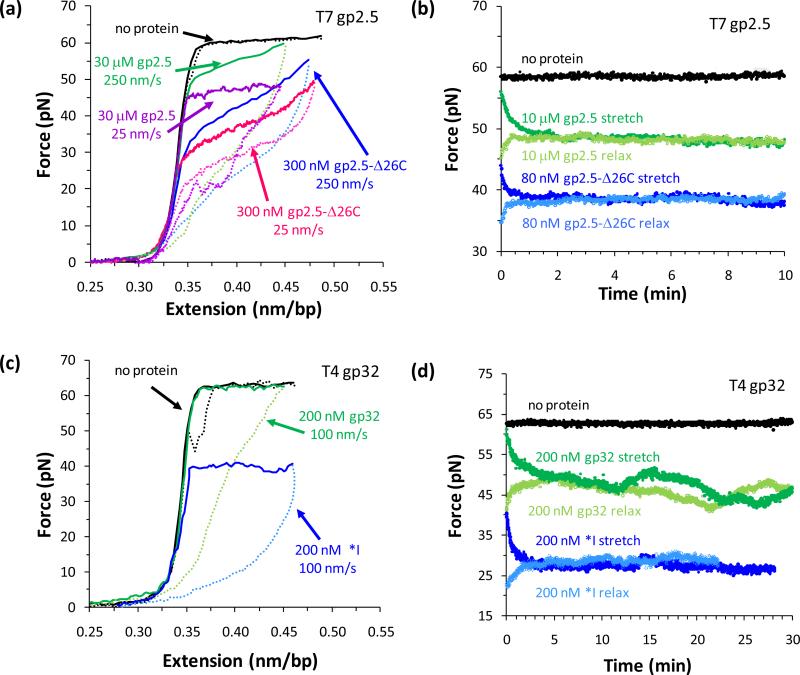
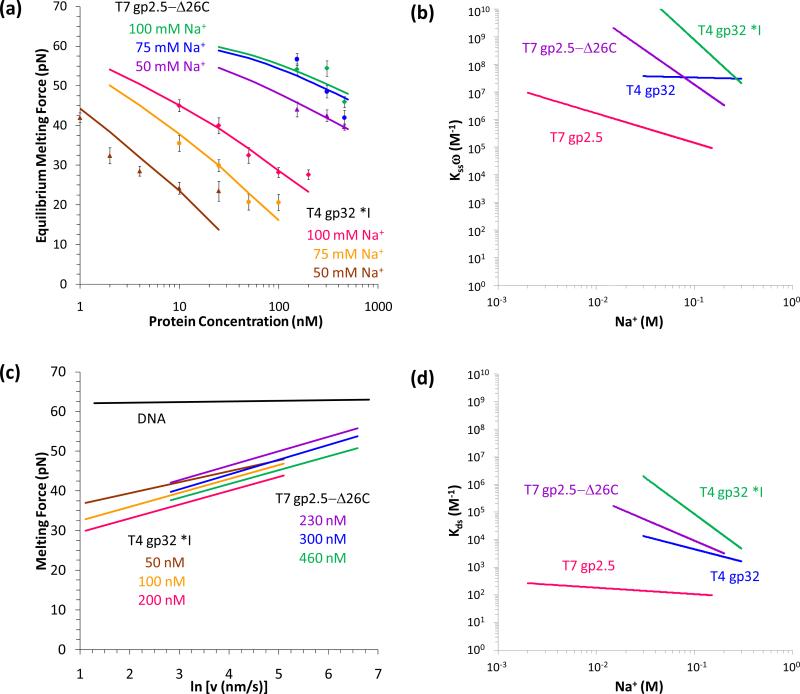
Although bacteriophage replication systems are well-studied models which have provided fundamental insights into the elaborate DNA replication process, the principal roles and interactions of each protein at the coordinated replication fork are not yet well-understood [172]. DNA replication in bacteriophage T7 is one of the simplest model systems, with a replisome comprised of T7 DNA polymerase, T7 helicase-primase, and SSB gene protein 2.5 (gp2.5). gp2.5 has 232 residues and preferentially binds ssDNA with a single OB fold in the core [166]. The protein forms a dimer in solution due to electrostatic binding of the highly acidic 26-residue C-terminus with the DNA binding site of another monomer, an interaction which occurs in the absence of DNA [166, 173-174]. gp2.5-Δ26C lacks this C-terminal tail, which is essential to gp2.5 interactions with the T7 DNA polymerase and helicase-primase at the replication fork [173, 175-176]. In addition to its predominant function as an SSB, gp2.5 facilitates complementary DNA strand annealing [177] and homologous base pairing [174, 177] more efficiently than SSBs from more complex replication systems such as those of bacteriophage T4 or E. coli [177-180].
Bacteriophage T4 is another excellent model replication system, and T7 gene 32 protein (gp32) is the most well-studied SSB. gp32 is a monomer which preferentially binds ssDNA and destabilizes the DNA duplex [164, 178]. The DNA binding site containing an OB fold is in the core of gp32 (residues 22-253), while the N-terminal domain (NTD, residues 1-21) is essential for highly cooperative binding to ssDNA, and the acidic C-terminal domain (CTD, residues 254-301) regulates DNA binding affinity and interacts with other replication proteins [181-183]. The C-terminal domain is absent in the mutant *I, which binds ssDNA with greater affinity than gp32. The preferential ssDNA binding revealed in bulk studies indicated that the dsDNA melting temperature (Tm) would decrease in the presence of both gp32 and *I, but thermal melting experiments measured this activity only for *I [178, 184-185]. Single molecule methods quantified the duplex destabilization activity of gp32 and *I, explaining these inconsistent DNA binding and thermal melting results [19-21].
4.1.1 T7 gp2.5 and T4 gp32 preferentially bind ssDNA
A single DNA molecule stretched in the presence of single-stranded binding proteins exhibits a decrease in melting force. Figure 11 shows representative DNA stretching curves in the presence of T7 gp2.5 and its C-terminal mutant gp2.5-Δ26C (a),along with T4 gp32 and its C-terminal truncate *I (c). All four SSB proteins cause a pulling rate-dependent decrease in DNA melting force and strong hysteresis. These two features of the DNA stretching curves indicate that the experimental conditions are non-equilibrium. That is, typical DNA extension velocities induce DNA melting on a faster timescale than protein association and dissociation occur, so the process is irreversible and strong hysteresis is observed. The measured melting force (Fk) at pulling rate v is therefore not the equilibrium melting force (Fm). Although reducing the pulling rate sufficiently to achieve equilibrium conditions such that Fk converges to Fm is unfeasible, time-dependent force measurements allow measurement of Fm [19, 21, 23]. When the DNA molecule is stretched to a fixed position at the midpoint of the melting transition, the force measured as a function of time decreases exponentially until it converges to the equilibrium melting force Fm (Figure 11b, d). This behavior reflects an increase in DNA melting on the timescale required for additional SSB proteins to bind and reach equilibrium [26]. If the DNA molecule is instead held fixed at the midpoint during relaxation, the observed force increases until it converges to Fm, corresponding to protein dissociation and subsequent DNA reannealing.
Figure 11.
Equilibrium melting forces for DNA with bacteriophage SSBs T7 gp2.5 (left) and T4 gp32 (right). (a) DNA stretching (solid line) and relaxation (dotted line) curves in the absence of protein (black) and in the presence of 30 μM T7 gp2.5 at DNA pulling rates of 250 nm/s (green) and 25 nm/s (purple). Force-extension curves in the presence of the C-terminal truncate 300 nM T7 gp2.5-Δ26C shown at DNA pulling rates of 25 nm/s (blue) and 250 nm/s (pink). Solid lines with dark colors show stretching data, and dotted lines with light colors represent relaxation curves. (b) Force as a function of time at extensions fixed during stretching (filled circles) and relaxation (open circles). Data in the absence of protein is shown in black, data in the presence of 10 μM T7 gp2.5 is green, and data in the presence of 80 nM gp2.5-Δ26C is blue. (c) DNA stretching (solid line) and relaxation (dotted line) curves in the absence of protein (black) and in the presence of 200 nM T4 gp32 (green) and its CTD deletion mutant *I (blue) at a DNA pulling rate of 100 nm/s. (d) Time-dependent force measured at fixed extension in the absence of protein (black) and in the presence of 200 nM gp32 (green filled circles) or 200 nM *I (blue filled circles). Forces measured at a fixed position during relaxation are in light colored open circles. Figure is adapted from [26].
T4 gp32 does not appear to lower the melting force at the DNA extension velocity 100 nm/s, while *I significantly lowers the melting force under identical experimental conditions (Figure 11c), an observation that is consistent with the results of bulk thermal melting experiments [184-185]. However, time-dependent force measurements reveal that the equilibrium melting force Fm is lower in the presence of gp32 relative to DNA without protein (Figure 11d), demonstrating that gp32 does have the duplex destabilization activity which was expected but not observed in thermal melting experiments [184-185]. gp32 leads to a significantly smaller decrease in Fm than *I, which can be explained by the requirement for a conformation change prior to DNA binding, described below [19-22, 26].
In a method similar to melting temperature Tm analysis, the equilibrium melting force Fm measured as a function of protein concentration c behaves according the thermodynamic model [23]:
| (13) |
where Fm0 is the equilibrium melting force for DNA without protein, Δx is the force-dependent length difference between ssDNA and dsDNA, and ω is the cooperativity parameter. T7 gp32 binds ssDNA with high cooperativity (ω ≈ 103), while T4 gp2.5 binds does not bind ssDNA cooperatively (ω ≈ 1) [174, 186-188] . Fits of Equation (13) to experimental data (Figure 12a) yield Kss, the ssDNA equilibrium binding constant, and nss, the protein binding site size. The binding site size for all four SSB proteins is ~7 nucleotides (nt), which agrees with bulk biochemical measurements [174, 184].
Figure 12.
Equilibrium DNA binding for T7 gp2.5 and T4 gp32 and their CTD truncates as a function of salt concentration. (a) Equilibrium melting force as a function of protein concentration for mutants T4 gp32 *I (green, blue, and purple,) and T7 gp2.5-Δ26C (pink, orange, and brown) in 100 mM, 75 mM and 50 mM Na+. Fits to Equation (13) are shown as solid lines. (b) Equilibrium association binding constants to ssDNA for mutants T4 gp32 *I (green) and T7 gp2.5-Δ26C (green), along with wild type SSBs T4 gp32 (red) and T7 gp2.5 (blue). (c) DNA melting force is only weakly dependant on pulling rate in the absence of protein (black). Fits to Equation (14) of DNA melting force as a function of pulling rate in the presence of CTD truncates T4 gp32 *I and T7 gp2.5-Δ26C at various protein concentration. (d) Equilibrium association binding constant to dsDNA as a function of protein concentration for mutants T4 gp32 *I (green) and T7 gp2.5-Δ26C (green), along with wild type SSBs T4 gp32 (red) and T7 gp2.5 (blue). Figure is reproduced with permission from [1].
Figure 12b shows the cooperative equilibrium binding constants Kssω obtained from fits to Equation (13) as a function of salt concentration for each SSB protein and its mutant. Kssω is significantly higher and more salt-dependent for gp2.5-Δ26C and *I than the wild type SSB proteins [19-21, 23, 25]. This increase in ssDNA binding affinity of the C-terminal truncated mutants agrees with bulk measurements [26], and is likely due to the highly acidic C-terminus occupying the cationic DNA binding site in both wild type proteins, such that DNA binding requires its removal. The DNA binding site of gp32 interacts with the CTD of the same monomer, while the gp2.5 C-terminus interacts with the DNA binding site of a different monomer to facilitate dimerization. Lack of a C-terminus exposes the DNA binding site of each mutant, facilitating ssDNA binding. The stronger salt dependence of the ssDNA equilibrium binding constants reflects the release of cations into the solution following binding by the C-terminal truncates, while in the presence of the C-terminus cations released upon DNA binding are subsequently bound by the C-terminus, such that the overall cation release is much smaller for the wild type SSBs.
4.1.2 T7 gp2.5 and T4 gp32 slide on dsDNA in search of exposed ssDNA
The strong hysteresis indicating irreversible protein binding (Figure 11a, c) revealed the non-equilibrium nature of typical DNA stretching experiments in the presence of SSB proteins, leading to time-dependent force measurements designed to probe the equilibrium melting force Fm for characterization of ssDNA binding. The non-equilibrium experiments, utilizing the pulling rate-dependence of the measured melting force Fk, also provide insight into protein binding kinetics [20-21, 25]. Application of force deceases the free energy of base pair opening, which corresponds to an increase in the rate at which fluctuations expose the number of nucleotides required for protein binding. In this model, the melting transition Fk(v) occurs when velocity of DNA extension v is equal to the base pair opening rate [20]:
| (14) |
where ka is the rate at which proteins encounter a binding site of length nss, and the factor of 2 accounts for the probability that both ends of the DNA molecule may have the appropriate number of exposed nucleotides. Figure 12c illustrates the linear dependence of Fk(v) on ln(v), which yields the same nss value as that obtained from equilibrium force measurements (Figure 12a and Equation (13)).
The condition Fk(v) = Fm0 yields ka, the rate at which proteins find ssDNA binding sites, which depends upon the square of protein concentration and exceeds the 3D diffusion limit. This remarkable result indicates that these SSB proteins do not need to diffuse from bulk solution to encounter their ssDNA binding sites. They instead bind weakly to dsDNA and diffuse along the DNA molecule until they reach exposed ssDNA [20, 25].
In the 1D sliding model, the sliding rate ks, which involves the one-dimensional diffusion constant of the protein along DNA, combines with the dsDNA binding site size nds and DNA lattice occupancy γds to describe the protein binding rate ka [20]:
| (15) |
which depends upon the square of the protein concentration, since the McGhee-von Hippel binding isotherm determines the fractional dsDNA lattice saturation γds (Equation (4)).
Theoretical calculations of the first passage time of proteins diffusing in one dimension confirm this experimental observation [189-190]. Fits to Equations (15) and (4) yield the equilibrium binding constant for dsDNA Kds, the dsDNA binding site size nds, and the sliding rate ks. Kds, shown in Figure 12d as a function of salt concentration, is four orders of magnitude weaker than Kssω (Figure 12b) for all four proteins [20-21, 25], which reveals similar equilibrium ssDNA destabilization capability for wild type SSBs and their C-terminal truncated mutants.
Although binding affinity is ~104 times stronger for ssDNA than dsDNA in both bacteriophage SSB proteins, the mechanism for ssDNA binding affinity is different for each. gp2.5 binds with negligible cooperativity, so ω ≈ 1 [174] and thus Kssω ≈ Kss, indicating that preferential ssDNA binding is due to non-electrostatic interactions such as aromatic residues stacking with DNA bases. In contrast, gp32 binds highly cooperatively to ssDNA only, with ω ≈ 103 [186-188] . Thus interactions with gp32 monomers bound in adjacent positions are predominantly responsible for preferential ssDNA binding, and non-electrostatic binding to ssDNA only contributes one order of magnitude to the 10,000-fold stronger affinity for ssDNA than dsDNA.
4.1.3 DNA binding mechanisms of bacteriophage SSB proteins
SSB proteins from both bacteriophage T7 and T4 have cationic DNA binding sites with an OB-fold consisting of aromatic residues, which participate in base stacking [166, 181-183], and a highly acidic C-terminus required for DNA replication [173, 175, 178]. The salt-dependence of equilibrium binding constants for wild type SSBs and their C-terminal truncates (Figure 12b, d) supports a model in which the acidic C-terminus interacts with the cationic DNA binding site in low salt, reducing the ability of ~7 nt of ssDNA to occupy the binding site [21, 23]. A quantitative model in terms of counterion condensation describes ssDNA interactions of both T7 gp2.5 [23] and T4 gp32 [22].
Single molecule experiments reveal that both SSBs also weakly bind dsDNA in order to slide in one dimension until they locate ssDNA, for which they have ~104 times greater affinity. This 1D diffusion along dsDNA is likely to be an integral binding mechanism for SSBs because the presence of dsDNA in vivo dominates that of ssDNA. The formation of ssDNA tends to be transient, and thus the probability of SSBs encountering their binding sites through diffusion from solution is significantly lower than that of sliding along dsDNA. Sequence-specific dsDNA binding proteins employ a similar search mechanism, in which they bind dsDNA weakly and diffuse in 1D until they encounter their particular dsDNA binding sites [191-199]. Recent optical tweezers experiments explored complex mechanisms of facilitated diffusion along dsDNA, demonstrating intersegmental jumping on the DNA molecule with EcoRV [200], a restriction enzyme for which a coiled DNA conformation enhances dsDNA target search efficiency [200], and thus protein association rate depends upon tension applied to the template strand [201].
T7 gp2.5 and T4 gp32 share comparable 1D diffusion search mechanisms, C-terminal interactions with their DNA binding sites, and ssDNA destabilization properties, characteristics which reflect their similar functions in bacteriophage replication. T7 gp2.5 activity matches that of T4 gp32 in DNA replication, a parity which was unclear because T7 gp2.5 binds ssDNA with ~102 times weaker affinity and without cooperativity. Single-molecule stretching studies reveal that gp2.5 binding affinity for dsDNA is also ~102 times weaker than that of gp32, and thus preferential binding to ssDNA is approximately the same for both proteins. That is, the ssDNA stabilization activity of both SSBs depends upon binding affinity for ssDNA relative to that for dsDNA. Non-electrostatic interactions such as aromatic residues of gp2.5 stacking with exposed bases confer sufficient ssDNA binding affinity to achieve ~104-fold preference over dsDNA. In contrast, the non-electrostatic interactions of gp32 with ssDNA are ~10 times weaker than those of gp2.5, and dsDNA affinity is ~102 times stronger. gp32 therefore binds ssDNA cooperatively, and interactions between adjacently-bound monomers provide the factor of 103 required to reach the same level preferential ssDNA binding as that of gp2.5. Thus T7 gp2.5 function in bacteriophage replication is as effective as that of T4 gp32 despite weaker, non-cooperative ssDNA binding [26].
Furthermore, this overall weaker ssDNA interaction allow faster dissociation from ssDNA, and therefore DNA stretching curves in the presence of gp2.5 exhibit smaller hysteresis at low forces than those with gp32 (Figure 11a, c). This rapid dissociation kinetics may be responsible for the ability of T7 gp2.5 to stabilize ssDNA for its SSB function and facilitate processes which require dsDNA stabilization, such as strand annealing [177] and homologous base pairing [174, 177]. Nucleic acid rearrangements require removal of protein bound to ssDNA, and faster dissociation from ssDNA relative to gp32 may allow gp2.5 to promote stand annealing more efficiently than gp32. Although gp32 binds ssDNA highly cooperatively with significantly slower dissociation kinetics, T4 recombination mediator protein UvsY destabilizes gp32-ssDNA complexes and stabilizes dsDNA, facilitating the nucleic acid rearrangements required in recombination [24].
Single molecule DNA stretching experiments provide a comprehensive description of T7 and T4 SSB interactions with both forms of DNA, leading to quantitative models of bacteriophage SSB binding mechanisms which are consistent with ensemble biochemical results. These models allow characterization of dsDNA binding affinity as well as ssDNA binding kinetics and thermodynamics. Further analysis also reveals the details of the mechanisms that may regulate binding of SSBs to both forms of dsDNA in vivo.
4.2 Bacterial SSB proteins
Although SSBs of bacteria are more complex than those of bacteriophages, they remain among the simpler SSBs, also composed of a single polypeptide with one or two OB folds [163]. Similar to bacteriophage SSBs, the SSB of Sulfolobus solfataricus binds DNA as a monomer [168]. However, all other known bacterial SSBs function as tetramers or dimers [167]. E. coli SSB is the most typical example of a bacterial SSB, and its role in bacterial DNA replication is relatively well-understood [163]. However, recent single molecule experiments clarify the DNA binding conformations of this well-studied ssDNA binding protein.
4.2.1 E. coli SSB binding modes to ssDNA are salt-dependent
The single-stranded binding protein of E. coli is a homotetramer, with one OB fold per monomer [202]. The SSB has multiple binding modes which depend highly on salt concentration [167, 203-208], and DNA stretching experiments with magnetic tweezers measured the ssDNA stabilization activity of E. coli SSB [209-210], demonstrating its salt-dependence [210]. In high salt solution conditions, E. coli SSB tetramers bind ~65 nt of ssDNA with low cooperativity, a mode known as (SSB)65 [167, 202]. In low salt, the SSB tetramers bind highly cooperatively (ω ≈ 102) [208, 211] with a binding site size of ~35 nt in the (SSB)35 configuration [211]. A recent single molecule study quantifies the dynamics of the transition between these modes and reveals a third binding mode, referred to as (SSB)35b, which also occludes 35 nt and arises from structural rearrangement of the (SSB)35 complex [212]. Although E. coli SSB is relatively well-studied [167], recent single molecule experiments quantify transitions between DNA binding modes and provide insight into transient ssDNA-protein conformations in solution.
4.2.2 B. subtilis DnaD preferentially binds ssDNA
B. subtilis DnaD is a key protein in bacterial DNA replication [213-221] that binds both ssDNA and dsDNA [218, 220, 222], although its function remains unknown [3]. The N-terminal domain (NTD) of DnaD is involved in interactions between monomers which form scaffolds responsible for DNA looping, while the C-terminal domain (CTD) includes the DNA binding domain [220]. Single molecule AFM experiments reveal that DnaD increases the contour length of dsDNA and preferentially binds ssDNA [3]. DNA stretching curves in the presence of DnaD exhibit hysteresis characteristic of SSBs (Figure 11a, c), indicating slow dissociation from ssDNA [3]. The results also show significant dsDNA aggregation in the presence of DnaD, an effect which is consistent with protein-induced DNA looping from scaffolding [218-219]. Although the NTD is responsible for this scaffolding in the absence of DNA [220], DNA stretching curves in presence of the NTD do not exhibit DNA aggregation or evidence of significant protein binding affinity [3]. In contrast, the substantial hysteresis in the presence of the CTD indicates the domain preferentially binds ssDNA and does not dissociate on the timescale of relaxation, and the increase in dsDNA contour length reflects dsDNA binding activity. Similar to the NTD, however, the CTD does not induce the dsDNA aggregation evident with full-length DnaD [3]. DNA stretching curves in the presence of both NTD and CTD also do not show dsDNA aggregation, which is consistent with lack of DNA scaffolding ability for the separated domains [220]. Single molecule stretching experiments reveal the preferential ssDNA binding of DnaD, which may be relevant in transcription initiation, such as interaction with DnaA to facilitate loading of the helicase DnaC [3], or indicate another function for the protein, elucidating its role in bacterial DNA replication.
5 PROTEINS THAT BIND BOTH DOUBLE AND SINGLE-STRANDED DNA
A key cellular process for genetic rearrangements during DNA replication and synthesis is homologous recombination, in which nucleic acid strand exchange occurs between similar molecules. Homologous recombination is the most frequent type of recombination in retroviruses and retrotransposons [223], which carry two genomic RNA molecules [224-226]. Retroviral recombination occurs predominantly between these two RNA strands during strand transfer events [223], which require nucleic acid rearrangements. Homologous recombination in more complex systems also requires nucleic acid strand exchange. Thus homologous recombination and DNA replication require proteins such as nucleic acid chaperones, DNA recombinases, and DNA polymerases, which bind both single- and double-stranded DNA in order to facilitate these processes. A comparable affinity for both forms of DNA results in complex binding properties, and single molecule force spectroscopy methods explore the DNA binding mechanisms of these proteins [1, 12].
5.1 Nucleic acid chaperones in retroviruses and retrotransposons
Retroviruses and retrotransposons store genetic information in a linear RNA strand, and their DNA polymerase reverse transcriptase converts the RNA template into dsDNA which will be integrated into cellular genomic DNA. Reverse transcription includes multiple stages which involve rearrangement of nucleic acid secondary structure. These processes require nucleic acid chaperone proteins, which aggregate nucleic acids, destabilize base pairing, and promote complementary strand annealing. The chaperone activity of these proteins requires strong binding affinity for both single- and double-stranded nucleic acids, a property that is not yet well-understood.
5.1.1 Retroviral nucleocapsid proteins
Nucleocapsid (NC) proteins are the nucleic acid chaperones in retroviruses. NC proteins tend to be small cationic proteins with minimal structure other than zinc-stabilized CCHC finger motifs [227-230]. Retroviral reverse transcription requires NC for primer annealing, strand transfer, and other instances of secondary structure rearrangement [223]. NC facilitates the annealing of a transfer RNA (tRNA) primer to the primer binding site (PBS) on a positive strand of viral genomic RNA [231-233], a process which involves destabilization of RNA base pairing. Reverse transcriptase then polymerizes the minus strand of complementary DNA (cDNA) as its RNaseH domain degrades the RNA template, proceeding through the long-terminal repeat (LTR) region at the end of the genomic RNA strand. Reverse transcription of the remaining viral RNA requires minus strand transfer, in which the newly synthesized cDNA must anneal to the LTR on the opposite end of the RNA genome. However, secondary structures within the cDNA and the LTR RNA are thermodynamically stable, and thus the energy barrier to strand annealing makes duplex formation unlikely. NC destabilizes these secondary structures and facilitates annealing of the complementary DNA-RNA duplex [234-236]. A second strand transfer, where the plus DNA strand anneals to the PBS region on the opposite end of the minus DNA strand, is also inefficient without NC [234-236]. The nucleic acid chaperone activity of NC may also be essential for genomic RNA dimerization during retroviral maturation [237-239].
In addition to its nucleic acid chaperone properties, NC also participates in genomic RNA packaging for retroviral assembly, during which it is a domain of the Gag polyprotein [224-226, 240], and viral dsDNA integration [241-242]. Multiple stages of the HIV-1 life cycle require NC, and antiviral drugs targeted its zinc fingers [144, 243-244] until non-specific toxicity concerns became clear [245]. The protein has little known structure other than these zinc fingers, and thus further drug targeting requires a detailed understanding of its function. Single molecule DNA stretching experiments have characterized the nucleic acid chaperone activity of several retroviral NC proteins [143, 246-248].
5.1.2 HIV-1 NC destabilizes dsDNA and dissociates rapidly from ssDNA
Human immunodeficiency virus type 1 (HIV-1) NC is a 55-residue protein with two zinc fingers and a basic N-terminal tail [249-251]. Single molecule DNA stretching experiments reveal that the nucleic acid chaperone aggregates DNA, destabilizes dsDNA, and facilitates duplex reannealing [143, 246, 252-253]. The presence of 10 nM HIV-1 NC reduces the free energy of DNA melting from ~2kBT (Figure 13a) to ~1kBT (Figure 13b) [252-253], destabilizing the DNA duplex. The increase in the width of the melting transition (ΔFm) also suggests weak duplex destabilization, and positively correlates with chaperone activity [18, 246, 253]. The DNA stretching and relaxation curves in the presence of HIV-1 NC (Figure 13c) do not exhibit the strong hysteresis that is characteristic of SSBs (Figure 11a, c). SSBs bind ssDNA and dissociate slowly, such that the DNA molecule cannot reanneal on the timescale of the relaxation cycle. In contrast, HIV-1 NC (or NCp7) rapidly dissociates from ssDNA, allowing the two strands to reanneal and form the DNA duplex [143, 246]. However, the NC precursor products Gag, NCp15, and NCp9 which have less chaperone activity than NCp7 induce more hysteresis, implying slower DNA binding kinetics [246]. Rapid kinetics of nucleic acid binding is an essential component of HIV-1 NC chaperone activity, and it requires a delicate balance between ssDNA binding and dissociation [143].
Figure 13.
HIV-1 NC lowers the free energy of DNA melting ΔG. (a) Force-extension curve of dsDNA (solid line), and the FJC model (Equation (2)) for ssDNA stretching (dotted line, Springer chapter ref 56). (b) Stretching curves of dsDNA (solid line) and ssDNA (dotted line, [253]) in the presence of 7 nM HIV-1 NC. (c) Stretching curves of dsDNA extension (solid lines) and relaxation (dotted lines) in the absence of protein (black) and in the presence of 10 nM HIV-1 NC (green) [258].
The nucleic acid interactions of HIV-1 NC involve aromatic residues in the zinc fingers which stack with ssDNA bases [254-255]. Interchanging the order of the zinc fingers forms mutant 2-1, which has significantly lower DNA binding affinity and lacks the rapid kinetics of wild type HIV-1 NC [143]. In mutant 1-1, a copy of the first zinc finger replaces the second zinc finger, also leading to lower binding affinity and slower interaction kinetics than HIV-1 NC [143]. However, the DNA binding and dissociation kinetics of this mutant are the least compromised among those studied, and it retains some ability to facilitate minus strand transfer [256]. Thus rapid kinetics relies on specific zinc finger architecture, both of which are crucial for the chaperone function of HIV-1 NC.
5.1.3 Rapid kinetics of retroviral NCs correspond to efficient nucleic acid chaperone activity
Single molecule stretching experiments have also characterized DNA binding of NC proteins from other retroviruses, elucidating the role of rapid kinetics in nucleic acid chaperone activity. NC proteins from Moloney murine leukemia virus (MLV), Rous sarcoma virus (RSV), and human T-cell lymphotropic virus type 1 (HTLV-1) differ predominantly in ssDNA binding affinities [247]. DNA stretching curves in the presence of HIV-1 NC (Figure 13c) and RSV NC both exhibit minimal hysteresis, which indicates rapid DNA binding kinetics for both proteins [247]. The clearest disparity between them is the effect on the DNA melting force. HIV-1 NC lowers the melting force, reflecting duplex destabilization, while RSV NC increases the melting force, suggesting duplex stabilization. This duplex stabilization activity of RSV NC is unique among the retroviral NC proteins examined. MLV NC and HTLV-1 NC (Figure 14a) both lower the DNA melting force, the magnitude of which implies moderate duplex destabilization activity [247]. However, DNA stretching curves in the presence of these two NC proteins exhibit significant hysteresis, reflecting their relatively slow dissociation from ssDNA. Furthermore, HTLV-1 is unable to aggregate nucleic acids, while the remaining retroviral NC proteins have similar strong aggregation abilities [247].
Figure 14.
The CTD of HTLV-1 NC regulates DNA dissociation kinetics. (a) DNA extension (solid line) and relaxation (dotted line) curves in the absence of protein (black) and in the presence of 700 nM HTLV-1 NC wild type (green). (b) DNA extension (solid line) and relaxation (dotted line) curves in the absence of protein (black) and in the presence of 200 nM ΔC29 HTLV-1 NC. (c) Force as a function of time at fixed extension during DNA relaxation (open circles) in the presence of 700 nM wild type HTLV NC (green) and 200 nM ΔC29 (blue), fit to single exponentials (solid black lines). Inset shows ΔC29 data on a shorter time scale. Figures adapted from [248].
These results reveal that nucleic acid chaperone activity correlates with rapid DNA interaction kinetics across a variety of retroviruses. HIV-1 NC and RSV NC are the most effective chaperones, MLV NC has moderate chaperone ability, and HTLV-1 is an extremely poor chaperone. This comparison suggests that nucleic acid chaperone activity may be related to thermodynamic stability of secondary structures formed in the LTR regions of the retroviral genomic RNA [247]. Only the complex secondary structure of the HTLV-1 LTR region is inconsistent with this observation [247], which suggests that the extremely slow dissociation rate of HTLV-1 NC may serve a specific biological function.
5.1.4 HTLV-1 NC binds cooperatively to ssDNA
Figure 14 shows representative DNA stretching curves in the presence of HTLV-1 NC (a) and ΔC29 HTLV-1 NC, a mutant lacking 26 residues of the acidic C-terminal (b). The significant hysteresis observed with the full-length protein is characteristic of SSBs (Figure 11a, c), while the lack of hysteresis with the C-terminal truncate resembles the behavior of nucleic acid chaperones such as HIV-1 NC (Figure 13c) and RSV NC. Time-dependent force measurements, in which the force is monitored as a function of time at fixed extension, reveal that wild type HTLV-1 NC lowers the equilibrium melting force Fm more than its CTD deletion mutant (Figure 14c). When the DNA molecule is held at the midpoint during relaxation, the force measured increases until it converges to Fm, which is consistent with protein dissociation from ssDNA. Equilibrium constants for DNA reannealing in the presence of the protein obtained from these force relaxation measurements indicate that wild type HTLV-1 NC dissociates from ssDNA an order of magnitude more slowly than its C-terminal truncate [248].
These results are consistent with a model in which the CTD regulates the chaperone activity of HTLV-1 NC. The strongly salt-dependent nature of the interaction between the anionic CTD and the cationic NTD (data in [248]) indicates that the CTD occupies the DNA binding site in low salt, reducing the DNA association rate of wild type HTLV-1 NC [248]. Lack of a CTD in the mutant leaves the NTD available for binding ssDNA and contributes to the on rate, an electrostatic mechanism of DNA binding regulation by the CTD that resembles the mechanism used by bacteriophage SSBs described above. Additionally, the off rate of ΔC29 HTLV-1 NC is significantly faster than that of the wild type protein [248]. This is consistent with a model in which HTLV-1 NC requires the CTD to bind ssDNA cooperatively [248]. The acidic CTD of each bound monomer interacts with the basic NTD of the adjacently bound monomer, and this cooperative interaction reduces the rate of dissociation from ssDNA [248]. In contrast, both T4 gp32 and its CTD truncate *I bind ssDNA cooperatively, which reflects the fact that cooperative binding does not require the gp32 CTD. Therefore the *I mutant has a higher on rate but the same off rate as wild type gp32, leading to an increase in ssDNA affinity. However, the ΔC29 HTLV-1 NC mutant has faster on and off rates than wild type HIV-1 NC, which results in rapid kinetics and thus confers efficient nucleic acid chaperone activity.
Cooperative binding to ssDNA is unexpected for a retroviral nucleocapsid protein such as that of HTLV-1, particularly since the resulting low off rate compromises its nucleic acid chaperone function. A biological function of more importance may require slow dissociation from ssDNA, superseding the need for efficient chaperone activity. Although that biological function remains unknown, one possibility is exclusion of human APOBEC3G proteins, for which HTLV-1 NC does not possess a degradation mechanism [257]. The rapid kinetics of HIV-1 NC allow APOBEC3G to bind ssRNA [258] and be co-packaged into HIV-1 virions [259-261]. In contrast, HTLV-1 virions have few APOBEC3G proteins [262-263], a reduction which requires the CTD of HTLV-1 NC [257]. It is possible that HTLV-1 NC binds ssRNA cooperatively to exclude binding of APOBEG3G [248]. Thus the slow ssDNA dissociation kinetics that reduce HTLV-1 NC chaperone activity may be responsible for reduced packaging of APOBEC3G proteins [248].
5.1.5 Target-site primed reverse transcription in the retrotransposon LINE-1 requires the ORF1 protein
Some retrotransposons, such as retroviruses, use LTR regions of genomic RNA for minus strand transfer during reverse transcription. Mobile genetic elements which also reproduce via an RNA intermediate but lack this repeat region are called non-LTR retrotransposons. Long interspersed nuclear element type 1 (LINE-1, or L1) is a non-LTR retrotransposon abundant in the human genome. L1 encodes an endonuclease, which cleaves cellular genomic DNA at the site of insertion. This DNA strand anneals to L1 genomic RNA and becomes the primer for reverse transcriptase, a process called target site-primed reverse transcription (TPRT) [264]. The L1 nucleic acid chaperone, open reading frame 1 protein (ORF1p), is essential for TPRT-based retrotranspositon, and it may destabilize the dsDNA duplex at the target site and facilitate annealing of the DNA-RNA hybrid [265]. The first cDNA strand transcribed from the RNA template is already joined to cellular DNA cleaved at the target site. The L1 endonuclease also cleaves the opposite strand of cellular dsDNA at the second target site, which anneals to the end of the first cDNA strand and serves as a primer for polymerization of the second cDNA strand [265]. This second strand exchange may also involve the nucleic acid chaperone activity of ORF1p, which may destabilize genomic dsDNA and stabilize the DNA-cDNA duplex [265].
5.1.6 ORF1p increases DNA melting transition width and aggregates ssDNA
The ORF1 protein from mouse L1 is 357 residues [266], which is significantly larger than NC proteins in LTR retroviruses and retrotransposons. L1 ORF1p also lacks the CCHC domain characteristic of NC proteins and ORF1 proteins from all other non-LTR retrotransposons [267]. The basic CTD contains the nucleic acid binding site, while the acidic NTD contains a 120-residue coiled-coil domain [268], which is required for the protein–protein interactions [266] that allow mouse L1 ORF1p to form a stable trimer [268]. Two arginine (R) residues in the basic CTD, at positions 297-298, are integral to nucleic acid binding and chaperone activity [269]. Single molecule DNA stretching experiments reveal that wild type ORF1p (RR) increases the transition width, or the change in force from the beginning to end of the DNA melting plateau, moderately relative to that for HIV-1 NC [269]. The transition width ΔFm reflects the cooperativity of the melting transition, and an increase in ΔFm correlates positively with nucleic acid chaperone activity [18, 246, 252-253]. The wild type RR protein also decreases the extension at which the ssDNA transition occurs, reflecting ssDNA aggregation [269]. That is, the protein induces effects that make ssDNA attracted to itself, decreasing the effective length of ssDNA at a particular force. Therefore moderate ssDNA aggregation may contribute to nucleic acid chaperone activity.
5.1.7 ORF1p mutants which induce strong DNA aggregation inhibit nucleic acid chaperone activity
Substituting alanine (A) for both arginine residues results in a mutant (AA) with minimal RNA binding affinity and no chaperone activity [269]. DNA stretching curves in the presence of the AA mutant reflect a significant decrease in DNA binding, minimal ssDNA aggregation, and almost no impact on the transition width ΔFm [269]. However, replacing one or both of the arginine residues with lysine (K) conserves charge, resulting in mutants (RK, KR, and KK) with nucleic acid binding affinity similar to that of wild type RR. The RK mutant shows a decrease in nucleic acid chaperone activity, and facilitates L1 retrotransposition less efficiently than wild type ORF1p. In contrast, the KR and KK mutants also show diminished nucleic acid chaperone activity and are unable to facilitate retrotransposition [269]. DNA stretching studies show that all three mutants increase ΔFm and aggregate ssDNA, reflecting some chaperone ability [269]. However, the mutants also significantly aggregate dsDNA, and KR and KK in particular form extremely stable protein-dsDNA aggregates at relatively low concentrations [269]. These results illustrate that mutations in ORF1p that result in severe nucleic acid aggregation inhibit chaperone activity.
The amino acid at position 159 of ORF1p is responsible for a 15-fold difference in retrotransposition efficiency in two variants of mouse L1, TFC and TFspa [270]. TFC ORF1p contains aspartic acid (D159), while the ORF1p of TFspa contains histidine (H159). Single molecule methods and bulk assays establish that D159 and H159 ORF1p have similar nucleic acid binding affinities [270]. However, D159 is a more efficient nucleic acid chaperone than H159 [270]. DNA stretching curves in the presence of D159 have a larger increase in transition width ΔFm than those with H159 [270], which reflects greater chaperone activity [18, 246, 252-253]. Although both variants of ORF1p induce similar hysteresis, indicating preferential binding to ssDNA, D159 aggregates ssDNA significantly less than H159. Smaller ssDNA aggregation, and thus faster dissociation from ssDNA, is consistent with bulk experiments which indicate D159 has more rapid nucleic acid binding kinetics than H159 [270]. This implies that ORF1 proteins possess optimal ssDNA aggregation properties to effectively direct nucleic acid rearrangements for L1 retrotransposition. Thus mutations that decrease ΔFm or induce strong DNA aggregation interfere with the precise balance of nucleic acid aggregation, duplex destabilization, and strand reannealing, which is characteristic of efficient nucleic acid chaperone activity.
5.2 DNA recombinases
Although nucleic acid chaperones mediate strand transfer in retroviruses and retrotransposons, strand exchange for homologous recombination in all three domains of life involves recombinase proteins [271]. Despite the sequence variation among recombinases RadA in archaea, RecA in bacteria [272], and Rad51 in eukaryotes, these proteins all assemble in similar filaments on both ssDNA and dsDNA [273]. Single molecule force spectroscopy methods elucidate the dynamics of protein-DNA filament formation [271].
5.2.1 Human recombinase Rad51 binds ssDNA and dsDNA dynamically
Human recombinase protein Rad51 assembles on DNA into helical nucleoprotein filaments. Scanning force microscopy images illustrate that Rad51 binds both ssDNA and dsDNA [271]. Monomers of Rad51 form irregular structures, suggesting dynamic assembly on ssDNA [271]. Single molecule experiments with magnetic tweezers probe dynamic filament formation, revealing the kinetics of Rad51 assembly and disassembly on dsDNA. In these experiments, the DNA molecule is tethered at a constant force, typically 1 pN [271]. As Rad51 binds along the length of the molecule for filament formation, the corresponding increase in extension is monitored as a function of time [271]. The exponential relaxation of the extension evokes time-dependent force measurements of equilibrium protein binding (Figure 11b, c). Conversely, Rad51 unbinding causes reformation of the dsDNA strand and subsequent decrease in extension as a function of time [271]. The exponential nature of these binding measurements suggests dynamic, simultaneous formation of filaments from multiple Rad51 association and dissociation sites along the DNA molecule.
Experiments which combine optical tweezers with fluorescence imaging show that Rad51 binds at multiple sites along a single DNA molecule [274]. Fluorescently-labeled Rad51 assembles onto dsDNA molecules, and the ends of a single filament are each tethered to a bead in an optical trap. Force-extension curves indicate a longer contour length and a shorter force-induced melting plateau for Rad51-coated dsDNA relative to bare DNA [274]. Fluorescent imaging shows that Rad51 coats the DNA molecule inhomogeneously. The force-extension curve of non-fluorescent regions of the dsDNA molecule, where Rad51 is not bound, is characteristic of bare dsDNA stretching curves. Regions of the molecule fully coated with Rad51 are contiguously fluorescent, with force-extension curves that exhibit a change in contour length and lack a force-induced melting transition. Partly-coated segments of the DNA molecule are intermittently fluorescent, and the stretching curves of this region exhibit a short melting plateau and a change in contour length, features which are consistent with a linear combination of rigid protein-bound segments and elastic bare DNA [274]. This elasticity analysis demonstrates that Rad51 binds DNA in a rigid, stable filament [274-275]. Scanning force microscopy and fluorescence experiments confirm that Rad51 nucleates at multiple sites on a single dsDNA molecule, forming tightly associated nucleoprotein filaments [276]. Recent work with optical tweezers and fluorescently-labeled Rad51 suggests that nucleoprotein filament disassembly proceeds from the ends, as ATP hydrolysis occurs on terminal Rad51 monomers [277]. Thus single molecule force spectroscopy provides insight into the binding activity of a protein essential for homologous recombination, a key process during chromosomal DNA replication.
5.3 DNA polymerases
Chromosomal DNA molecules are millions of base pairs in length, and the vast amount of information they carry must be replicated with high accuracy in order to sustain life. The enzymes which are largely responsible for this are known as DNA polymerases, and they are intricate complexes with multiple subunits [278-281]. The polymerase subunits of these holoenzymes synthesize a complementary dsDNA strand based upon an ssDNA template [278-281]. This catalytic activity requires that DNA polymerases bind both double- and single-stranded DNA.
5.3.1 E. coli DNA polymerase alpha binds both ssDNA and dsDNA
The α subunit is the 1160-residue DNA polymerase of E. coli Pol III, a complex 10-subunit asymmetric dimer which coordinates simultaneous leading and lagging strand synthesis at the replication fork [278-281]. Although a crystal structure of residues 1-197 demonstrates that α subunit folds into the right hand shape characteristic of DNA polymerases [282], the absence of significant sequence similarities between Pol III α and well-known DNA polymerases leaves DNA binding domains unclear [283]. The partial crystal structure reveals a dual helix-hairpin-helix motif (HhH)2, at residues 833-889 which is the predicted dsDNA binding domain [284-285]. Although the E. coli Pol III α crystal structure does not include the C-terminal residues 917-1160, sequence homology modeling suggests an OB fold at residues 978-1078 which may be the ssDNA binding site [170, 282, 286-289]. The full-length crystal structure of Thermus aquaticus Pol III reveals similarities to E. coli Pol III [286], and a recent structure of the T. aquaticus α subunit shows the OB fold bound to DNA [290]. Single molecule DNA stretching experiments characterized the dsDNA and ssDNA binding activity of E. coli Pol III α, quantifying binding to both forms of DNA by distinct regions of the protein [291].
DNA stretching curves in the presence of full-length α (Figure 15a) show significant hysteresis, indicating that α remains bound to the single DNA strands, such that the DNA molecule cannot reanneal on the timescale of the relaxation cycle. Although the initial stretch with α follows the stretching curve of DNA without protein up to the melting plateau, subsequent stretches reflect that some protein remains bound, stabilizing a fraction of ssDNA over the length of the molecule. Thus all of the bound protein does not dissociate upon complete relaxation, and the DNA molecule reaches a saturated combination of roughly half ssDNA and half dsDNA [291]. In contrast with SSBs and HTLV-1 NC, Pol III α demonstrates a strong preference for ssDNA without decreasing the DNA melting force. This indicates that the protein does not destabilize existing dsDNA, but binds ssDNA available due to force-stabilized fluctuations. Furthermore, DNA stretching experiments reveal that Pol III α stabilizes the DNA duplex, increasing the DNA melting force in a salt-independent manner (data available in [291]). This is due to protein binding to dsDNA, an interaction which must be disrupted prior to force-induced melting of the duplex.
Figure 15.
E. coli DNA polymerase alpha binds ssDNA and dsDNA at distinct binding sites. (a) Extension (solid lines) and relaxation (dotted lines) cycles in the presence of 100 mM α. Although the initial extension curve (solid green line) is similar to that of DNA without protein (black), the relaxation (dotted green line) exhibits significant hysteresis, indicating that α remains bound to ssDNA and prevents reannealing. Subsequent stretches reflect that some protein remains bound, stabilizing a fraction of ssDNA over the length of the molecule. (b) Increase in DNA melting force characterizes dsDNA binding, and solid lines are fits to the McGhee-von Hippel binding isotherm (Equation (4)) which yield equilibrium association constants Kds for each construct. (c) Relaxation data fit to the WLC and FJC polymer models yields equilibrium association constants Kss for each construct. Full-length α (green) has strong binding affinity for both single- and double-stranded DNA. Both the α1-917 (blue) and α1-835 (purple) fragments show strong dsDNA binding, and no affinity for ssDNA. In contrast, α917-1160 (pink) binds ssDNA but not dsDNA. Figures reproduced with permission from [1].
5.3.2 (HhH)2 domain binds dsDNA and the predicted OB fold binds ssDNA
DNA stretching curves in the presence of the 917-residue N-terminal fragment (α1-917) also exhibit a higher melting force, but no hysteresis. This indicates that α1-917 binds dsDNA, stabilizing the duplex, but does not bind ssDNA. Thus the 917 N-terminal residues do not contain the ssDNA binding site, but do contain the dsDNA binding site [291]. Single molecule experiments with residues 1-835 (α1-835) did not detect a change in melting force, hysteresis, or any other protein-induced effects, indicating that the fragment does not bind DNA. Therefore, the dsDNA binding site lies predominantly within residues 836-917, a region of Pol III α which includes the (HhH)2 domain at residues 833-889.
DNA stretching curves with the C-terminal fragment α917-1160 exhibit the type of hysteresis evident with full-length α, but lack the higher melting force. Similar features are evident with a fragment of residues 978-1160 (α917-1160), which indicates that both proteins bind ssDNA but not dsDNA [291]. In contrast, stretching experiments with α1076-1160, a fragment which does not include the predicted OB fold at residues 978-1078, do not show any evidence of DNA binding activity [291]. These results are consistent with an ssDNA binding site at the OB fold.
5.3.3 Quantitative binding affinities for double- and single-stranded DNA
Equilibrium binding constants for the full-length α protein and the three fragments α1-917, α1-835, and α917-1160 were obtained for both dsDNA and ssDNA [291]. Measurements of the melting force Fm as a function of protein concentration determine fractional binding to DNA αds (Equation (12)). Fits to the McGhee-von Hippel binding isotherm (Equation (4)) yield Kds for the four proteins. Furthermore, protein binding to ssDNA stabilizes a fraction of ssDNA, and fits of this relaxation data to a linear combination of the contour lengths of dsDNA (bds) and ssDNA (bss) yield the force-dependent contour length b, which is proportional to the fraction of protein-stabilized ssDNA:
| (16) |
Fits of αds to the McGhee-von Hippel binding isotherm (Equation (4)) determine Kss for each protein [291]. The equilibrium binding constants determined (Figure 15b-c) indicate that full-length α has strong binding affinity for both single- and double-stranded DNA. Both the α1-917 and α1-835 fragments show strong dsDNA binding, and no affinity for ssDNA. In contrast, α917-1160 binds ssDNA but not dsDNA [291]. These quantitative results demonstrate that the (HhH)2 motif is primarily responsible for dsDNA binding, while ssDNA binding arises from the region containing the predicted OB fold.
6 CONCLUSIONS
Force spectroscopy can be used to characterize single- and double-stranded DNA binding affinities and binding kinetics for a large range of binding ligands and proteins, including small molecules with a single binding mode to complex proteins with multiple binding modes. Ethidium and ruthenium complexes intercalate within the base pairs of dsDNA, revealing an increase in length that can be used to quantify the thermodynamics and kinetics of intercalation. Nuclear proteins such as eukaryotic histones and prokaryotic histone-like proteins package and compact dsDNA through specific and non-specific binding. Compaction can be characterized as these higher order structures are disrupted and unfolded by force. HMG proteins, found in high concentration in the nucleus, introduce bends in the DNA backbone, which can be characterized by measuring the DNA persistence length as a function of concentration, revealing DNA binding affinity as well as DNA bending angles induced upon protein binding. Bacteriophage SSBs such as T4 gp32 bind weakly to dsDNA, allowing them to slide along the DNA in search of an interface with ssDNA, which they preferentially bind stabilize. The equilibrium ssDNA binding results in overall destabilization of DNA, which allows determination of ssDNA binding affinity by measuring a reduction in the DNA melting force as a function of SSB concentration. Rate-dependent DNA force-induced melting in the presence of SSBs also allows measurement of dsDNA binding affinity and the rate at which these proteins slide on dsDNA. In contrast to proteins that exhibit a primary binding mode to either ssDNA or dsDNA, retroviral nucleic acid chaperones such as HIV-1 NC bind both dsDNA and ssDNA with similar binding affinity, and they also exhibit rapid binding kinetics, allowing them to rapidly switch between ssDNA and dsDNA binding modes. This capability allows nucleic acid chaperones to facilitate nucleic acid rearrangements in reverse transcription and other key replication processes. Substantially larger proteins such as the DNA polymerase subunit of E. coli have distinct binding sites for dsDNA and ssDNA, which can be distinguished by examining mutant proteins lacking the domains responsible for each binding mode. The results described here illustrate how single molecule stretching experiments can provide crucial insight into the DNA binding activity and functional mechanism of a diverse range of molecules and proteins with multiple DNA binding mechanisms.
7 ACKNOWLEDGEMENTS
We acknowledge funding from US National Institutes of Health (R01GM072462) and the US National Science Foundation (MCB-0744456). Kathy Chaurasiya was supported by IGERT (DGE-0504331).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8 REFERENCES
- 1.McCauley MJ, Williams MC. Review: Optical Tweezers Experiments Resolve Distinct Modes of DNA-Protein Binding. Biopolymers. 2009;91:265–282. doi: 10.1002/bip.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gosse C, Croquette V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophysical Journal. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Zhang W, Machon C, Orta A, Phillips N, Roberts CJ, Allen S, Soultanas P. Single-molecule atomic force spectroscopy reveals that DnaD forms scaffolds and enhances duplex melting. Journal of Molecular Biology. 2008;377:706–714. doi: 10.1016/j.jmb.2008.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clausen-Schaumann H, Rief M, Tolksdorf C, Gaub HE. Mechanical stability of single DNA molecules. Biophysical Journal. 2000;78:1997–2007. doi: 10.1016/S0006-3495(00)76747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasas S, Thomson NH, Smith BL, Hansma HG, Zhu XS, Guthold M, Bustamante C, Kool ET, Kashlev M, Hansma PK. Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry. 1997;36:461–468. doi: 10.1021/bi9624402. [DOI] [PubMed] [Google Scholar]
- 6.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic Effects On the Elasticity Of Single DNA Molecules. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 8.Odijk T. Stiff chains and filaments under tension. Macromolecules. 1995;28:7016–7018. [Google Scholar]
- 9.Podgornik R, Hansen PL, Parsegian PA. Elastic moduli renormalization in self-interacting stretchable polyelectrolytes. Journal of Chemical Physics. 2000;113:9343–9350. [Google Scholar]
- 10**.Wenner JR, Williams MC, Rouzina I, Bloomfield VA. Salt Dependence of the Elasticity and Overstretching Transition of Single DNA Molecules. Biophysical Journal. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith SB, Finzi L, Bustamante C. Direct Mechanical Measurements of the Elasticity of Single DNA Molecules by Using Magnetic Beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 12.McCauley MJ, Williams MC. Mechanisms of DNA binding determined in optical tweezers experiments. Biopolymers. 2007;85:154–168. doi: 10.1002/bip.20622. [DOI] [PubMed] [Google Scholar]
- 13**.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. Entropy and heat capacity of DNA melting from temperature dependence of single molecule stretching. Biophysical Journal. 2001;80:1932–9. doi: 10.1016/S0006-3495(01)76163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. Effect of pH on the overstretching transition of double-stranded DNA: evidence of force-induced DNA melting. Biophysical Journal. 2001;80:874–81. doi: 10.1016/S0006-3495(01)76066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Harris SA, Sands ZA, Laughton CA. Molecular dynamics simulations of duplex stretching reveal the importance of entropy in determining the biomechanical properties of DNA. Biophysical Journal. 2005;88:1684–91. doi: 10.1529/biophysj.104.046912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Piana S. Structure and energy of a DNA dodecamer under tensile load. Nucleic Acids Research. 2005;33:7029–38. doi: 10.1093/nar/gki1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. The electromechanics of DNA in a synthetic nanopore. Biophysical Journal. 2006;90:1098–106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams MC, Rouzina I, Bloomfield VA. Thermodynamics of DNA interactions from single molecule stretching experiments. Accounts of Chemical Research. 2002;35:159–166. doi: 10.1021/ar010045k. [DOI] [PubMed] [Google Scholar]
- 19*.Pant K, Karpel RL, Williams MC. Kinetic regulation of single DNA molecule denaturation by T4 gene 32 protein structural domains. Journal of Molecular Biology. 2003;327:571–578. doi: 10.1016/s0022-2836(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 20.Pant K, Karpel RL, Rouzina L, Williams MC. Mechanical measurement of single-molecule binding rates: Kinetics of DNA helix-destabilization by T4 gene 32 protein. Journal of Molecular Biology. 2004;336:851–870. doi: 10.1016/j.jmb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 21**.Pant K, Karpel RL, Rouzina I, Williams MC. Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: Single molecule force spectroscopy measurements. Journal of Molecular Biology. 2005;349:317–330. doi: 10.1016/j.jmb.2005.03.065. [DOI] [PubMed] [Google Scholar]
- 22.Rouzina I, Pant K, Karpel RL, Williams MC. Theory of electrostatically regulated binding of T4 gene 32 protein to single- and double-stranded DNA. Biophysical Journal. 2005;89:1941–1956. doi: 10.1529/biophysj.105.063776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. Journal of Biological Chemistry. 2006;281:38689–38696. doi: 10.1074/jbc.M608460200. [DOI] [PubMed] [Google Scholar]
- 24.Pant K, Shokri L, Karpel RL, Morrical SW, Williams MC. Modulation of T4 gene 32 protein DNA binding activity by the recombination mediator protein UvsY. Journal of Molecular Biology. 2008;380:799–811. doi: 10.1016/j.jmb.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shokri L, Marintcheva B, Eldib M, Hanke A, Rouzina I, Williams MC. Kinetics and thermodynamics of salt-dependent T7 gene 2.5 protein binding to single- and double-stranded DNA. Nucleic Acids Research. 2008;36:5668–5677. doi: 10.1093/nar/gkn551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Shokri L, Rouzina I, Williams MC. Interaction of bacteriophage T4 and T7 single-stranded DNA-binding proteins with DNA. Physical Biology. 2009;6 doi: 10.1088/1478-3975/6/2/025002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: An extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 28.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 29.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–12. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 30.Konrad MW, Bolonick JI. Molecular dynamics simulation of DNA stretching is consistent with the tension observed for extension and strand separation and predicts a novel ladder structure. Journal of the American Chemical Society. 1996;118:10989–10994. [Google Scholar]
- 31.Lebrun A, Lavery R. Modelling extreme stretching of DNA. Nucleic Acids Research. 1996;24:2260–2267. doi: 10.1093/nar/24.12.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosikov KM, Gorin AA, Zhurkin VB, Olson WK. DNA stretching and compression: large-scale simulations of double helical structures. Journal of Molecular Biology. 1999;289:1301–26. doi: 10.1006/jmbi.1999.2798. [DOI] [PubMed] [Google Scholar]
- 33.Storm C, Nelson PC. Theory of high-force DNA stretching and overstretching. Physical Review E. 2003;67 doi: 10.1103/PhysRevE.67.051906. [DOI] [PubMed] [Google Scholar]
- 34.Cocco S, Yan J, Leger JF, Chatenay D, Marko JF. Overstretching and force-driven strand separation of double-helix DNA. Physical Review E. 2004;70 doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 35.Ho D, Zimmermann JL, Dehmelt FA, Steinbach U, Erdmann M, Severin P, Falter K, Gaub HE. Force-Driven Separation of Short Double-Stranded DNA. Biophysical Journal. 2009;97:3158–3167. doi: 10.1016/j.bpj.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leger JF, Romano G, Sarkar A, Robert J, Bourdieu L, Chatenay D, Marko JF. Structural transitions of a twisted and stretched DNA molecule. Physical Review Letters. 1999;83:1066–1069. [Google Scholar]
- 37.Whitelam S, Pronk S, Geissler PL. There and (Slowly) back again: Entropy-driven hysteresis in a model of DNA overstretching. Biophysical Journal. 2008;94:2452–2469. doi: 10.1529/biophysj.107.117036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitelam S, Pronk S, Geissler PL. Stretching chimeric DNA: A test for the putative S-form. Journal of Chemical Physics. 2008;129 doi: 10.1063/1.3009266. [DOI] [PubMed] [Google Scholar]
- 39.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 40.Strick TR, Dessinges MN, Charvin G, Dekker NH, Allemand JF, Bensimon D, Croquette V. Stretching of macromolecules and proteins. Reports on Progress in Physics. 2003;66:1–45. [Google Scholar]
- 41***.Shokri L, McCauley MJ, Rouzina I, Williams MC. DNA overstretching in the presence of glyoxal: structural evidence of force-induced DNA melting. Biophysical Journal. 2008;95:1248–55. doi: 10.1529/biophysj.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutton JR, Wetmur JG. Effect of chemical modification on the rate of renaturation of deoxyribonucleic acid. Deaminated and glyoxalated deoxyribonucleic acid. Biochemistry. 1973;12:558–63. doi: 10.1021/bi00727a032. [DOI] [PubMed] [Google Scholar]
- 43.Danilowicz C, Limouse C, Hatch K, Conover A, Coljee VW, Kleckner N, Prentiss M. The structure of DNA overstretched from the 5' 5' ends differs from the structure of DNA overstretched from the 3' 3' ends. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13196–13201. doi: 10.1073/pnas.0904729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44***.van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc Natl Acad Sci U S A. 2009;106:18231–6. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rye HS, Yue S, Wemmer DE, Quesada MA, Haugland RP, Mathies RA, Glazer AN. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and applications. Nucleic Acids Research. 1992;20:2803–12. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C, Curth U, Urbanke C, Kang CH. Crystal structure of human mitochondrial single stranded DNA binding protein at 2.4 angstrom resolution. Nature Structural Biology. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 47.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 48.Williams MC, Rouzina I, McCauley MJ. Peeling back the mystery of DNA overstretching. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18047–18048. doi: 10.1073/pnas.0910269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nature Structural Biology. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 50.Krautbauer R, Clausen-Schaumann H, Gaub HE. Cisplatin changes the mechanics of single DNA molecules. Angewandte Chemie-International Edition. 2000;39:3912–+. doi: 10.1002/1521-3773(20001103)39:21<3912::AID-ANIE3912>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 51*.Krautbauer R, Pope LH, Schrader TE, Allen S, Gaub HE. Discriminating small molecule DNA binding modes by single molecule force spectroscopy. Febs Letters. 2002;510:154–158. doi: 10.1016/s0014-5793(01)03257-4. [DOI] [PubMed] [Google Scholar]
- 52.Lerman LS. Structural considerations in the interaction of DNA and acridines. J Mol Biol. 1961;3:18–30. doi: 10.1016/s0022-2836(61)80004-1. [DOI] [PubMed] [Google Scholar]
- 53.Anselmetti D, Fritz J, Smith B, Fernandez-Busquets X. Single Molecule DNA Biophysics with Atomic Force Microscopy. Single Molecules. 2000;1:53–58. [Google Scholar]
- 54**.Vladescu ID, McCauley MJ, Rouzina I, Williams MC. Mapping the phase diagram of single DNA molecule force-induced melting in the presence of ethidium. Physical Review Letters. 2005;95 doi: 10.1103/PhysRevLett.95.158102. [DOI] [PubMed] [Google Scholar]
- 55***.Vladescu ID, McCauley MJ, Nunez ME, Rouzina I, Williams MC. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nature Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- 56.Krautbauer R, Fischerlander S, Allen S, Gaub HE. Mechanical fingerprints of DNA drug complexes. Single Molecules. 2002;3:97–103. [Google Scholar]
- 57.Eckel R, Ros R, Ros A, Wilking SD, Sewald N, Anselmetti D. Identification of binding mechanisms in single molecule-DNA complexes. Biophysical Journal. 2003;85:1968–1973. doi: 10.1016/S0006-3495(03)74624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tessmer I, Baumann CG, Skinner GM, Molloy JE, Hoggett JG, Tendler SJB, Allen S. Mode of drug binding to DNA determined by optical tweezers force spectroscopy. Journal of Modern Optics. 2003;50:1627–1636. [Google Scholar]
- 59.Sischka A, Toensing K, Eckel R, Wilking SD, Sewald N, Ros R, Anselmetti D. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophysical Journal. 2005;88:404–411. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W, Barbagallo R, Madden C, Roberts CJ, Woolford A, Allen S. Progressing single biomolecule force spectroscopy measurements for the screening of DNA binding agents. Nanotechnology. 2005;16:2325–2333. doi: 10.1088/0957-4484/16/10/055. [DOI] [PubMed] [Google Scholar]
- 61.Bustamante C, Marko JF, Siggia ED, Smith S. ENTROPIC ELASTICITY OF LAMBDA-PHAGE DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 62.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: Cooperative and non-cooperative binding of large ligands to a one-dimensional homogeneous lattice. Journal of Molecular Biology. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 63.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 64.Rocha MS, Ferreira MC, Mesquita ON. Transition on the entropic elasticity of DNA induced by intercalating molecules. Journal of Chemical Physics. 2007;127 doi: 10.1063/1.2768945. [DOI] [PubMed] [Google Scholar]
- 65.Mihailovic A, Vladescu L, McCauley M, Ly E, Williams MC, Spain EM, Nunez ME. Exploring the interaction of ruthenium(II) polypyridyl complexes with DNA using single-molecule techniques. Langmuir. 2006;22:4699–4709. doi: 10.1021/la053242r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66**.Murade CU, Subramaniam V, Otto C, Bennink ML. Interaction of Oxazole Yellow Dyes with DNA Studied with Hybrid Optical Tweezers and Fluorescence Microscopy. Biophysical Journal. 2009;97:835–843. doi: 10.1016/j.bpj.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Husale S, Grange W, Hegner M. DNA mechanics affected by small DNA interacting ligands. Single Molecules. 2002;3:91–96. [Google Scholar]
- 68.Rocha MS, Viana NB, Mesquita ON. DNA-psoralen interaction: A single molecule experiment. Journal of Chemical Physics. 2004;121:9679–9683. doi: 10.1063/1.1806817. [DOI] [PubMed] [Google Scholar]
- 69.Lincoln P, Norden B. Binuclear ruthenium(II) phenanthroline compounds with extreme binding affinity for DNA. Chemical Communications. 1996:2145–2146. [Google Scholar]
- 70**.Paramanathan T, Westerlund F, McCauley MJ, Rouzina I, Lincoln P, Williams MC. Mechanically manipulating the DNA threading intercalation rate. Journal of the American Chemical Society. 2008;130:3752–3. doi: 10.1021/ja711303p. [DOI] [PubMed] [Google Scholar]
- 71.Rye HS, Yue S, Wemmer DE, Quesada MA, Haugland RP, Mathies RA, Glazer AN. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and applications. Nucleic Acids Res. 1992;20:2803–12. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bennink ML, Scharer OD, Kanaar R, Sakata-Sogawa K, Schins JM, Kanger JS, de Grooth BG, Greve J. Single-molecule manipulation of double-stranded DNA using optical tweezers: Interaction studies of DNA with RecA and YOYO-1. Cytometry. 1999;36:200–208. doi: 10.1002/(sici)1097-0320(19990701)36:3<200::aid-cyto9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 73**.Kleimann C, Sischka A, Spiering A, Tonsing K, Sewald N, Diederichsen U, Anselmetti D. Binding kinetics of bisintercalator Triostin a with optical tweezers force mechanics. Biophysical Journal. 2009;97:2780–4. doi: 10.1016/j.bpj.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olins AL, Olins DE. Spheroid Chromatin Units (Q Bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 75.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–71. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 76.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–8. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 77.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 78.Alberts B. Molecular biology of the cell. Garland Science; New York: 2008. [Google Scholar]
- 79.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 81.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 82.Van Holde KE. Chromatin. Springer-Verlag; New York: 1989. [Google Scholar]
- 83.Leuba SH, Bustamante C. Analysis of chromatin by scanning force microscopy. Methods Mol Biol. 1999;119:143–60. doi: 10.1385/1-59259-681-9:143. [DOI] [PubMed] [Google Scholar]
- 84.Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci U S A. 1994;91:11621–5. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8872–7. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86***.Bennink ML, Leuba SH, Leno GH, Zlatanova J, de Grooth BG, Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol. 2001;8:606–10. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 87***.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Claudet C, Angelov D, Bouvet P, Dimitrov S, Bednar J. Histone octamer instability under single molecule experiment conditions. J Biol Chem. 2005;280:19958–65. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- 89.Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leuba SH, Bennink ML, Zlatanova J. Single-molecule analysis of chromatin. Methods Enzymol. 2004;376:73–105. doi: 10.1016/S0076-6879(03)76006-6. [DOI] [PubMed] [Google Scholar]
- 91.Pope LH, Bennink ML, van Leijenhorst-Groener KA, Nikova D, Greve J, Marko JF. Single chromatin fiber stretching reveals physically distinct populations of disassembly events. Biophys J. 2005;88:3572–83. doi: 10.1529/biophysj.104.053074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zlatanova J. Forcing Chromatin. Journal of Biological Chemistry. 2003;278:23213–23216. doi: 10.1074/jbc.R300007200. [DOI] [PubMed] [Google Scholar]
- 93***.Kruithof M, Chien FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nature Structural & Molecular Biology. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 94.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–68. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 95.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 96.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–7. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Staynov DZ. The controversial 30 nm chromatin fibre. BioEssays. 2008;30:1003–1009. doi: 10.1002/bies.20816. [DOI] [PubMed] [Google Scholar]
- 98.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kruithof M, van Noort J. Hidden Markov Analysis of Nucleosome Unwrapping Under Force. Biophysical Journal. 2009;96:3708–3715. doi: 10.1016/j.bpj.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility group chromosomal proteins. Molecular and Cellular Biology. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends in Biochemical Sciences. 2001;26:152–3. doi: 10.1016/s0968-0004(00)01777-1. [DOI] [PubMed] [Google Scholar]
- 102.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Progress in Nucleic Acid Research and Molecular Biology. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 103.Johns E, editor. HMG Chromosomal Proteins. Academic Press; New York: 1982. [Google Scholar]
- 104.Agresti A, Bianchi ME. HMGB proteins and gene expression. Current Opinion in Genetics & Development. 2003;13:170–8. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 105.Bianchi ME, Beltrame M. Flexing DNA: HMG-box proteins and their partners. American Journal of Human Genetics. 1998;63:1573–7. doi: 10.1086/302170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vijayanathan V, Thomas T, Shirahata A, Thomas TJ. DNA condensation by polyamines: a laser light scattering study of structural effects. Biochemistry. 2001;40:13644–13651. doi: 10.1021/bi010993t. [DOI] [PubMed] [Google Scholar]
- 107.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes & Development. 1998;12:462–72. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitsouras K, Wong B, Arayata C, Johnson RC, Carey M. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Molecular and Cellular Biology. 2002;22:4390–401. doi: 10.1128/MCB.22.12.4390-4401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paull TT, Carey M, Johnson RC. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes & Development. 1996;10:2769–81. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 110.Kruppa M, Moir RD, Kolodrubetz D, Willis IM. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Molecular Cell. 2001;7:309–18. doi: 10.1016/s1097-2765(01)00179-4. [DOI] [PubMed] [Google Scholar]
- 111.Laser H, Bongards C, Schuller J, Heck S, Johnsson N, Lehming N. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13732–7. doi: 10.1073/pnas.250400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci U S A. 2008;105:10320–5. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P. HMGB1: guiding immunity from within. Trends Immunol. 2005;26:381–7. doi: 10.1016/j.it.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 114.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–42. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 115.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes & Development. 1993;7:1521–34. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 116.Ross ED, Hardwidge PR, Maher LJ., 3rd HMG proteins and DNA flexibility in transcription activation. Molecular and Cellular Biology. 2001;21:6598–605. doi: 10.1128/MCB.21.19.6598-6605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117***.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectual roles of HU: Formation of flexible hinges and rigid filaments. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–8. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 120.LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 121.Ragab A, Travers A. HMG-D and histone H1 alter the local accessibility of nucleosomal DNA. Nucleic Acids Research. 2003;31:7083–9. doi: 10.1093/nar/gkg923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klass J, Murphy FI, Fouts S, Serenil M, Changela A, Siple J, Churchill ME. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Research. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Murphy FVI, Sweet RM, Churchill ME. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. European Molecular Biology Organization Journal. 1999;18:6610–8. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stott K, Tang GSF, Lee K-B, Thomas JO. Structure of a complex of tandem HMG boxes and DNA. Journal of Molecular Biology. 2006;360:90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 125.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA With Optical Tweezers. Biophysical Journal. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bouchiat C, Wang MD, Allemand JF, Strick TR, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Molecular Cell. 2004;14:355–62. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 128.Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A. Cyclization of short DNA fragments and bending fluctuations of the double helix. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5397–402. doi: 10.1073/pnas.0500983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wiggins PA, van der Heijden T, Moreno-Herrero F, Spakowitz A, Phillips R, Widom J, Dekker C, Nelson PC. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat Nanotechnol. 2006;1:137–41. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 130.Rouzina I, Bloomfield VA. DNA bending by small, mobile multivalent cations. Biophysical Journal. 1998;74:3152–64. doi: 10.1016/S0006-3495(98)78021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Churchill ME, Changela A, Dow LK, Krieg AJ. Interactions of high mobility group box proteins with DNA and chromatin. Methods in Enzymology. 1999;304:99–103. doi: 10.1016/s0076-6879(99)04009-4. [DOI] [PubMed] [Google Scholar]
- 132.Schellman JA. Flexibility of DNA. Biopolymers. 1974;13:217–26. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- 133.Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill ME, Crane-Robinson C, Privalov PL. DNA binding and bending by HMG boxes: Energetic determinants of specificity. Journal of Molecular Biology. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 134.Hardwidge PR, Kahn JD, Maher LJ, 3rd, Ross ED. Dominant effect of protein charge rather than protein shape in apparent DNA bending by engineered bZIP domains; HMG proteins and DNA flexibility in transcription activation. Biochemistry. 2002;41:8277–88. doi: 10.1021/bi020185h. [DOI] [PubMed] [Google Scholar]
- 135.Hardwidge PR, Parkhurst KM, Parkhurst LJ, Maher LJ., 3rd Reflections on apparent DNA bending by charge variants of bZIP proteins. Biopolymers. 2003;69:110–7. doi: 10.1002/bip.10321. [DOI] [PubMed] [Google Scholar]
- 136.Bustamante C, Rivetti C. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annual Review of Biophysics and Biomolecular Structure. 1996;25:395–429. doi: 10.1146/annurev.bb.25.060196.002143. [DOI] [PubMed] [Google Scholar]
- 137.Janicijevic A, Sugasawa K, Shimizu Y, Hanaoka F, Wijgers N, Djurica M, Hoeijmakers JH, wyman C. DNA bending by the human damage recognition complexes XPC-HR23B. DNA Repair (Amst.) 2003;2:325–336. doi: 10.1016/s1568-7864(02)00222-7. [DOI] [PubMed] [Google Scholar]
- 138.Mysiak ME, Bleijenberg MH, Wyman C, Holthuizen PE, van der Vliet PC. Bending of adenovirus origin DNA by nuclear factor I as shown by scanning force microscopy is required for optimal DNA replication. J. Virol. 2004;78:1928–1935. doi: 10.1128/JVI.78.4.1928-1935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seong GH, Kobatake E, Miura K, Nakazawa A, Aizawa M. Direct atomic force microscopy visualization of integration host factor induced DNA bending structure of the promoter regulatory region on the Pseudomonas TOL plasmid. Biochem.Biophys.Res.Commun. 2002;291:361–366. doi: 10.1006/bbrc.2002.6443. [DOI] [PubMed] [Google Scholar]
- 140.Rivetti C, Guthold M, Bustamante C. Scanning force microscopy of DNA deposited onto mica: Equilibration versus kinetic trapping studied by statistical polymer chain analysis. Journal of Molecular Biology. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 141.Dame RT, van Mameren J, Luijsterburg MS, Mysiak ME, Janicijevic A, Pazdzior G, van der Vliet PC, Wyman C, Wuite GJL. Analysis of scanning force microscopy images of protein-induced DNA bending using simulations. Nucleic Acids Research. 2005;33 doi: 10.1093/nar/gni073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang JY, McCauley MJ, Maher LJ, Williams MC, Israeloff NE. Mechanism of DNA flexibility enhancement by HMGB proteins. Nucleic Acids Research. 2009;37:1107–1114. doi: 10.1093/nar/gkn1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143***.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. Journal of Molecular Biology. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 144.Cruceanu M, Stephen AG, Beuning PJ, Gorelick RJ, Fisher RJ, Williams MC. Single DNA molecule stretching measures the activity of chemicals that target the HIV-1 nucleocapsid protein. Analytical Biochemistry. 2006;358:159–170. doi: 10.1016/j.ab.2006.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Crothers DM, Metzger H. The influence of polyvalency on the binding properties of antibodies. Immunochemistry. 1972;9:341–357. doi: 10.1016/0019-2791(72)90097-3. [DOI] [PubMed] [Google Scholar]
- 146.Cui T, Wei S, Brew K, Leng F. Energetics of binding the mammalian high mobility group protein HMGA2 to poly(dA-dT)2 and poly(dA)-poly(dT) Journal of Molecular Biology. 2005;325:629–645. doi: 10.1016/j.jmb.2005.07.048. [DOI] [PubMed] [Google Scholar]
- 147.Grasser KD, Teo S-H, Lee K-B, Broadhurst RW, Rees C, Hardman CH, Thomas JO. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1) European Journal of Biochemistry. 1998;253:787–795. doi: 10.1046/j.1432-1327.1998.2530787.x. [DOI] [PubMed] [Google Scholar]
- 148*.McCauley M, Hardwidge PR, Maher LJ, 3rd, Williams MC. Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys J. 2005;89:353–64. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grainger DC, Hurd D, Goldberg MD, Busby SJW. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Research. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Williams RM, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. The Federation of European Microbiological Societies Microbiology Letters. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 151.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nature Reviews Microbiology. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 152.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Current Opinion in Structural Biology. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 153.Swinger KK, Rice PA. Structure-based analysis of HU-DNA binding. Journal of Molecular Biology. 2007;365:1005–16. doi: 10.1016/j.jmb.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 155.Ali BMJ, Amit R, Braslavsky I, Oppenheim AB, Gileadi O, Stavans J. Compaction of single DNA molecules induced by binding of integration host factor (IHF) Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10658–10663. doi: 10.1073/pnas.181029198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bonnefoy E, Rouviere-Yaniv J. HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 1992;11:4489–96. doi: 10.1002/j.1460-2075.1992.tb05550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157**.McCauley MJ, Zimmerman J, Maher LJ, 3rd, Williams MC. HMGB binding to DNA: single and double box motifs. J Mol Biol. 2007;374:993–1004. doi: 10.1016/j.jmb.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Salomo M, Keyser UF, Kegler K, Gutsche C, Struhalla M, Immisch C, Hahn U, Kremer F. Kinetics of TmHU binding to DNA as observed by optical tweezers. Microsc Res Tech. 2007;70:938–43. doi: 10.1002/jemt.20498. [DOI] [PubMed] [Google Scholar]
- 159.Salomo M, Kroy K, Kegler K, Gutsche C, Struhalla M, Reinmuth J, Skokov W, Immisch C, Hahn U, Kremer F. Binding of TmHU to single dsDNA as observed by optical tweezers. J Mol Biol. 2006;359:769–76. doi: 10.1016/j.jmb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 160.Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 161.Wiggins PA, Dame RT, Noom MC, Wuite GJL. Protein-Mediated Molecular Bridging: A Key Mechanism in Biopolymer Organization. Biophysical Journal. 2009;97:1997–2003. doi: 10.1016/j.bpj.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chase JW, Williams KR. Single-Stranded DNA Binding Proteins Required for DNA Replication. Annual Review of Biochemistry. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 163.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Critical Reviews in Biochemistry and Molecular Biology. 2009;44:98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 164.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an Organizer/Mobilizer of Genome Maintenance Complexes. Critical Reviews in Biochemistry and Molecular Biology. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Haseltine CA, Kowalczykowski SC. A distinctive single-stranded DNA-binding protein from the Archaeon Sulfolobus solfataricus. Molecular Microbiology. 2002;43:1505–1515. doi: 10.1046/j.1365-2958.2002.02807.x. [DOI] [PubMed] [Google Scholar]
- 166.Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein - Multiple DNA binding modes and cooperativities. Annual Review of Biochemistry. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 168.Kerr ID, Wadsworth RIM, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. European Molecular Biology Organization Journal. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annual Review of Biochemistry. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 170.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. European Molecular Biology Organization Journal. 1993;12:861–7. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Philipova D, Mullen JR, Maniar HS, Lu JA, Gu CY, Brill SJ. A hierarchy of SSB protomers in replication protein A. Genes & Development. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 172.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annual Review of Biochemistry. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 173.Kim YT, Richardson CC. Acidic carboxyl-terminal domain of gene-2.5 protein of bacteriophage-T7 is essential for protein-protein interactions. Journal of Biological Chemistry. 1994;269:5270–5278. [PubMed] [Google Scholar]
- 174.Kim YT, Tabor S, Bortner C, Griffith JD, Richardson CC. Purification and characterization of the bacteriophage T7 gene 2.5 protein. A single-stranded DNA-binding protein. Journal of Biological Chemistry. 1992;267:15022–31. [PubMed] [Google Scholar]
- 175.Kim YT, Tabor S, Churchich JE, Richardson CC. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. Journal of Biological Chemistry. 1992;267:15032–40. [PubMed] [Google Scholar]
- 176.Notarnicola SM, Mulcahy HL, Lee J, Richardson CC. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. Journal of Biological Chemistry. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 177.Kong DC, Richardson CC. Single-stranded DNA binding protein and DNA helicase of bacteriophage T7 mediate homologous DNA strand exchange. European Molecular Biology Organization Journal. 1996;15:2010–2019. [PMC free article] [PubMed] [Google Scholar]
- 178.Alberts BM, Frey L. T4 bacteriophage gene 32: A structural protein in the replication and recombination of DNA. Nature. 1970;227:1313–8. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 179.Christiansen C, Baldwin RL. Catalysis of DNA reassociation by the Escherichia coli DNA binding protein: A polyamine-dependent reaction. Journal of Biological Chemistry. 1977;115:441–54. doi: 10.1016/0022-2836(77)90164-4. [DOI] [PubMed] [Google Scholar]
- 180.Rezende LF, Willcox S, Griffith JD, Richardson CC. A single-stranded DNA-binding protein of bacteriophage T7 defective in DNA annealing. Journal of Biological Chemistry. 2003;278:29098–29105. doi: 10.1074/jbc.M303374200. [DOI] [PubMed] [Google Scholar]
- 181.Giedroc DP, Khan R, Barnhart K. Overexpression, purification, and characterization of recombinant T4 gene 32 protein22-301 (g32P-B) Journal of Biological Chemistry. 1990;265:11444–55. [PubMed] [Google Scholar]
- 182.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal-structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 183.Wu M, Flynn EK, Karpel RL. Details of the nucleic acid binding site of T4 gene 32 protein revealed by proteolysis and DNA T-m depression methods. Journal of Molecular Biology. 1999;286:1107–1121. doi: 10.1006/jmbi.1999.2541. [DOI] [PubMed] [Google Scholar]
- 184.Jensen DE, Kelly RC, von Hippel PH. DNA “melting” proteins. II. Effects of bacteriophage T4 gene 32-protein binding on the conformation and stability of nucleic acid structures. Journal of Biological Chemistry. 1976;251:7215–28. [PubMed] [Google Scholar]
- 185.Waidner LA, Flynn EK, Wu M, Li X, Karpel RL. Domain effects on the DNA-interactive properties of bacteriophage T4 gene 32 protein. Journal of Biological Chemistry. 2001;276:2509–2516. doi: 10.1074/jbc.M007778200. [DOI] [PubMed] [Google Scholar]
- 186.Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, von Hippel PH. On the thermodynamics and kinetics of the cooperative binding of bacteriophage T4-coded gene 32 (helix destabilizing) protein to nucleic acid lattices. Biophysical Journal. 1980;32:403–18. doi: 10.1016/S0006-3495(80)84964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. Journal of Molecular Biology. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 188.Lonberg N, Kowalczykowski SC, Paul LS, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. III. Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III) Journal of Molecular Biology. 1981;145:123–38. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 189.Sokolov IM, Metzler R, Pant K, Williams MC. First passage time of N excluded-volume particles on a line. Physical Review E. 2005;72 doi: 10.1103/PhysRevE.72.041102. [DOI] [PubMed] [Google Scholar]
- 190*.Sokolov IM, Metzler R, Pant K, Williams MC. Target search of N sliding proteins on a DNA. Biophysical Journal. 2005;89:895–902. doi: 10.1529/biophysj.104.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Winter RB, Berg OG, Von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor-operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 192.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Research. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophysical Journal. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Blainey PC, van Oijent AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hu T, Grosberg AY, Shklovskii BI. How proteins search for their specific sites on DNA: The role of DNA conformation. Biophysical Journal. 2006;90:2731–2744. doi: 10.1529/biophysj.105.078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang YM, Austin RH, Cox EC. Single molecule measurements of repressor protein 1D diffusion on DNA. Physical Review Letters. 2006;97 doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 197.Kim JH, Larson RG. Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules. Nucleic Acids Research. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cherstvy AG, Kolomeisky AB, Kornyshev AA. Protein-DNA interactions: Reaching and recognizing the targets. Journal of Physical Chemistry B. 2008;112:4741–4750. doi: 10.1021/jp076432e. [DOI] [PubMed] [Google Scholar]
- 199.Blainey PC, Luo GB, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nature Structural & Molecular Biology. 2009;16:1224–U34. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200***.van den Broek B, Lomholt MA, Kalisch SMJ, Metzler R, Wuite GJL. How DNA coiling enhances target localization by proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15738–15742. doi: 10.1073/pnas.0804248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.van den Broek B, Noom MC, Wuite GJL. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Research. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nature Structural Biology. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 203.Ruyechan WT, Wetmur JG. Studies on the noncooperative binding of the Escherichia coli DNA unwinding protein to single-stranded nucleic acids. Biochemistry. 1976;15:5057–64. doi: 10.1021/bi00668a017. [DOI] [PubMed] [Google Scholar]
- 204.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. Journal of Biological Chemistry. 1985;260:3594–603. [PubMed] [Google Scholar]
- 205.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 206.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. Journal of Biological Chemistry. 1988;263:4629–40. [PubMed] [Google Scholar]
- 207.Lohman TM, Bujalowski W, Overman LB. E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends in Biochemical Sciences. 1988;13:250–5. [PubMed] [Google Scholar]
- 208.Overman LB, Bujalowski W, Lohman TM. Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry. 1988;27:456–71. doi: 10.1021/bi00401a067. [DOI] [PubMed] [Google Scholar]
- 209.Hatch K, Danilowicz C, Coljee V, Prentiss M. Direct measurements of the stabilization of single-stranded DNA under tension by single-stranded binding proteins. Physical Review E. 2007;76 doi: 10.1103/PhysRevE.76.021916. [DOI] [PubMed] [Google Scholar]
- 210.Hatch K, Danilowicz C, Coljee V, Prentiss M. Measurement of the salt-dependent stabilization of partially open DNA by Escherichia coli SSB protein. Nucleic Acids Research. 2008;36:294–299. doi: 10.1093/nar/gkm1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ferrari ME, Bujalowski W, Lohman TM. Cooperative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)(35) binding mode. Journal of Molecular Biology. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 212.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. Journal of Molecular Biology. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bruand C, Ehrlich SD, Janniere L. Primosome assembly site in Bacillus-subtilis. European Molecular Biology Organization Journal. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bruand C, Sorokin A, Serror P, Ehrlich SD. Nucleotide sequence of the Bacillus-subtilis DnaD gene. Microbiology-Uk. 1995;141:321–322. doi: 10.1099/13500872-141-2-321. [DOI] [PubMed] [Google Scholar]
- 215.Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Molecular Microbiology. 2001;42:245–255. doi: 10.1046/j.1365-2958.2001.02631.x. [DOI] [PubMed] [Google Scholar]
- 216.Li Y, Kurokawa K, Matsuo M, Fukuhara N, Murakami K, Sekimizu K. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Molecular Genetics and Genomics. 2004;271:447–457. doi: 10.1007/s00438-004-0996-6. [DOI] [PubMed] [Google Scholar]
- 217.Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Molecular Microbiology. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- 218.Turner IJ, Scott DJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD protein: a putative link between DNA remodeling and initiation of DNA replication. Federation of European Biochemical Societies Letters. 2004;577:460–464. doi: 10.1016/j.febslet.2004.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhang WK, Carneiro M, Turner IJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. Journal of Molecular Biology. 2005;351:66–75. doi: 10.1016/j.jmb.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Carneiro M, Zhang WK, Ioannou C, Scott DJ, Allen S, Roberts CJ, Soultanas P. The DNA-remodelling activity of DnaD is the sum of oligomerization and DNA-binding activities on separate domains. Molecular Microbiology. 2006;60:917–924. doi: 10.1111/j.1365-2958.2006.05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang WK, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis primosomal protein DnaD untwists supercoiled DNA. Journal of Bacteriology. 2006;188:5487–5493. doi: 10.1128/JB.00339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. Early steps of Bacillus subtilis primosome assembly. Journal of Biological Chemistry. 2001;276:45818–45825. doi: 10.1074/jbc.M101996200. [DOI] [PubMed] [Google Scholar]
- 223.Negroni M, Buc H. Mechanisms of retroviral recombination. Annual Review of Genetics. 2001;35:275–302. doi: 10.1146/annurev.genet.35.102401.090551. [DOI] [PubMed] [Google Scholar]
- 224.Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag Polyproteins during RNA packaging in-vivo. Journal of Virology. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur AO. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: Characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 226.Zhang YQ, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. Journal of Virology. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Berg J. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 228.Covey SN. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Research. 1986;14:623–33. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Green LM, Berg JM. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6403–7. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Henderson LE, Copeland TD, Sowder RC, Smythers GW, Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. Journal of Biological Chemistry. 1981;256:8400–6. [PubMed] [Google Scholar]
- 231.Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, Gruninger-Leitch F, Barre-Sinoussi F, LeGrice SF, Darlix JL. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. European Molecular Biology Organization Journal. 1989;8:3279–85. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6472–6. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Li X, Quan Y, Arts EJ, Li Z, Preston BD, de Rocquigny H, Roques BP, Darlix JL, Kleiman L, Parniak MA, Wainberg MA. Human immunodeficiency virus Type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA(Lys3) in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. Journal of Virolology. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse-transcriptase catalyzed DNA strand transfer reactions and modulates RNase-H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 235.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. Journal of Biological Chemistry. 1995;270:15005–11. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 236.You JC, McHenry CS. Human-immunodeficiency-virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. Journal of Biological Chemistry. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 237.Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. Journal of Biological Chemistry. 1990;216:689–99. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 238.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7577–81. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sakaguchi K, Zambrano N, Baldwin ET, Shapiro BA, Erickson JW, Omichinski JG, Clore GM, Gronenborn AM, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5219–23. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang YQ, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation (vol 69, pg 5716, 1995) Journal of Virology. 1997;71:5712–5712. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: Stimulation by the viral nucleocapsid protein. Journal of Virology. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gao K, Gorelick RJ, Johnson DG, Bushman F. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. Journal of Virology. 2003;77:1598–1603. doi: 10.1128/JVI.77.2.1598-1603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mely Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini-Reviews in Medicinal Chemistry. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 244.McDonnell NB, DeGuzman RN, Rice WG, Turpin JA, Summers MF. Zinc ejection as a new rationale for the use of cystamine and related disulfide-containing antiviral agents in the treatment of AIDS. Journal of Medicinal Chemistry. 1997;40:1969–1976. doi: 10.1021/jm970147+. [DOI] [PubMed] [Google Scholar]
- 245.Rice WG, Turpin JA. Virus-encoded zinc fingers as targets for antiviral chemotherapy. Reviews in Medical Virology. 1996;6:187–199. doi: 10.1002/(SICI)1099-1654(199612)6:4<187::AID-RMV176>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 246*.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Research. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247**.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. Retroviral Nucleocapsid Proteins Display Nonequivalent Levels of Nucleic Acid Chaperone Activity. Journal of Virology. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248**.Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, Williams MC, Musier-Forsyth K. C-terminal Domain Modulates the Nucleic Acid Chaperone Activity of Human T-cell Leukemia Virus Type 1 Nucleocapsid Protein via an Electrostatic Mechanism. Journal of Biological Chemistry. 2010;285:295–307. doi: 10.1074/jbc.M109.051334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Darlix JL, Lapadattapolsky M, Derocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. Journal of Molecular Biology. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 250.Levin JG, Guo JH, Rouzina I, Musier-Forsyth K. Progress in Nucleic Acid Research and Molecular Biology. Elsevier Academic Press Inc; San Diego: 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism; pp. 217–286. [DOI] [PubMed] [Google Scholar]
- 251.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends in Biochemical Sciences. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 252*.Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Psi-RNA packaging signal. Implications for genome recognition. Journal of Molecular Biology. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 255.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 256.Guo JH, Wu TY, Kane BF, Johnson DG, Henderson LE, Gorelick RJ, Levin JG. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. Journal of Virology. 2002;76:4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258*.Iwatani Y, Chan DSB, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Research. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Goila-Gaur R, Strebel K. HIV-I Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5 doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends in Biochemical Sciences. 2007;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 261.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philosophical Transactions of the Royal Society B-Biological Sciences. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 263.Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, Imada K, Tanaka Y, Uchiyama T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2 doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2BM RNA is primed by a nick at the chromosomal target shite - A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 265.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Molecular and Cellular Biology. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Martin SL, Li JF, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. Journal of Molecular Biology. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 267.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Molecular Biology and Evolution. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 268.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in Ll retrotransposition. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269**.Martin SL, Cruceanu M, Branciforte D, Li PWI, Kwok SC, Hodges RS, Williams MC. LINE-1 retrotransposition chaperone activity of the requires the nucleic acid ORF1 protein. Journal of Molecular Biology. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 270*.Martin SL, Bushman D, Wang F, Li PWL, Walker A, Cummiskey J, Branciforte D, Williams MC. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic Acids Research. 2008;36:5845–5854. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Research. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yu X, VanLoock MS, Yang S, Reese JT, Egelman EH. What is the structure of the RecADNA filament? Current Protein & Peptide Science. 2004;5:73–79. doi: 10.2174/1389203043486883. [DOI] [PubMed] [Google Scholar]
- 273.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.van Mameren J, Modesti M, Kanaar R, Wyman C, Wuite GJL, Peterman EJG. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophysical Journal. 2006;91:L78–L80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.van Mameren J, Peterman EJG, Wuite GJL. See me, feel me: methods to concurrently visualize and manipulate single DNA molecules and associated proteins. Nucleic Acids Research. 2008;36:4381–4389. doi: 10.1093/nar/gkn412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJG, Wuite GJL, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 277.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJG, Wuite GJL. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kornberg A, Baker TA. DNA Replication. W.H. Freeman & Company; New York: 1992. [Google Scholar]
- 279.Kelman Z, O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annual Review of Biochemistry. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 280.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annual Review of Biochemistry. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 281.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Advances in Protein Chemistry. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 282.Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–92. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 283.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Research. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Research. 1996;24:2488–97. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Shao X, Grishin NV. Common fold in helix-hairpin-helix proteins. Nucleic Acids Research. 2000;28:2643–50. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 287.Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure, and function. Current Opinion in Structural Biology. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 288.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annual Review of Biophysics and Biomolecular Structure. 2003;32:115–33. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Zhao X-Q, Hu J-F, Yu J. Comparative analysis of eubacterial DNA polymerase III alpha subunits, Genomics. Proteomics and Bioinformatics. 2006;4:203–211. doi: 10.1016/S1672-0229(07)60001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wing RA, Bailey S, Steitz TA. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III alpha-subunit. Journal of Molecular Biology. 2008;382:859–869. doi: 10.1016/j.jmb.2008.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291***.McCauley MJ, Shokri L, Sefcikova J, Venclovas C, Beuning PJ, Williams MC. Distinct double- and single-stranded DNA binding of E. coli replicative DNA polymerase III alpha subunit. ACS Chemical Biology. 2008;3:577–587. doi: 10.1021/cb8001107. [DOI] [PMC free article] [PubMed] [Google Scholar]