Abstract
Autophagy is a conserved catabolic stress response pathway that is increasingly recognized as an important component of both innate and acquired immunity to pathogens. The activation of autophagy during infection not only provides cell-autonomous protection through lysosomal degradation of invading pathogens (xenophagy), but also regulates signaling by other innate immune pathways. This review will focus on recent advances in our understanding of three major areas of the interrelationship between autophagy and innate immunity, including how autophagy is triggered during infection, how invading pathogens are targeted to autophagosomes, and how the autophagy pathway participates in “tuning” the innate immune response.
1. Introduction
The evolution of the endomembrane system was a critical event that “emancipated” primordial unicellular eukaryotes from the need to be in continuous contact with their food sources by allowing the storage of nutrients that could be used during periods of starvation [1]. The process of autophagy, which is conserved from yeast to metazoans, involves the coordinated actions of dozens of autophagy genes (ATG) that mediate membrane rearrangements to permit catabolism of these stored energy sources [2]. Not surprisingly, this complex and evolutionarily ancient pathway has evolved to respond to many other stressors besides starvation, including hypoxia, high temperature, overcrowding, reactive oxygen species, and endoplasmic reticulum (ER) stress [3]. Of note, at the beginning of eukaryotic life, the endomembrane system also permitted the internalization of microorganisms that could be harmful to the cell. Accordingly, the autophagy pathway may have also evolved as a multi-pronged system to control pathogens, both in a cell-autonomous manner, and, in metazoan organisms, in the coordinated regulation of innate and adaptive immunity [4–6]. In this review, we will highlight recent advances in our understanding of the roles of autophagy in innate immunity, focusing on the triggering of autophagy during infection, the targeting of pathogens to the autophagic machinery, and the emerging roles for autophagy in “tuning” the innate immune response.
1.1. Autophagy as a conserved host defense response to infection
In the past several years, substantial evidence has accumulated indicating that autophagy represents a conserved host defense response against diverse intracellular pathogens [4–6]. ATG genes and the host response to pathogen infection were linked in the first report describing a mammalian ATG gene [7]. In this study, enforced neuronal expression of beclin 1, an ortholog of yeast ATG6/VPS30, protected mice from lethal CNS alphavirus infection [7]. ATG genes have since been shown to play a protective role in vivo against diverse pathogens in a wide variety of animal models. For example, ATG genes control the spread of the hypersensitive response to tobacco mosaic virus in plants [8], protect Drosophila against infection with Listeria monocytogenes [9] and vesicular stomatitis virus (VSV) [10], and protect mice against infection with L. monocytogenes, Toxoplasma gondii [11], herpes simplex virus 1 (HSV-1) [12] and Sindbis virus [13]. In addition, several in vitro studies have described important roles for autophagy in the control of many bacteria, viruses, and parasites [4]. There have also been reports of pathogens utilizing components of the host autophagic machinery to promote their own replication, and in the case of pathogens that are themselves eukaryotes, of utilizing their own autophagic machinery as part of their own intracellular survival or virulence strategies [4]. However, it is important to note that all published studies to date with non-fungal pathogens (i.e. bacteria, viruses, and parasites) in in vivo infection model systems have demonstrated a protective function of ATG genes.
1.2. Orchestration of the initial defense against infection by the innate immune system
In 1989, Janeway first postulated that a class of pattern recognition molecules must exist that function as an initial defense against infection by rapidly detecting conserved molecular features shared by pathogens [14]. This prediction was confirmed by the subsequent discovery of several classes of pattern recognition receptors (PRRs), including families of Toll-like receptors (TLRs), RIG-I like receptors (RLRs), some members of Nod-like receptor (NLR) family and C-type lectins, and the double-stranded RNA binding protein kinase PKR. These PRRs recognize conserved components of pathogens (or products of their replication) that are collectively termed pathogen-associated molecular patterns (PAMPs).
Another seminal hypothesis in the field of innate immunity was the “Danger Model” first put forth in 1994 by Matzinger [15], which proposed that cellular damage was a critical factor underlying immune activation. Indeed, over the past several years, a growing repertoire of danger-associated molecular patterns (DAMPs) have been identified that result in the activation of host stress response pathways [16–18]. These include products of necrotic cells (extracellular ATP and DNA, monosodium urate crystals), indicators of environmental stress (hypoxia, cold), perturbation of intracellular ion gradients, generation of reactive oxygen species (ROS) and accumulation of misfolded proteins.
Upon stimulation, PRRs and danger receptors activate signaling pathways that constitute the front lines of host defense against pathogen infection. These range from cell-autonomous innate immune responses, such as PKR-mediated activation of translational control programs that restrict viral replication [19], to induction of proinflammatory cytokine and chemokine production via activation of MAPK, NF-κB, IRF and IL-1β pathways, resulting in local and systemic inflammation [17, 20]. In turn, the milieu of proinflammatory signals secreted in response to PRR and DAMP receptor activation helps direct the adaptive immune response [21].
1.3. Interaction of autophagy and the innate immune system during infection
Given the crucial roles of autophagy and the innate immune system in front line defense against infection, it is reasonable to speculate that intricate cross-talk exists between the two. Although there is still much to be learned, it is becoming clear that a subset of PRRs and DAMPs activate autophagy and that ATG genes are intimately involved in tailoring the response triggered by many PRRs and DAMP receptors. There is also increasing evidence that the intracellular recognition and targeting of pathogens to autophagosomes is a central element of innate immunity. Moreover, autophagy can positively or negatively regulate PRR and DAMP receptor signaling in a cell-type and context-dependent manner to shape inflammatory responses and adaptive immunity. Further knowledge of these complex interrelationships will be critical to fully understand host-pathogen interactions and to develop new antimicrobial therapies that exploit this understanding.
2. Triggering autophagy during pathogen infection
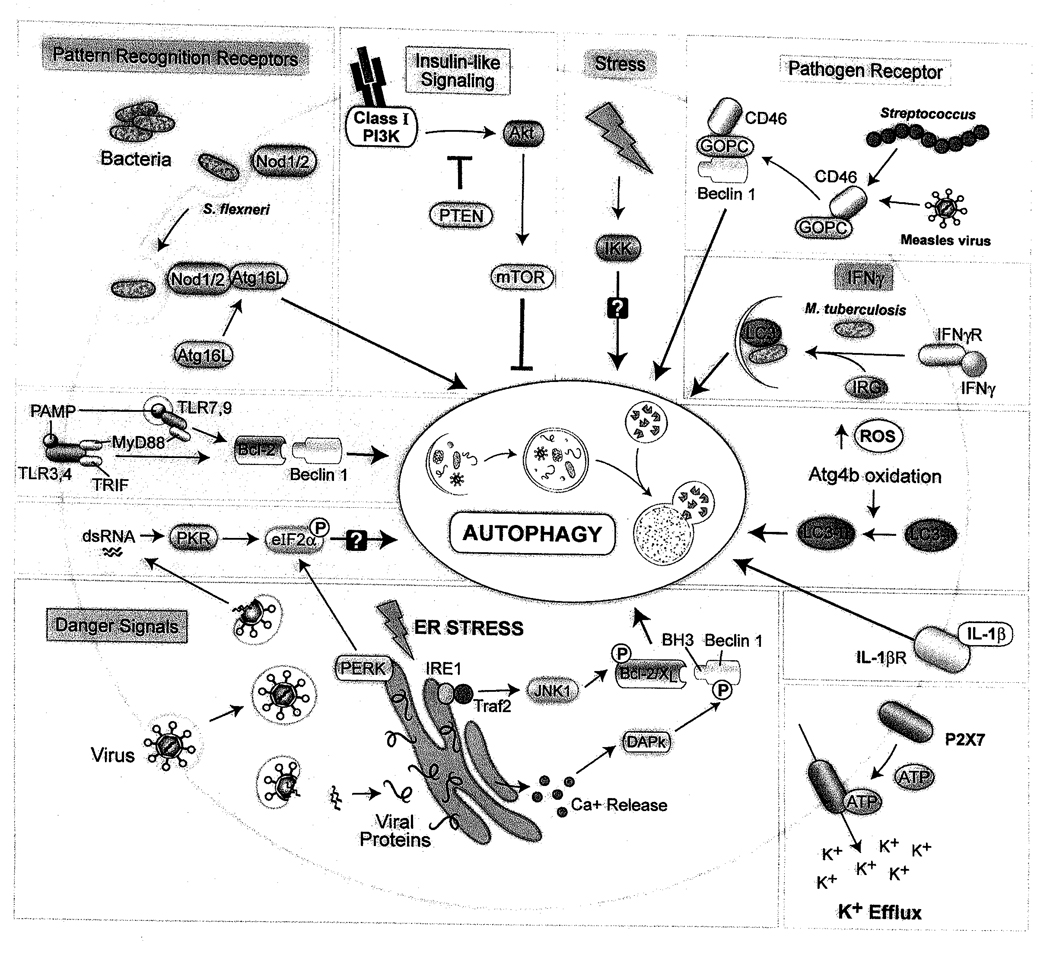
The sensing of PAMPs by PRRs or the activation of stress response pathways by DAMPs can trigger autophagy in a cell-autonomous manner. In addition, several pathogens trigger autophagy by mechanisms that may be independent of PAMPs or DAMPs or at least not yet known to involve these sensors. Moreover, autophagy induction via engagement of a pathogen receptor has recently been described [22]. Cytokines such as IFNγ produced during infection can also regulate autophagy in target cells to control intracellular pathogens. In this section, we provide an overview of innate immunity-associated pathways that have been linked with activation of autophagy (Figure 1).
Figure 1.
Cellular pathways that function in triggering autophagy during infection.
2.1. Pattern recognition receptors (PRRs)
An important clue that PRRs may exert antimicrobial effects in vivo through autophagy induction was provided by studies with L. monocytogenes infection in Drosophila [9]. Mutation of the PRR, PGRP-LE, or hemocyte-specific deletion of Atg5 increased animal lethality, suggesting a critical role for both PGRP-LE and autophagy in host defense against Listeria in vivo. The PRR, PGRP-LE, mediated autophagy induction in response to the Listeria PAMP, diaminopimelic acid-containing peptidoglycan (DAP-PGN), and controlled intracellular bacterial growth in cultured primary hemotocytes or a macrophage-like cell line through an autophagy-dependent mechanism. Thus, although not formally proven, it is reasonable to speculate that autophagy induction by the PRR PGRP-LE may be important for controlling Listeria infection in vivo. Listeria-induced autophagy was independent of imd and Toll, the two major known PAMP signal transduction pathways in Drosophia [23], suggesting that autophagy induction by PRRs may utilize downstream components that are distinct from other innate immune signaling pathways. In addition, lysine-type PGN induced autophagy in Drosophila cells lacking PRGP-LE, suggesting the existence of additional PRRs that are capable of activating autophagy.
2.2. Toll-like receptors (TLRs)
TLRs are a family of Type I integral membrane proteins that recognize PAMPs via their leucine-rich regions (LRR) to activate proinflammatory cytokine and Type I interferon (IFN) production via NF-κB-, MAPK- and interferon-regulatory factor (IRF)-dependent pathways [20]. The function of TLRs is phylogenetically conserved and to date, 12 members (TLR1-12) have been identified in mammals. Several of the mammalian TLRs have now been linked to autophagy induction [24].
In the first report of TLR-induced autophagy [25], the Gram-negative bacterial cell wall component lipopolysaccharide (LPS) stimulated autophagy (as defined by the formation of GFP-LC3 punctae, LC3-II conversion, and electron microscopy) in primary human alveolar macrophages and a murine macrophage cell line. In the murine macrophage cell line, LPS-induced GFP-LC3 punctae formation was decreased by TLR4 siRNA or the expression of dominant negative mutant TLR4. LPS-induced autophagy also required the TLR adaptor TRIF and the downstream TLR signaling components RIP1 and p38 MAPK, but not the TLR adaptor MyD88. Moreover, incubation of macrophages with LPS caused increased colocalization of Mycobacterium tuberculosis (which does not contain LPS) with autophagosomes, resulting in decreased mycobacterial survival.
The role of specific TLRs in autophagy induction was further examined by Delgado et al [26]. The authors found that TLR3, TLR4, and TLR7 agonists, but not TLR1/2, TLR5, or TLR9 agonists, induced autophagy in murine macrophages. Similar to the findings of Xu et al. with TLR4, they found that TLR7 ligand-dependent autophagy decreased the intracellular survival of the BCG vaccine strain of M. tuberculosis (although in this study they found that such autophagy required the adaptor MyD88). An interesting question is therefore, whether heterologous infections (or specific treatment with TLR ligands), through TLR-dependent activation of autophagy, may augment host defense against mycobacteria and other intracellular infections. Another open question is whether the differential role of MyD88 in TLR-dependent autophagy in different studies reflects different TLR family member specific signaling pathways or other factors.
In addition to these two studies, there are also other more recent reports that demonstrate that LPS (a TLR4 ligand) and PAM3CYS4 (a TLR1/2 ligand) induce autophagy in primary dendritic cells [27] and that LPS induces autophagy in bone marrow-derived macrophages (BMDMs) [28]. Although the role of TLR signaling was not specifically investigated in these studies and the authors only measured autophagy induction using the GFP-LC3 punctae formation assay, taken together, these studies are consistent with the model that diverse TLRs may function in autophagy activation. One potential biochemical mechanism has been proposed by which TLR signaling may activate autophagy; upon TLR stimulation, the TLR adaptors, TRIF and MyD88, interact with the autophagy protein, Beclin 1, resulting in the disruption of its interaction with Bcl-2 [29], which normally functions to suppress autophagy [30, 31]. Further studies are required to define additional mechanisms by which TLRs activate the autophagy pathway.
Despite the growing consensus that TLR signaling activates autophagy, not all studies are consistent with this conclusion. For example, Saitoh et al. found that LPS fails to induce autophagy in primary macrophages [32]. The basis for the discrepancy between their findings and other reports is unknown. Another interesting finding is that ATG genes may be important in controlling cellular responses to TLR agonists in a manner that this does not involve classical macroautophagy. Sanjuan et al [33] used latex beads coated with TLR agonists to model phagocytosis in macrophages. They observed TLR agonist-dependent LC3 lipidation, recruitment of GFP-LC3 and GFP-Beclin 1 to the site of phagocytosis, and more rapid phagosomal maturation in a manner that required the essential ATG genes Atg5 and Atg7 and was reversed by PI3K inhibitors. However, EM studies failed to reveal the presence of classical double-membraned autophagosomes, leading the authors to postulate a unique role for the autophagic machinery in TLR agonist-induced phagocytosis. As discussed in more detail below, although Saitoh et al did not find that TLR agonists such as LPS induce autophagy [32], they found that ATG genes were important for modulating TLR-activated proinflammatory cytokine production. Finally, another interesting point regarding TLR signaling and autophagy is that, not only may TLR signaling activate autophagy, but autophagy may also function to activate TLR signaling. Lee et al. found that autophagy was essential for delivery of viral nucleic acids to endosomal TLRs and Type I IFN production in plasmacytoid dendritic cells infected with vesicular stomatitis virus [34]. Thus, there appears to be extensive cross-talk between TLRs, autophagy, and innate immune responses.
2.3. The Nod-like receptor (NLR) family
Like TLRs, the NLR family is evolutionarily ancient, with homology in domain organization to the plant resistance genes that sense PAMPs and DAMPS during pathogen infection [17]. The mammalian NLR family contains >20 members; all members contain a central NACHT domain, most members contain a C terminal LRR domain, and NLRs primarily differ in their N-terminal domains, which mediate interactions with downstream signaling molecules. NLRs can be further subdivided into three distinct subfamilies based on their domain structure: the Nods (which primarily activate signaling via NF-κB and MAPK), and the NALPs and IPAF/NAIP (which are components of the “inflammasome”) [17]. In recent years, links between the autophagy pathway and members of each NLR subfamily have emerged that implicate autophagy in important executioner and regulatory roles in NLR signaling.
Two recent studies have shown that Nod1 and Nod2 proteins are PRRs that function in autophagy induction in response to bacterial PGN. Travassos et al [28] found that Nod1 agonists induce autophagy in epithelial cells and that Nod2 agonists induce autophagy in myeloid cells (including in wild-type but not Nod2−/− mouse peritoneal macrophages). Nod1-deficient MEFs were defective in targeting Shigella flexneri ΔIcsB (a mutant that does not inhibit autophagy) and L. moncytogenes to GFP-LC3 -positive structures (presumably autophagosomes) and had increased intracellular S. flexneri ΔIcsB bacterial proliferation. Similarly, bone marrow-derived monocytes from mice with a frameshift mutation in Nod2 (1007insC) associated with Crohn’s disease were deficient in bacterial autophagy. The authors postulate that, unlike Nod1- and Nod2-dependent activation of inflammatory signaling, Nod-dependent activation of bacterial autophagy is independent of the adaptor kinase RIP2 and of NF-κB transcription factor activity since RIP2-deficient macrophages display normal S. flexneri ΔIcsB-induced autophagy (but impaired NF-κB-dependent chemokine secretion).Instead, they propose the interesting model that the Nod proteins may act as nucleating factors for the initiation of bacterial autophagy by recruiting the autophagy protein, ATG16L1, to the bacterial entry site. However, it not yet clear how Nod1 or Nod2 activation leads to ATG16L1 recruitment to the sites of bacterial entry, whether such recruitment is sufficient for the initiation of Nod-dependent autophagy or other as of yet unidentified signals are required, and precisely how Nod-ATG16L1 interactions regulate bacterial autophagy. Of note, while cells from donors homozygous for the Crohn’s disease risk-associated ATG16L1 T300A allele show impaired autophagy following stimulation with Nod ligands, they do not show a deficiency in their interaction with Nod1 or Nod2. Thus, although this polymorphism may affect Nod-dependent autophagy, such an effect is likely to be independent of direct interactions with Nod proteins.
Cooney et al [27] found that muramyldipeptide (MDP), a breakdown product of PGN that functions as a NoD2 and NALP3 ligand, induced autophagy in human dendritic cells. MDP-induced autophagy in dendritic cells was abrogated by siRNA against NOD2, RIP2, and the ATG genes, ATG5, ATG7, and ATG16L1, but not by siRNA against NALP3 or MyD88. Interestingly, the authors report that MDP-induced autophagy enhanced surface MHC Class II antigen expression, and found that NOD2 or ATG16L1 knockdown reduced antigen-specific CD4 T cell proliferative responses to dendritic cells infected with a Salmonella vector expressing tetanus toxin. Moreover, dendritic cells from Crohn’s disease patients with NOD2 or ATG16L1 risk alleles had impaired induction of autophagy in response to MDP, but not to the TLR2 ligand PAM3CYS4. In addition, dendritic cells with Crohn’s disease-associated NOD2 mutations had decreased colocalization of S. typhimurium and adherent invasive E. coli with lysosomes, and increased survival of intracellular adherent invasive E. coli. Thus, the authors propose that NOD2 stimulation induces autophagy in dendritic cells, which in turn, enhances MHC Class II antigen presentation and bacterial targeting for lysosomal degradation. As a corollary, they propose that defects in NOD2 and/or ATG16L1-dependent autophagy could contribute to features of Crohn’s disease, such as bacterial persistence and altered immune regulation.
Taken together, these studies provide convincing evidence that Nod1/2 induce autophagy upon sensing bacterial PAMPs. Given the broad expression pattern of Nod1 (in epithelial cells and fibroblasts) and Nod2 (in myeloid cells and Paneth cells) and given that PGN is common to many bacterial pathogens, these findings will likely have broad implications for our understanding of how autophagy is triggered by bacterial infection. Moreover, it is reasonable to speculate that Nod1/2-dependent autophagy may function to restrict intracellular bacterial multiplication, but formal proof of this concept will require evidence that Nod1/2 restriction of intracellular bacteria requires classical macroautophagy, at both a genetic and cellular level. Although perhaps unlikely, it is theoretically possible that GFP-LC3-positive structures may colocalize with intracellular bacteria in a manner that is independent of classical macroautophagy, and it is also possible that NLRs activation may enhance bacterial targeting to lysosomes in an autophagy-dependent manner. It is also reasonable to speculate that Crohn’s disease-associated NOD2 or ATG16L1 risk alleles may contribute to Crohn’s disease through impaired autophagy activation by bacterial PAMP stimulation. However, it should be noted that there are conflicting results in the literature regarding whether the ATG16L1 T300A risk allele results in impaired bacterial autophagy [35, 36] and this question has not been examined in cells from Crohn’s disease patients with the ATG16L1 T300A risk allele. Thus, further studies are required to more clearly elucidate the role that deregulation of autophagy plays in the pathogenesis of Crohn’s disease with NOD2 or ATG16L1 polymorphisms. Also, given the discrepant results discussed above, it remains uncertain whether RIP2 is involved in Nod-mediated autophagy activation, and it is yet unknown what other downstream signaling molecules are involved in triggering bacterial PAMP-induced Nod1/2-dependent autophagy.
Another interesting question is whether Nod2 may also mediate virus-induced autophagy. Sabbah et al [37] recently reported the surprising finding that Nod2 interacts with viral single stranded RNA (ssRNA) to induce IFN-β production utilizing the adaptor protein MAVS (rather than RIP2). Nod2-deficient mice produced less IFN-β in response to respiratory syncytial virus (RSV) or H1N1 influenza infection and were more susceptible to lethal RSV infection. Although the relationship between Nod2 function and autophagy was not examined in this study, the question arises whether, in addition to bacterial ligand-induced autophagy, Nod2 may also function in virus-induced autophagy. As a corollary, it will be interesting to determine whether bacterial ligands, similar to viruses, may also signal through MAVS via Nod2.
2.4. dsRNA-dependent protein kinase (PKR)
The dsRNA-dependent protein kinase (PKR) is a ubiquitously expressed cytosolic serine/threonine kinase that, along with GCN2, PERK and HRI, is one of the four mammalian eIF2α kinases that regulate protein translation in response to cellular stresses [38]. PKR was the first antiviral effector protein discovered [39] and, as its name implies, it is a PRR that binds dsRNA PAMPs produced during viral infection [19]. In yeast, GCN2 is the sole eIF2α kinase and induces translational arrest during periods of nutrient deprivation [38]. Talloczy et al [40] found that GCN2 was also required for starvation-induced autophagy and that PKR can functionally complement starvation-induced autophagy in GCN2-null yeast. Moreover, MEFs deficient in PKR or containing a non-phosphorylatable knock-in mutation in eIF2α were deficient in virus-induced autophagy. Of note, the HSV-1 essential neurovirulence gene product, ICP34.5, which recruits protein phosphatase lα to dephosphorylate eIF2α [41, 42], inhibited starvation- and virus-induced autophagy [40], and an HSV-1 mutant virus lacking ICP34.5, but not wild-type HSV-1, was efficiently recruited to autophagosomes in cultured cells [43]. Interestingly, the HSV-1 ICP34.5 protein was later shown to target autophagy at an additional level by directly binding to Beclin 1 and HSV-1 lacking the 20 amino acid Beclin 1-binding domain is highly attenuated in a mouse model of herpes simplex encephalitis [12]. The neurovirulence of this mutant virus, but not that of viruses that are neuroattenuated by virtue of mutations outside the ICP34.5 gene, was fully restored in pkr−/− mice, providing genetic proof that PKR lies upstream of the autophagy execution pathway in vivo. The precise mechanisms by which PKR activation and eIF2α phosphorylation trigger autophagy during viral infection remain unknown and represent an important avenue for further investigation.
2.5. The IKK complex
In canonical NF-κB signaling, the IKK complex, consisting of the kinases IKKα and IKKβ as well as the regulatory subunit IKKγ (NEMO), is activated by the kinase TAK1 and transduces NFκB-activating signals by phosphorylating IκB. This induces its polyubiquitination and degradation, thus permitting the translocation of the p50/p65 NF-κB transcription factor to the nucleus, where it participates in the transcriptional response to a wide variety of stresses [44]. In noncanonical IKK signaling, the kinase NIK specifically activates the IKKα kinase, which phosphorylates p100, leading to its proteolytic processing to the transcription factor p52 and heterodimerization with Rel-B to activate gene transcription [44]. Signaling through the noncanonical pathway appears to be relatively restricted to signaling via members of the TNF superfamily of receptors such as BAFF and CD40, while the canonical pathway is activated by most of the PAMP sensing pathways, including all TLRs, RLRs, Nod1 and Nod2 [17, 20].
Earlier studies demonstrated that both the canonical and noncanonical NF-κB pathways could be negatively regulated by targeting IKK and NIK for autophagic degradation [45–47]. Djavaheri et al also found that NF-κB transcription factor activity could suppress TNFα-mediated autophagy [48], suggesting crosstalk between the two pathways. Recently, Criollo et al [49] reported that constitutive activation of the IKK pathway (using either transfected constitutively active IKK mutants or membrane-localized NEMO) results in autophagy induction. Furthermore, they showed that known autophagy-inducing stimuli such as starvation and rapamycin induce IKK activation (although they did not examine the effects of infection or microbial PAMPs). Interestingly, IKK induction of autophagy was associated with certain common triggers of autophagy, such as p53 depletion, mTOR inhibition, AMPK and JNK1 activation, and disruption of the Bcl-2/Beclin 1 complex, but was genetically independent of NF-κB.
Collectively, these studies provide evidence of crosstalk between the NF-κB or IKK and autophagy pathways and suggest that induction of IKK may represent an important common pathway for induction of autophagy by many stress signals. It will be critical in future studies to precisely define the specific contexts in which IKK activation can induce autophagy as well as upstream and downstream components of the pathway. For example, all of the PRR pathways discussed above are known to activate IKK signaling [19, 20], raising the intriguing question of whether this may be a crucial mechanism by which they activate autophagy during infection.
2.6. Endoplasmic reticulum (ER) stress, the unfolded protein response, and calcium release
Viral pathogens usurp the host translational machinery to produce large quantities of virally-encoded proteins, in many cases overwhelming the folding capacity of the ER [50]. The ER serves as sentinel for viral infection by activating the unfolded protein response (UPR), which involves signaling pathways initiated by three sensor proteins: IRE1, PERK and ATF6, that act to decrease protein translation and translocation into the ER and to increase its folding capacity [16]. Preliminary evidence indicates that at least two of the three UPR sensor proteins, IRE1 and PERK, may play a role in autophagy induction.
In response to ER stress, IRE1 activates chaperone genes and also becomes autophosphorylated and recruits TRAF2, which initiates a signaling cascade that results in JNK activation [51]. Ogata et al found that activation of the UPR with ER stressors induced autophagy, and this effect was abrogated in IRE1-deficient cells or in cells treated with a JNK inhibitor, but not in PERK-deficient cells or cells with ATF6 knockdown [52]. The role of IRE1 and JNK in virus-induced autophagy has not yet been formally studied, but the prediction is that ER stress induced by virus infection might mediate autophagy in an IRE1- and JNK-dependent manner. Although PERK was not required for autophagy in the study by Ogata et al., PERK has been shown to be required for autophagy induced by polyglutamine expansion proteins and hypoxia [53, 54]. A proposed mechanism is PERK-dependent transcriptional upregulation of certain autophagy genes, such as LC3 and Atg5, which may be required to maintain the pool of proteins needed for autophagosome expansion during periods of sustained stress and autophagy activation. As with IRE1, it is not yet known whether PERK is required for autophagy induction during viral infection.
Although the mechanism remains poorly understood, stimuli that result in activation of the UPR also result in the release of calcium stored within the ER [55]. Increased intracellular calcium levels are sensed by CaMKKβ, which downregulates mTOR signaling by activating AMPK [56]. Increased intracellular calcium levels also result in the activation of DAPk, which phospohorylates Beclin 1 within its BH3 domain, resulting in decreased affinity for the Bcl-2 antiapoptotic family member Bcl-XL and Bcl-2 and induction of autophagy [57, 58]. Taken together, these data provide additional evidence that the ER is a central transducer of stress signals that activate autophagy. Additional studies are warranted to examine the potential roles of the ER, UPR, and calcium release in pathogen-triggered autophagy.
2.7. Insulin-like signaling
The insulin-like, Class I PI3K, Akt/Tor signaling pathway plays an evolutionarily conserved role in negatively regulating autophagy during development and starvation in diverse eukaryotic organisms, including flies, worms, and mammals [3, 59–61]. Two recent studies in model organisms suggest that this signaling pathway also negatively regulates pathogen-induced autophagy. In a Drosophila model of VSV infection, Shelly et al. found that suppression of the PBK/Akt/Tor signaling pathway (by overexpression of PTEN, a phosphatase that blocks PI3K signaling, expression of a deleted form of an adaptor that couples the insulin receptor to the catalytic subunit of PI3K, or RNAi targeting of Akt) suppresses VSV replication in a manner that requires ATG genes [10]. Similiarly, in a C elegans model of S. typhimurium infection, Jia et al. found that pathogen resistance conferred by a loss-of-function mutation in the insulin-like tyrosine kinase receptor, daf-2, or by overexpression of the forkhead transcription factor, DAF-16, requires several key components of the autophagic machinery [62]. Together, these studies suggest that the inactivation of insulin-like/class I PI3K/Akt signaling may represent a crucial host defense response that fights viral and intracellular bacterial infections through autophagy activation. Further studies are needed to examine this hypothesis in mammalian models of infection.
2.8. Interferon-γ(IFNγ)
IFNγ is a member of the type I cytokine family with pleiotropic effects on innate and adaptive immunity [63]. IFNγ signaling is critical for the control of intracellular pathogens [64] in part through its ability to regulate the expression and function of the immunity-related GTPases (IRGs), which represent a family of ~20 members in mice while only a single IRG (IRGM) is found in humans [65]. The protective effects of IFNγ and IRGs were initially linked to autophagy in vitro by the observation that treatment of macrophages with IFNγ or enforced expression of the murine IRG LRG-47 or human IRGM results in the targeting of mycobacteria-containing vacuoles for autophagic degradation [66, 67].
The mechanisms by which IFNγ or IRGs function to activate autophagy is not yet understood, and the role of IFNγ and IRG-mediated autophagy activation has yet to be evaluated in vivo. It is possible that defects in autophagy may underlie the markedly enhanced susceptibility to lethal M. tuberculosis infection observed in LRG-47−/− mice [68]. In humans, mutations in the IFNγ receptor result in increased susceptibility to infection with intracellular pathogens, especially mycobacteria [64] and a deletion in the promoter of IRGM (leading to decreased mRNA levels) is associated with increased susceptibility to Crohn’s disease [69]. Although reductions in IRGM1 levels are associated with decreased autophagy and impaired elimination of intracellular mycobacteria in vitro [67, 69] it is not yet known whether patients with the Crohn’s disease-associated IRGM deletion polymorphism have altered autophagy or susceptibility to intracellular bacterial infections. Adding to the complexity, autophagy proteins have also been found to interact with IRGs in ways that may not involve their role in classical macroautophagy. For instance, Atg5 is required to target Irga6 to the damaged parasitophorous vacuoles in macrophages infected with T. gondii [11]. Future studies will be important to further elucidate the precise roles of IFNγ and the IRGs in pathogen-induced autophagy and in protection against Crohn’s disease.
2.9. Activation of autophagy by binding to a pathogen receptor
Recently, a ubiquitous cell surface receptor, CD46, that binds several pathogens, has been shown to trigger autophagy [22]. Joubert et al. found that engagement of CD46 with cross-linking antibodies induces Atg5-, Atg7- and PI3K-dependent autophagy in HeLa cells. Moreover, they found that the adaptor protein GOPC (also known as PIST, FIG or CAL) interacted with the cytoplasmic tail of a CD46 splice variant (Cyt-1), was required for CD46-mediated activation of autophagy, and interacted with Beclin 1 via its coiled-coil domain. Interestingly, a murine neuron-specific isoform of GOPC, nPIST, was previously shown to interact with Beclin 1 [70], and GOPC has been implicated in intracellular trafficking of cell surface receptors [71]. The knockdown of CD46 or GOPC blocked autophagy induction during in vitro infection with measles virus and the knockdown of GOPC reduced Group A Streptococcus targeting to autophagosomes without affecting bacterial invasion (while the knockdown of CD46 reduced both autophagic targeting of Group A Streptococcus and bacterial invasion) Based on these findings, the authors conclude that CD46 can trigger autophagy upon pathogen recognition through a mechanism that involves GOPC and downstream ATG genes.
It is as yet unknown whether CD46 induces autophagy in response to pathogen recognition in primary cells or in vivo. It is also not yet known whether the knockdown of CD46 and GOPC blocks measles-induced autophagy through a direct mechanism or through preventing uptake of the virus and subsequent recognition by other PRRs. Furthermore, murine cells lack CD46 but still are capable of inducing autophagy in response to Group A Streptococcus [72]. Thus, it will be interesting to determine whether other cell surface pathogen receptors besides CD46, in addition to other families of PRRs described above, trigger bacterial-induced autophagy in murine cells.
2.10. Microbes or microbial proteins that trigger autophagy by an unknown mechanism
There are several examples of microbes or microbial proteins that induce autophagy for which the mechanism underlying autophagy induction is not known. For example, toxins encoded by diverse bacteria, including Vibrio cholerae cytolysin [73], Bacillus anthracis lethal factor [74] and Helicobacter pylori vacuolating cytotoxin [75] induce autophagy. Although the mechanism(s) by which these toxins induce autophagy is not known, it could be due to the triggering of PRRs that recognize protein PAMPs or the activation of DAMP receptors (i.e. due to pore formation by toxins such as Vibrio cholerae cytolysin). The HIV envelope glycoprotein induces autophagy in uninfected bystander CD4 T cells [76]. This requires CD4 and CXCR4 cell surface expression, but is independent of CD4 and CXCR4 signaling and may somehow, through an as-of-yet undefined mechanism, involve the fusigenic activity of HIV gp41 [77] Unlike VSV infection in Drosophila cells [10], a recent study showed that only live, and not UV-inactivated, Sindbis virus induced autophagy in MEFs and neuronal cells [13], indicating that a pre-formed PAMP is not invariably sufficient for virus-induced autophagy. Rather, for certain viruses, it seems likely that cytoplasmic sensors of viral nucleic acids may play a heretofore unrecognized role in autophagy activation.
3. Targeting of pathogens to autophagosomes
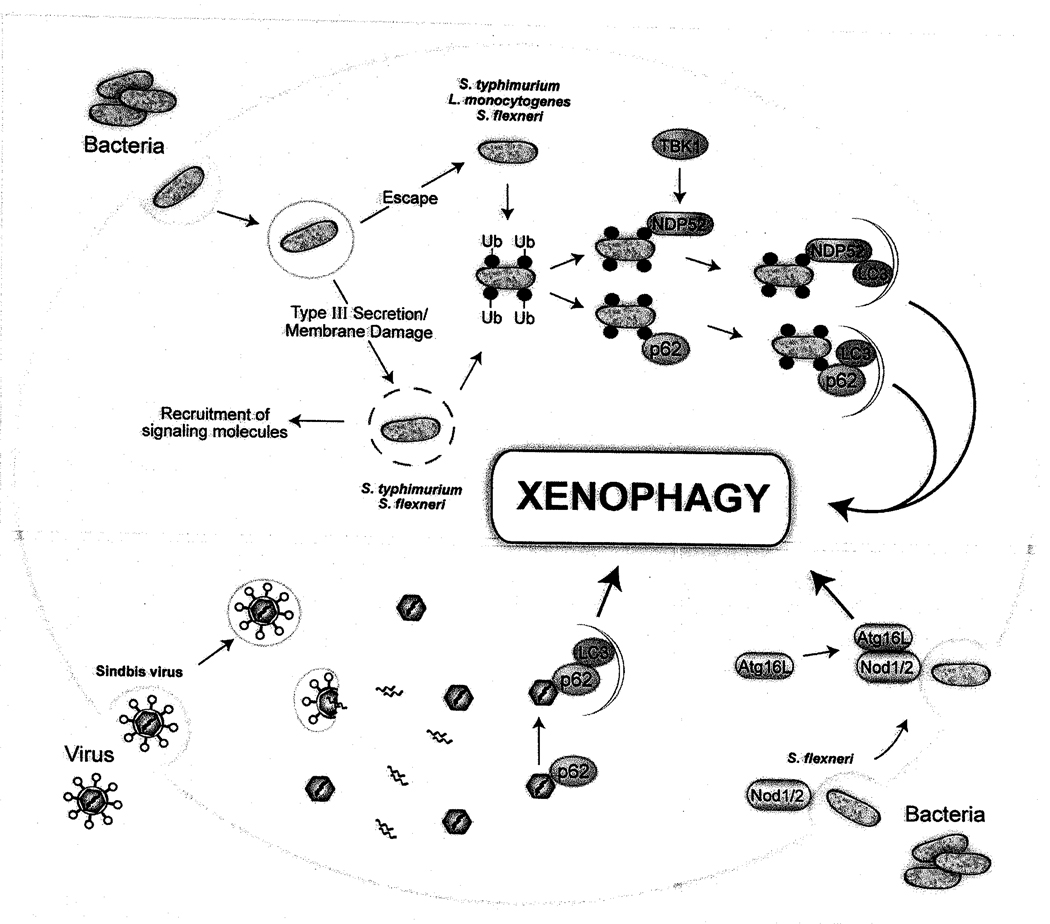
In 1966 De Duve wondered “whether this intriguing process is essentially blind and accidental or discriminating and directed” [78]. It is now becoming clear that, while autophagy under basal conditions may indeed be stochastic, under many forms of cellular stress, autophagy is a selective process that can target specific damaged organelles, aberrant protein aggregates, and intracellular pathogens [79]. Recent studies provide some important clues as to how this may be accomplished, namely (but not exclusively) by utilizing adaptor molecules that act as bridges between ubiquitin-tagged cargoes and the autophagic machinery. In addition, these adaptor molecules may also serve as scaffolds for assembling not only the autophagic machinery but also other stress-activated host signaling molecules to integrate the innate immune response. In this section, we will review recent studies that have elucidated potential molecular mechanisms by which intracellular bacteria and viral nucleocapsids can be targeted to autophagosomes (Figure 2).
Figure 2.
Mechanisms of targeting pathogens to autophagosomes.
3.1. Bacterial targeting
A link between ubiquitin and autophagy first emerged with the discovery that intracellular ubiquitin-positive protein aggregates accumulate in Atg5- and Atg7-deficient mice, resulting in neurodegeneration [80−82]. The targeting of ubiquitinated cargoes for autophagic degradation has since been linked to recognition by adaptor proteins such as p62 and NBR1, both of which can simultaneously bind ubiquitin via a UBA domain and LC3 via a LC3-interacting region (LIR) domain in order to target ubiquitinated cargoes for degradation [83–85]. Early on, there were also some hints that ubiquitination might play a role in the control of intracellular bacterial infection. For example, Perrin et al found that S. typhimurium, which normally resides in intracellular vacuoles, became coated with ubiquitin upon escape into the cytoplasm of the host cell and that the cytosol-adapted pathogen, L. monocytogenes, avoided recognition by the ubiquitin system by using actin-based motility [86]. Subsequently, it was found that disruption of the vacuolar membrane by the bacterial S. typhimurium Type III secretion system (T3SS) was necessary for ubiquitin association [87]. In this study, the authors also showed that ubiquitin-associated S. typhimurium often colocalized with LC3 and that intracellular bacterial survival was increased in Atg5−/− MEFs [87].
Evidence from several recent studies now links the accumulation of ubiquitin around the intracellular bacteria S typhimurium, S. flexneri and L. monocytogenes with autophagy adaptor proteins, suggesting a mechanism by which bacteria may be targeted for xenophagic degradation. In the first study, Zheng et al showed that cytosolic S. typhimurium becomes coated by a multilammellar membrane that is positive for p62, ubiquitin and LC3 [88]. Mutation of either the LIR or ubiquitin-binding domains of p62 reduced the association of LC3 with ubiquitin-associated bacteria, and knockdown of p62 resulted in decreased bacterial-LC3 colocalization and increased bacterial replication. Finally, increased numbers of p62-associated bacteria were observed in Atg5-deficient cells.
In the second study, Thurston et al [89] expanded on previous work that implicated the IKK-related kinase TBK1 in the control of bacteria-containing vacuolar integrity and intracellular Salmonella, enteropathogenic E. coli, and S. pyogenes replication in a manner that was independent of TBK1 effects on interferon regulatory faetor-3 and Type I IFN production [90, 91]. Thurston et al. found that the TBK1 adaptors Nap1 and SINTBAD (but not TANK) colocalized with ubiquitin-associated Salmonella, and that their in vitro binding to ubiquitin required an additional cellular factor, NDP52 (nuclear dot protein 52). The authors then demonstrated that NDP52 is a novel autophagy adaptor protein that, like p62, contains LIR and ubiquitin-binding domains and restricts intracellular bacterial replication.
In the third study, Yoshikawa et al found that L. monocytogenes ActA mutants (that are defective in actin-based motility and fail to avoid recognition by ubiquitin) were targeted to the autophagosomes by a mechanism that involves ubiquitination, and recruitment of p62 and LC3 [92]. This process required the ATG gene Atg5 and resulted in decreased bacterial survival, whereas in p62-deficient MEFs, the survival of wild-type and ActA bacterial mutants were equivalent. Interestingly, by studying the effects of mutations in different domains of ActA on interactions with LC3, the authors found that motility was not necessary for Listeria to escape autophagic recognition. Rather, the ability of ActA to recruit the Arp2/3 complex (which catalyzes actin nucleation) and Ena/VASP was necessary for avoidance of ubiquitination and autophagic targeting. Thus, not only are cytosolic bacteria targeted by ubiquitin/p62-dependent-mechanisms for autophagic degradation, but they have evolved specific mechanisms to antagonize such targeting. Perhaps possession of a successful strategy to antagonize autophagic targeting is a requirement for successful cytosolic bacterial replication. It remains unknown precisely how the recruitment of Arp2/3 and Ena/VASP allows Listeria to escape ubiquitination and autophagic recognition, but Yoshikawa et al. propose the interesting theory that by exploiting the ability of Act to recruit host cell cytoskeletal proteins, the bacterium may disguise itself as a host cell organelle.
Finally, Dupont et al. demonstrated that Shigella phagocytic vacuolar membrane remnants (that are generated by bacterial T3SS-dependent membrane damage) are polyubiquitinated, recruit the p62 adaptor and autophagy protein LC3, and are targeted for autophagic degradation. Interestingly, these membrane remnants are also associated with galectin 3 [93], a member of the lectin family that has been proposed to have roles both as a PRR and as a DAMP [94], and numerous other molecules involved in sensing and transduction of PAMP and DAMP signals including Nod1, Ipaf, Asc, caspase-1, TRAF6 and NEMO. In cells deficient in autophagy (either by virtue of dominant negative Atg4b expression or Atg5 mutation), polyubiquitinated proteins and p62 accumulated on membrane remnants, which was associated with an increase in early NF-κB-dependent cytokine production, reactive oxygen species production, and necrotic cell death. Thus, the paradigm that emerges from these studies is that the ubiquitin, p62-dependent autophagic targeting of ruptured membranes that occurs during pathogen phagocytosis helps to control detrimental downstream signaling during bacterial invasion into host cells.
A few major themes are noteworthy in the studies of bacterial targeting to the autophagosome. First, the cellular machinery involved in such targeting may parallel that which is involved in specific targeting of host proteins and organelles, at least with respect to the prominent role for ubiquitin and p62 in autophagic targeting. It is not yet known whether novel autophagic adaptors such as NDP52 identified in studies of bacterial targeting are also involved in the targeting of host cargo, and whether there are “bacterial specific targeting factors”. Second, cytosolic bacteria such as Listeria escape autophagic recognition, perhaps by “masquerading as host organelles”. Third, intracellular membranes that are disrupted by bacterial secretion systems somehow serve as triggers for autophagy and perhaps act as sites of assembly of signaling cascades that regulate other innate immunity pathways.
Several important unanswered questions remain regarding bacterial targeting. It is unclear what signals alert the cell to the presence of membrane damage or cytosolic bacteria and how such signals are transmitted to initiate the recruitment of ubiquitin, adaptor molecules and the autophagy machinery. It is also unclear whether bacterial products are ubiquitinated directly or whether they are associated with ubiquitinated host proteins, and the identity of the host ubiquitin ligases remains to be determined. Furthermore, while the utilization of components of the autophagy pathway appears to be conserved, it is unclear whether the same molecular events are involved in bacterial-specific autophagy as general autophagy.
3.2. Viral targeting
By comparison to bacteria, very little is presently known about the targeting of viruses for xenophagy. However, Orvedahl et al. recently demonstrated an interaction between Sindbis virus capsid protein and p62, and found that p62, as well as autophagy execution genes, were required for the colocalization of Sindbis virus proteins to autophagosomes [13]. Genetic knockdown of p62 resulted in increased Sindbis virus capsid accumulation and accelerated cell death, without affecting viral replication. Moreover, in vivo, genetic inactivation of Atg5 in virally-infected mouse neurons resulted in the delayed clearance of Sindbis virus capsid, the accumulation of p62 aggregates, increased neuronal death, and increased animal mortality. Taken together, these results suggest that p62 adaptor-mediated autophagic viral protein clearance promotes cell survival. Interestingly, Sindbis virus capsid is not ubiquitinated, and virally-infected Atg5-deficient neurons that display p62 aggregates do not have ubiquitin aggregates, suggesting that, at least certain viruses, may be targeted by the p62 adaptor for autophagic degradation via molecular tags other than ubiquitin. It will be important to define the precise identity of such tags, as well as the signaling events that trigger viral targeting to autophagosomes.
Another important question is whether p62 serves a more general role as an adaptor for autophagic targeting of other virus families. Along these lines, even before p62 was known to be an autophagy adaptor protein, naturally occurring mutations in the Drosophila melanogaster p62 homolog Rel(2)P were associated with varying susceptibility to infection with the sigma rhabdovirus in vivo [95, 96] and Rel(2)P was shown to co-immunoprecipitate with the sigmavirus nucleocapsid (N) and phosphoprotein (P) in infected cells [97]. Thus, further studies are warranted to explore the role of p62 as an autophagic adaptor protein that functions more broadly in antiviral host defense.
4. Tuning innate immunity by the autophagic machinery
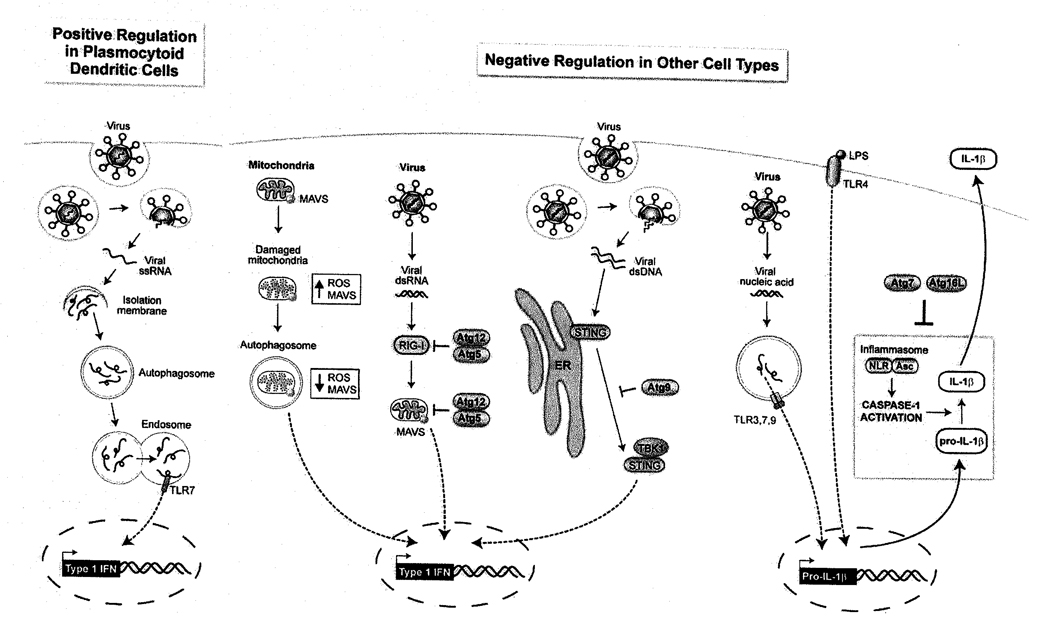
A general theme is that activation of the autophagy machinery is one of many branches of a conserved host response pathway. To date, no “autophagy-specific” immunological stimuli have been found; rather, the induction of autophagy is one part of a multifaceted host response that acts in conjunction with other innate immune pathways. Not only is autophagy induced as part of a multifaceted host response, but increasing evidence suggests that autophagy may “fine-tune” other aspect of innate immune responses, including Type I IFN production, inflammasome signaling, and RLR signaling (Figure 3).
Figure 3.
Roles of the autophagy machinery in “tuning”innate immunity.
4.1. Autophagy proteins and Type I IFN production
Type I IFN production is a central component of the innate immune response to viral pathogens and is important for survival during viral infections, as genetic deletion of the Type I IFN receptor renders mice highly susceptible to lethal infection with many viruses [98]. Production of Type I IFN is induced by PRRs that detect viral nucleic acid and results in the transcriptional regulation of hundreds of interferon-stimulated genes (ISGs) that collectively produce an “antiviral state” [99]. In most cell types, RLRs activate Type I IFN production in response to RNA virus infection [20]. However, a small subset of specialized dendritic cells called plasmacytoid dendritic cells (pDCs) primarily utilize TLR7- and TLR9-mediated signaling and produce large quantities of Type I IFN upon virus infection [100].
In the first study to describe “tuning” of the innate immune response by the autophagy pathway, Lee et al found that ATG genes can positively regulate Type I IFN production, as they are required for delivery of viral ssRNA to endosomal TLR7 in pDCs both in vitro and in vivo during VSV infection [34]. In contrast, ATG genes can negatively regulate RLR-mediated induction of Type I IFN in MEFs both via conjugation of Atg5-Atg12 to CARD domains of RLR signaling molecules including RIG-I and MAVS [101] and by controlling ROS and MAVS levels through elimination of dysfunctional mitochondria [102]. Atg9 has also recently been discovered to downregulate interferon-stimulatory DNA (ISD)-mediated Type I IFN production in MEFs, but this effect was independent of the essential ATG gene Atg7 [103]. Thus, ATG genes can positively or negatively regulate Type I IFN signaling in a cell-type and stimulus-dependent manner, and in at least some cases this involves non-canonical roles of ATG genes. Further studies are required to determine the importance of these interactions in influencing the outcome of viral infections in vivo. It should also be noted that in each of these studies, the authors observed parallel effects on pro-inflammatory cytokine production, indicating that “tuning” of innate antiviral immunity by the autophagy machinery is not limited to Type I IFN production.
4.2. Autophagy proteins and the inflammasome
Several members of the first two NLR subfamilies, namely NALP1, NALP3 and PAF, are PRRs that respond to PAMPs or DAMPs by assembling into macromolecular “inflammasome” complexes that process procaspase-1 to its active form, caspase-1, resulting in the cleavage of pro-IL-1β and pro-IL-18 to their active, secreted forms (Martinon and Tschopp).
Several lines of observation suggest that the autophagy and inflammasome pathways may functionally intersect. On the one hand, IL-1β, one of the main products of inflammasome activation, is an inducer of autophagy [104], and DAMPs that trigger autophagy such extracellular ATP/K+ efflux [105] and reactive oxygen species (ROS) [106] are also known to activate inflammasome signaling [107]. On the other hand, certain components of the inflammasome may negatively regulate autophagy. For example, Suzuki et al found that autophagy induction is enhanced in S. flexneri-infected macrophages deficient in Ipaf or caspase-1 but not in the inflammasome adaptor Asc [108]. Intriguingly, there is also strong genetic evidence that ATG genes may negatively regulate the secretion of the inflammasome products, the pro-inflammatory cytokines, IL-1β and IL-18. Saitoh et al found that deletion of Atg16L1, Atg7 or pharmacologic inhibition of autophagy in primary murine embryonic fibroblasts (MEFs) resulted in dramatically increased IL-1β secretion in response to stimulation with LPS or viral nucleic acid PAMPs without alterations in transcriptional induction of IL-lβ, suggesting a specific role for basal autophagy in the downregulation of caspase-1-mediated pro-IL-1β conversion in primary MEFs [32]. The authors also found that ROS generation and K+ efflux were required for enhanced IL-1β secretion, and that the levels of ROS were higher in Atg16L1-deficient MEFs, as has been seen with other autophagy-deficient cell lines [102]. Moreover, they found that mouse chimeras engrafted with Atg16l1−/− fetal liver hematopoietic progenitors have higher serum concentrations of IL-1β and IL-18 after treatment with dextran sodium sulfate. While the precise mechanisms by which ATG genes negatively regulate pro-inflammatory secretion remain unknown, these data provide important evidence that the autophagy machinery plays a crucial role in titrating inflammatory responses in vivo.
4.3. Autophagy proteins and RIG-I-like receptor (RLR) signaling
The RIG-I-like receptor (RLR) family includes the RNA helicases RIG-I and MDA-5, which activate IRF3, NF-κB and MAPK signaling in response to RNA virus infection [20, 109]. RIG-I is also activated by dA:dT rich dsDNA via the generation of RNA intermediates by RNA pol III [110, 111]. In contrast to TLRs 3, 7, 8 and 9, which sense viral nucleic acids within endosomal compartments, RLRs are thought to participate in cytoplasmic antiviral surveillance [112].
While no studies have proven a direct role for RLRs in autophagy activation, several reports have found that autophagy down-regulates RLR signaling, albeit via distinct (though not necessarily mutually exclusive) mechanisms. Jounai et al found that the CARD domains of RIG-I, MDA-5 and their downstream adaptor MAVS are conjugated by Atg5-Atg12, which results in the downregulation of type I IFN and NF-κB-dependent cytokine production induced by VSV infection [101]. Tal et al. showed that defective clearance of mitochondria led to increased ROS production, resulting in the amplification of RLR signaling (and increased type I IFN production) in autophagy-deficient MEFs [102]. These studies contrast with the role played by ATG genes in the delivery of TLR7 ligands to endosomes in plasmacytoid dendritic cells which resulted in increased VSV-induced Type IFN production [34], suggesting that autophagy may regulate virus-induced Type I IFN production in a cell-type and PRR pathway-dependent manner. Data from in vivo model systems of viral infection are very limited, but recently Orvedahl et al. found that neuronal deletion of the ATG gene Atg5 had no effect on CNS Type I IFN production in neonatal mice with Sindbis virus infection [13].
4.4. Autophagy proteins and STING
STING (also known as MITA, MPYS, ERIS) is a recently discovered transmembrane protein that is required in vitro for efficient activation of type I IFN and pro-inflammatory cytokine production in response to transfected interferon-stimulatory DNA (ISD) or infection with VSV, Sendai virus, HSV-1 and L. monocytogenes [113] [114] [115] [116]. STING-deficient mice are defective in their CD4 and CD8 T cell responses to DNA vaccines and are highly susceptible to lethal infection with HSV-1 [117]. While STING is typically localized to the mitochondria or ER in unstimulated cells, after transfection with ISD or infection with HSV-1, it redistributes to perinuclear punctate structures, where it colocalizes with TBK1 and the exocyst component Sec5 [117].
A recent study suggests a novel potential link between components of the autophagic machinery and regulation of STING function. In response to ISD transfection, Saitoh et al found that STING translocated from the ER to cytoplasmic punctate structures (that lack double membranes ultrastructurally) where it assembled with TBK, and certain components of the autophagy pathway, including LC3, Atg9a, and p62, but not with other components of the autophagy pathway, such as ULK1 or Atg14. Genetic disruption of Atg9a, but not the ATG gene, Atg7, resulted in increased dsDNA-induced STING translocation from the Golgi to cytoplasmic punctate structures, increased assembly of STING and TBK, and aberrant innate immune activation, including increased IRF3 phopshorylation, increased transcription of IFNβ, IL-6, and Cxcl10, and increased IFNβ prodcution. Thus, the autophagy protein, Atg9a, appears to have a unique function in controlling dsDNA-mediated, STING-dependent innate immune signaling. Together with the data suggesting a role for Atg5 in controlling RLR signaling and for Atg16L1 in controlling inflammasome activity, these data suggest that multiple different autophagy proteins act to “tune” innate immune signaling, functioning in a counterregulatory role. These functions of autophagy proteins appear to be independent of “classical macroautophagy”, as they are not conserved across all autophagy proteins that are essential for macroautophagy and do not seem to involve the formation of autophagosomes. The elucidation of the precise mechanisms by which autophagy proteins regulate innate immune signaling in an autophagy-independent manner is an important area of future investigation.
5. Conclusion
The phylogenetic conseveration of the autophagy pathway attests to its fundamental importance to eukaryotic life and, in its role in xenophagy, it may represent one of the most evolutionarily ancient forms of host defense. It is not surprising to find that, later in evolution, with the emergence of multifacted immune responses to pathogens, there also emerged complex interrelationships between the autophagy pathway and other innate immune pathways. To date, all cellular sensors and pathways that are implicated in triggering pathogen-induced autophagy have had previously characterized roles in innate immune pathways. Recent advances discussed in this review indicate that, in addition to these previously characterized roles in innate immunity, a crucial conserved biological function of pattern recognition molecules, damage sensors, and cellular stress signaling pathways may be the activation of autophagy during infection with intracellular pathogens.
There is also increasing evidence that cells use a conserved set of molecular machinery to target unwanted “self-constituents” that they use to target unwanted “microbial invaders”. This machinery of selective autophagy centers largely upon ubiquitination, a signal already known to be important in regulate innate immune responses, as well as adaptor proteins such as p62 and NDP52 that bridge ubiquitin-tagged substrates to the autophagosomal protein, LC3. However, as further research on targeting unfolds, it seems likely that distinct forms of selective autophagy will be found to exist for different classes of pathogens. Moreover, the autophagic targeting of different classes of pathogens may have different biological outcomes, perhaps in cell type-specific fashions, with respect to pathogen degradation, regulation of innate and adaptive immune responses, and the consequences of such targeting for cell and organismal survival.
Finally, there is growing evidence that autophagy is involved in “tuning” the innate immune response and that such “tuning” may exert either positive or negative regulatory effects depending on the cell-type and stimulus. This paradigm may be operative in the pathogenesis of Crohn’s disease, as recent studies suggest that a central pathology may be a lack of autophagy protein-mediated downregulation of pro-inflammatory cytokine production in response to environmental innate immune stimuli. An interesting general emerging theme is that autophagy proteins may exert negative regulatory control on inflammatory signaling through autophagy-independent mechanisms. Thus, the conserved autophagy machinery may have dually evolved to both target pathogens for lysosome degradation through classical autophagy, as well as to prevent, through mechanisms not yet fully understood, detrimental inflammatory responses triggered by such pathogens.
In summary, great strides have been made in the past few years in our understanding of autophagy and innate immunity. Recent discoveries offer tantalizing clues as to how the autophagy pathway and other innate immune pathways are integrated to produce a concerted host defense. Yet, we are only beginning to appreciate the sophistication and complexity of such integration. A more precise understanding of cross-talk between autophagy and other aspects of innate immunity will be required before we can develop specific therapies that trigger, target or tune the innate immune response to combat pathogens and control auto-inflammatory diseases.
Acknowledgements
We thank Kathryn Sumpter for critical reading of the manuscript and Angela Diehl for expert medical illustration. The work in the authors’ own laboratory was supported by the Ellison Medical Foundation Senior Scholars Award in Infectious Diseases (B.L.), NIH RO1 AI151367 (B.L), and NIH T32 AI070116 (R.S.).
References
- 1.de Duve C. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He C, Klionsky D. Regulation mechanisms and signaling pathways of autophagy. In Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Münz C. Enhancing immunity through autophagy. In Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 6.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang X, Kleeman L, Jiang H, Gordon G, Goldman J, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;8:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host & Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 16.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 17.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 18.Benko S, Philpott DJ, Girardin SE. The microbial and danger signals that activate Nod-like receptors. Cytokine. 2008;43:368–373. doi: 10.1016/j.cyto.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 19.García MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Molecular Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joubert PE, Meiffren G, Grégoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain P-O, Vidal M, Lotteau V, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Kurata S. Extracellular and intracellular pathogen recognition by Drosophila PGRP-LE and PGRP-LC. International Immunology. 2010 doi: 10.1093/intimm/dxp128. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delgado MA, Deretic V. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009;16:976–983. doi: 10.1038/cdd.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delgado M, Elmaoued R, Davis A, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson D, Campbell B, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2009;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 28.Travassos L, Carneiro L, Ramjeet M, Hussey S, Kim Y, Magalhães J, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 29.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 33.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Cornell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 34.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 35.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem. 2009;284:32602–32609. doi: 10.1074/jbc.M109.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts WK, Hovanessian A, Brown RE, Clemens MJ, Kerr IM. Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature. 1976;264:477–480. doi: 10.1038/264477a0. [DOI] [PubMed] [Google Scholar]
- 40.Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ1 34.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 42.He B, Gross M, Roizman B. The γ1 34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talloczy Z, Virgin HW, IV, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 44.Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006 doi: 10.1126/stke.3572006re13. rel3. [DOI] [PubMed] [Google Scholar]
- 45.Qing G, Yan P, Qu Z, Liu H, Xiao G. Hsp90 regulates processing of NF-κB2 p100 involving protection of NF-κB-inducing kinase (NIK) from autophagy-mediated degradation. Cell Res. 2007;17:520–530. doi: 10.1038/cr.2007.47. [DOI] [PubMed] [Google Scholar]
- 46.Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IκB kinase (IKK) Cell Res. 2006;16:895–901. doi: 10.1038/sj.cr.7310109. [DOI] [PubMed] [Google Scholar]
- 47.Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 48.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, Pierron G, Codogno P. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 49.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2009;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 51.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 52.Ogata M, Hino S-i, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 54.Rouschop KMA, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 56.Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 57.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of Beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- 59.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 60.Mcphee CK, Baehrecke EH. Autophagy in Drosophila melanogaster. Biochim Biophys Acta. 2009;1793:1452–1460. doi: 10.1016/j.bbamcr.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meléndez A, Levine B. WormBook. 2009. Aug 24, Autophagy in C. elegans; pp. 1–26. [DOI] [PubMed] [Google Scholar]
- 62.Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci USA. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 64.van de Vosse E, van Dissel JT, Ottenhoff THM. Genetic deficiencies of innate immune signalling in human infectious disease. Lancet Infect Dis. 2009;9:688–698. doi: 10.1016/S1473-3099(09)70255-5. [DOI] [PubMed] [Google Scholar]
- 65.Taylor GA, Feng CG, Sher A. Control of IFN-γ-mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases) Microbes Infect. 2007;9:1644–1651. doi: 10.1016/j.micinf.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 67.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 68.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 69.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N. A novel protein complex linking the δ2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35:921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 71.Cheng J, Wang H, Guggino WB. Modulation of mature cystic fibrosis transmembrane regulator protein by the PDZ domain protein CAL. J Biol Chem. 2004;279:1892–1898. doi: 10.1074/jbc.M308640200. [DOI] [PubMed] [Google Scholar]
- 72.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 73.Gutierrez MG, Saka HA, Chinen I, Zoppino FCM, Yoshimon T, Bocco JL, Colombo MI. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V. cholerae. Proc Natl Acad Sci USA. 2007;104:1829–1834. doi: 10.1073/pnas.0601437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan YK, Kusuma CM, St John LJ, Vu HA, Alibek K, Wu A. Induction of autophagy by anthrax lethal toxin. In Biochemical and Biophysical Research Communications. 2009;379:293–297. doi: 10.1016/j.bbrc.2008.12.048. [DOI] [PubMed] [Google Scholar]
- 75.Terebiznik MR, Raju D, Vázquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimon T, Colombo MI, Jones NL. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370–379. doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- 76.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008;4:998–1008. doi: 10.4161/auto.6880. [DOI] [PubMed] [Google Scholar]
- 78.de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 79.van der Vaart A, Mari M, Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–289. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 80.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 81.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 82.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 84.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 85.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun J-A, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 86.Perrin AJ, Jiang X, Birmingham CL, So NSY, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 87.Birmingham C, Smith A, Bakowski M, Yoshimori T, Brumell J. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 88.Zheng Y, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell J. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 89.Thurston T, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 90.Radtke AL, O'Riordan MXD. Homeostatic maintenance of pathogen-containing vacuoles requires TBK1-dependent regulation of aquaporin-1. Cell Microbiol. 2008;10:2197–2207. doi: 10.1111/j.1462-5822.2008.01199.x. [DOI] [PubMed] [Google Scholar]
- 91.Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MXD. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 93.Paz I, Sachse M, Dupont N, Cederfur C, Enninga J, Leffler H, Poirier F, Prevost MC, Lafont F, Sansonetti P. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010 doi: 10.1111/j.1462-5822.2009.01415.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 94.Sato S, St-Pierre C, Bhaumik P, Nieminen J. Galectins in innate immunity: dual functions of host soluble beta-galactoside-binding lectins as damage-associated molecular patterns (DAMPs) and as receptors for pathogen-associated molecular patterns (PAMPs) Immunol Rev. 2009;230:172–187. doi: 10.1111/j.1600-065X.2009.00790.x. [DOI] [PubMed] [Google Scholar]
- 95.Dru P, Bras F, Dezélée S, Gay P, Petitjean AM, Pierre-Deneubourg A, Teninges D, Contamine D. Unusual variability of the Drosophila melanogaster ref(2)P protein which controls the multiplication of sigma rhabdovirus. Genetics. 1993;133:943–954. doi: 10.1093/genetics/133.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gay P. [Drosophila genes which intervene in multiplication of sigma virus (author's transl)] Mol Gen Genet. 1978;159:269–283. doi: 10.1007/BF00268263. [DOI] [PubMed] [Google Scholar]
- 97.Wyers F, Dru P, Simonet B, Contamine D. Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J Virol. 1993;67:3208–3216. doi: 10.1128/jvi.67.6.3208-3216.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 100.Liu Y-J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 101.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin K, Ishii K, Kawai T, Akira S, Suzuki K, et al. The Atg5-Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tal M, Sasai M, Lee H, Yordy B, Shadel G, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat\ Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Biswas D, Qureshi OS, Lee WY, Croudace JE, Mura M, Lammas DA. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol. 2008;9:35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: a sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Inohara N, Sasakawa C, Nunez G. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baum A, García-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids. 2009 doi: 10.1007/s00726-009-0374-0. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase Ill-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 113.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 115.Jin L, Waterman PM, Jonseher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]