Abstract
The ability to genetically modify mesenchymal stem cells (MSCs) seeded inside synthetic hydrogel scaffolds would offer an alternative approach to guide MSC differentiation. In this report, we explored gene transfer to MSCs seeded on top, or inside matrix metalloproteinase (MMP) degradable hydrogels that were loaded with DNA/poly(ethylene imine) (PEI) polyplexes. DNA/PEI polyplexes were encapsulated inside poly(ethylene glycol) (PEG) hydrogels crosslinked with MMP degradable peptides via Michael Addition chemistry. Gene transfer was visualized and quantified through using a vector encoding for green fluorescent protein and luciferase. We found that gene transfer to MSCs was possible for cells seeded both in two and three dimensions. The amount of luciferase expression was similar for cells seeded in two and three dimensions even though the number of cells in three dimensions is significantly higher, indicating that gene transfer to cells seeded in two dimensions is more efficient than for cells seeded in three dimensions. The use of hydrogel scaffolds that allow cellular infiltration to deliver DNA may result in long lasting signals in vivo, which are essential for the regeneration of functional tissues.
Introduction
Tissue regeneration aims to promote the healing of diseased or injured tissue through the use of a biodegradable scaffold that supports cellular infiltration and contains bioactive signals that guide invading cells through tissue formation [1, 2]. During morphogenesis, extracellular matrix (ECM) proteins, cytokines, growth factors, cell-cell contacts and mechanical stimuli provide signals to cells that result in cell fates that, when orchestrated, result in tissue or organ formation [3]. The goal when designing scaffolds for tissue regeneration is to design an environment that remodels or changes as the cells infiltrate and proliferate so that all stages of tissue formation can be guided and that the end product is a completely natural tissue. Although peptides and growth factors are generally utilized as the bioactive signals in tissue engineering scaffolds, others and we have investigated the use of DNA delivery as an alternative or complementary approach to introduce bioactive signals into tissue engineering scaffolds [4–9]. In this approach the cells themselves produce the protein signals through the delivery of DNA that encodes for the desired protein.
Gene delivery can be achieved with the use of modified viruses (viral delivery) or polymers (non-viral delivery) that encapsulate or condense plasmid DNA into particles that can transport the DNA inside the cell. Although viral delivery is generally more efficient than non-viral approaches, it has limitations due to its potential immunogenicity and insertion mutagenesis [10]. Because of the mentioned safety concerns, non-viral approaches have been investigated. Poly(ethylene imine) (PEI) is a cationic polymer that has been widely utilized for non-viral gene delivery. PEI is able to condense DNA through electrostatic interactions between the positively charged amines in the PEI and the negatively charged phosphates on the DNA, forming particles in the range of 50 to 200 nm [11]. DNA/PEI polyplexes enter the cell through endocytosis and are believed to be able to escape the endosome through endosomal buffering (the proton sponge effect, [12, 13]). PEI has been successfully used in vitro and in vivo delivering DNA or siRNA to the brain [14, 15], lungs [16–19], abdomen [20], and tumors [21–23].
DNA delivery from tissue engineering hydrogel scaffolds is a versatile approach to promote the expression of tissue inductive factors locally to be used as signals to promote tissue formation. Naked DNA, as well as complexed DNA, has been incorporated into hydrogel scaffolds including collagen [5], pluronic-hyaluronic acid [24], PEG-poly(lactic acid)-PEG [25], engineered silk elastin [26], fibrin [9, 27], and PEG-hyaluronic acid [28] hydrogels. However, gene transfer efficiency remains a major limitation. We are interested in investigating gene transfer to MSCs in matrix metalloproteinase (MMP) degradable hydrogels due to their potential use to transfect MSC-like cells in vivo as they infiltrate the hydrogel scaffold. MMP-degradable hydrogels have been shown to allow cell migration through proteolytic degradation [29, 30] and to support cell growth in vivo [31, 32] and in vitro [29, 30, 33–35]. Thus, we believe that these hydrogels are interesting materials to explore the delivery of genes to MSCs as they infiltrate the hydrogel scaffold.
We previously reported on gene transfer to mouse MSCs inside MMP degradable PEG hydrogels using secreted reported plasmids [36]. However, 2D gene transfer and location of gene transfer within the hydrogel were not studied. In this report, we explored the delivery of PEI/DNA polyplexes from MMP degradable PEG hydrogels to MSCs grown on top or inside the hydrogel scaffolds using a vector encoding for both green fluorescent protein and luciferase. We wanted to determine if gene transfer could be achieved to cells seeded on top of DNA-loaded hydrogels and how long the expression lasted for. The ability to transfect cells slowly as the gel degrades and achieve long term transgene expression would increase the applicability of non-viral gene transfer for the genetic modification of cells. In the current report, we used previously established MMP-degradable hydrogels [30–35] and the methods we developed previously to incorporate DNA/PEI polyplexes through the bulk of the matrix with high activity [36].
Methods
Materials
Peptides GCRDGPQGIWGQDRCG (MMPxl) and Ac-GCGWGRGDSPG-NH2 (RGD) were obtained from NEOMPS (Genescript, Piscataway, NJ). ECM Gel is a comparable product to matrigel was purchased from Sigma-Aldrich (St. Louis, MO). Green fluorescent protein-luciferase expression vector (pEGFP-LUC, BD Biosciences, Palo Alto, CA) was expanded using an endotoxin free Giga Prep kit from Qiagen following the manufacturer’s instructions. Linear poly(ethylene imine) (25 kg/mole, PEI) was purchased from Polysciences (Warrington, PA). All other products were purchased from Fisher Scientific unless noted otherwise.
Cell culture
Mouse bone marrow cloned mesenchymal stem cells (mMSCs, D1, CRL12424) were purchased from ATCC (Manassas, VA, USA) and cultured in DMEM (Sigma-Aldrich) medium supplemented with 10% bovine growth serum (BGS, Hyclone, Logan, Utah) and 1% penicillin/streptomycin (Invitrogen, Grand Island, NY) at 37 °C and 5% CO2. The cells were split using trypsin following standard protocols.
PEG-VS synthesis
Four-armed PEG-OH (20 kDa, Nektar, Aerogen Ltd., Galway, IE) was modified with divinyl sulfone as previously described [36]. PEG-OH was dissolved in tetrahydrofluran (THF, Aldrich/Fluka, Basel, Switzerland) under inert atmosphere and heated to reflux in a Soxhlet apparatus filled with molecular sieves for 3–4 hours. The solution was allowed to cool to the touch and sodium hydride (NaH, Fluka, Basel, Switzerland), at 5-fold molar excess over OH groups, was added followed by the addition of divinyl sulfone, at 30-fold molar excess over OH groups. The reaction was carried out at room temperature (RT) under argon atmosphere with constant stirring for 3-days. Afterwards, the reaction solution was neutralized with acetic acid, filtered, concentrated and precipitated in ice-cold diethyl ether (Acros Organics/Chemie Brunschwig AG, Basel, Switzerland). Precipitation was repeated three times to remove unreacted divinyl sulfone. The final product was dried under vacuum and stored under argon at −20°C. The degree of functionalization was confirmed with 1H NMR (in CDCl3): 3.6ppm (PEG backbone), 6.1 ppm (d, 1H, CH2), 6.4 ppm (d, 1H, CH2), and 6.8ppm (dd, 1H, −SO2CH). The degree of end group conversion, as shown by NMR, was found to be 99%.
PEG hydrogel formation and DNA/PEI polyplex encapsulation
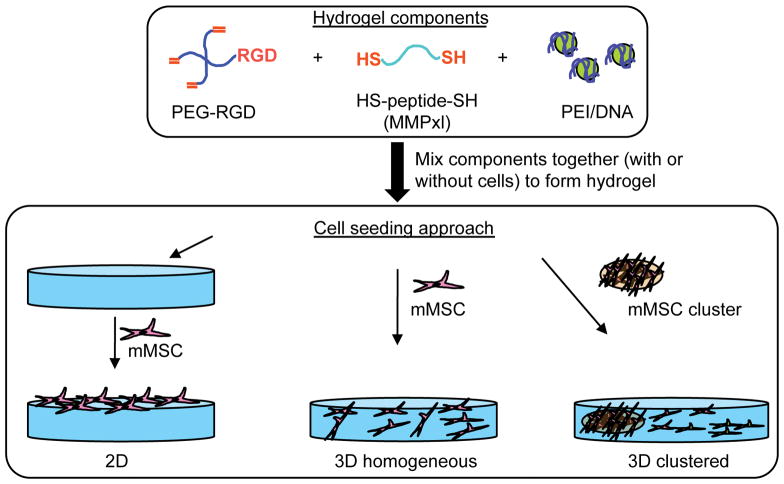
PEG hydrogels were formed by Michael-type addition of bis-cysteine containing MMPxl peptides onto four-armed poly(ethylene glycol)-vinyl sulfone (PEG–VS) functionalized with cell adhesion peptides (RGD peptides, 300μM) (Figure 1). Lyophilized aliquots of the crosslinker (0.91 mg MMPxl) were dissolved in 10 μL of 0.3 M triethanol amine (TOEA, pH=8.0) buffer immediately before mixing with a mixture of DNA/PEI polyplexes and PEG-VS. DNA/PEI polyplexes were formed by mixing plasmid DNA with PEI in DI water, vortexing for 15-s and incubating for 15-min at room temperature before adding 6.5 μL 3 M TOEA (final volume 65 μL) to the complex solution. A lyophilized aliquot of PEG-VS (6.5 mg) was dissolved in 25 μL of 0.3 M TOEA, mixed with 65 μL of DNA/PEI polyplex solution and 10 μL of MMPxl. The gel precursor solution was then placed either as a 100 μL drop or three 30 μL drops between two Teflon sheets (McMaster Carr, Elmhurst, IL) for 30-min at 37°C to allow for gelation. The final gel was swelled in DMEM medium before being placed inside 96-well plates for long-term culture.
Figure 1.
PEG hydrogels were formed through Michael addition of cysteine-containing matrix metalloproteinase sensitive peptides (HS-peptide-SH, MMPxl) with 4-armed PEG-vinyl sulfone pre-modified with cell adhesion peptides (PEG-RGD). Polyplexes (DNA/PEI) were encapsulated into hydrogel matrix by mixing with the precursor solution prior to gelation. Cells were seeded on top of the gel (2D), or inside the hydrogel matrix as single cells (3D homogeneous) or as a cluster of cells (3D clustered).
Cell Seeding
mMSCs cells were encapsulated using two protocols: homogeneous encapsulation, resulting in single cells throughout the hydrogel, and clustered encapsulation, resulting in a single cluster of cells inside a ECM gel clot (Figure 1). The cells were used between passages 3 and 10. For the homogeneously plated cells, the cells were encapsulated during gelation by mixing 45,000 cells in a 30 μL gel. The cells were mixed with the PEG-VS/polyplex solution before the crosslinker was added. For the clusters of cells, ECM gel clots were formed by resuspending D1 cells (2×104 cells) in 10 μL of DMEM and mixed with 10 μL ECM gel. The clusters were made by dropping 2.5 μL of the suspension onto a Teflon plate and incubating at 25 °C for 5 minutes. The clusters were incorporated into the PEG gel by placing them inside 30 μL of gel precursor solution. The resulting gel was pressed between two Teflon plates and gelled at 37°C for 30 minutes. The hydrogels were swelled in DMEM for two hours and cultured for up to 20 days in DMEM supplemented with 10% BGS and 1% PS. For cells cultured on top of the hydrogel (2D), the polyplex-loaded hydrogels were placed at the bottom of non-binding 96-well plates and mMSCs were plated (8000 cells/gel).
Gene Transfer Characterization
Gene transfer was characterized using a reporter plasmid encoding for green fluorescent protein (GFP) and luciferase. To assess the expression of GFP, inverted fluorescence microscopy was used at the indicated time points. For luciferase expression of cells cultured inside the hydrogel, cells were first collected from the hydrogel materials by degrading the gel with trypsin and then centrifuging the solution to pellet the cells. Hydrogels were degraded with trypsin by incubating the gels in a trypsin solution (0.25%) for 5 minutes and cells were recovered by centrifugation at 350g for 5 minutes. The collected cells were lysed and the luciferase activity was measured with Promega luciferase assay kit using the manufacturer’s instructions and a Luminometer (Modulus, Turner Biosystems, Sunnyvale, CA). Protein concentration was measured using the Bradford reagent, Coomassie Plus (Pierce, Rockford, IL), following the manufacture’s instructions.
Results and Discussion
The ability to genetically modify MSCs seeded inside synthetic hydrogel scaffolds would offer an alternative approach to protein delivery to guide their differentiation into functional tissues ex vivo. Additionally, MSC-like progenitor cells are believed to reside in most adult tissues and to be responsible for adult tissue regeneration. Therefore, the design of hydrogel materials that allow for cellular infiltration and deliver genes to infiltrating cells would be ideal to guide regeneration in vivo. In this report, we explored gene transfer to MSCs infiltrating MMP-degradable hydrogels that were loaded with DNA/PEI polyplexes. Cloned mouse bone marrow derived MSCs (D1) were used in this study as a model MSC cell type. D1 cells have similar characteristics to human MSCs and can be passaged up to 25 times under regular tissue culture conditions [37–41]. Cellular migration inside MMP degradable hydrogels has been demonstrated to occur primarily by proteolytic degradation of the scaffold [30]. Further, MMP activity inside MMP-degradable hydrogels has been found to be highest at the cell surfaces with MMP activity dramatically decreasing away from the cell surfaces [29]. Thus, inside MMP-degradable hydrogels cells can only degrade the gel immediately surrounding them and do not contribute to hydrogel degradation far away from their surface [29]. Gene transfer inside MMP-degradable PEG gels is, therefore, expected to occur as the cells proteolytically migrate through the hydrogel. Polyplexes will be internalized as the cells encounter them during the migration.
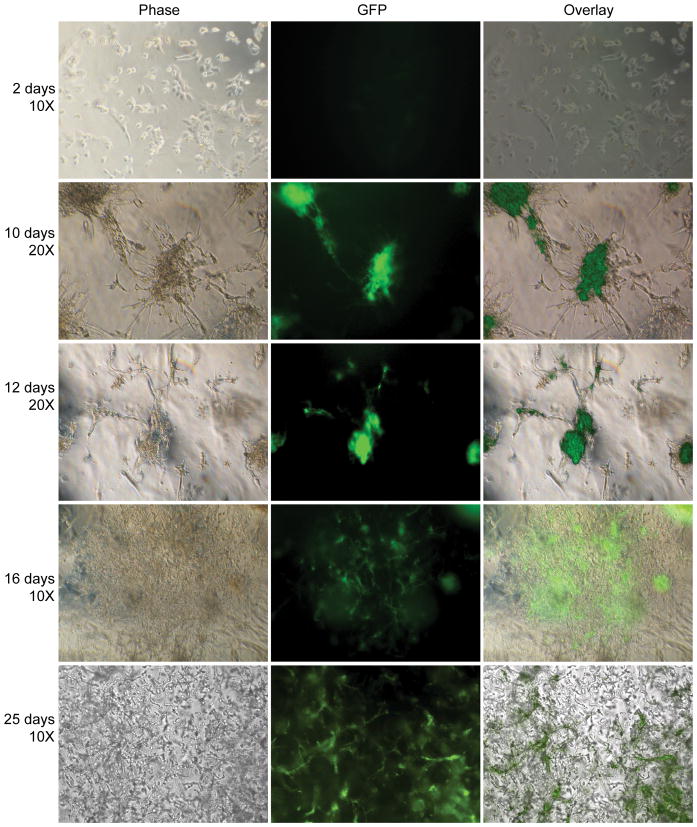
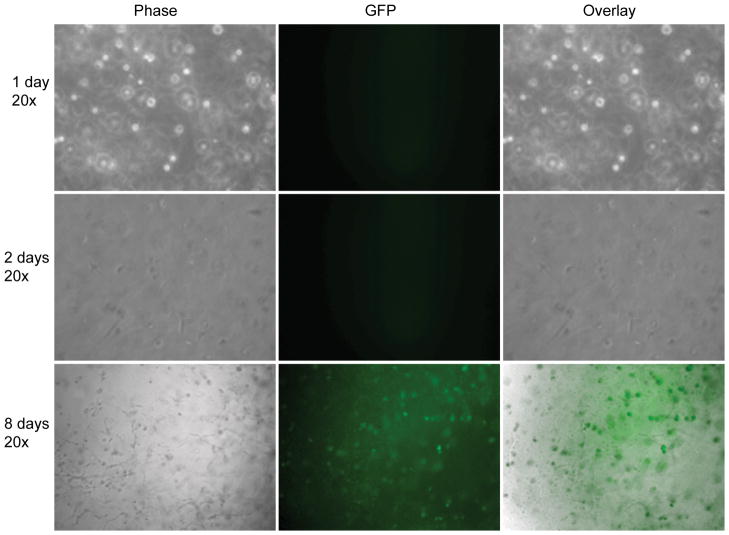
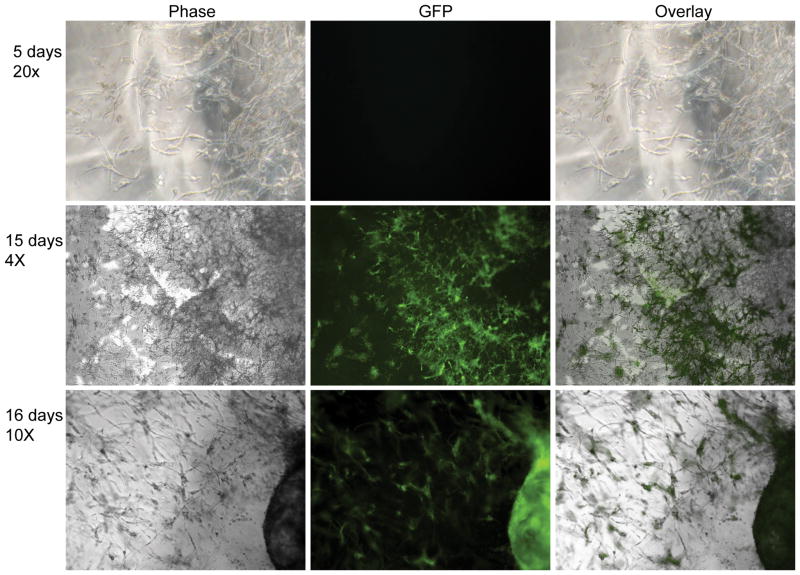
Two dimensional gene transfer from hydrogel scaffolds to mMSCs was assessed by plating cells directly on DNA/PEI loaded MMP degradable PEG hydrogels and gene expression was assayed for 25 days either through fluorescence microscopy (Figure 2) or through luciferase activity (Figure 5A). We have previously shown that the encapsulated polyplexes are not released unless the hydrogel is degraded [36]. Thus, transfection only occurs after the cells degrade the scaffold and the polyplexes are released. From phase and fluorescence images it was found that at early time point the cells spreaded over the entire surface and that GFP expression was weak and not easily observed with standard fluorescence microscopy (Figure 2, 2 days). However, as time progresses, cells became increasingly aggregated with cells binding to each other rather than the hydrogel, and with GFP gene expression being observed in the cell aggregates (Figure 2, 10 and 12 days). Interestingly, as the cells degraded the hydrogel, at later time points, the cells were able to spread through the entire surface and GFP gene expression was visible in a large number of cells (Figure 2, 16 and 25-days). These results indicate that initial cell attachment results through the RGDs presented at the hydrogel surface. However, as the cells proliferate the RGDs presented at the hydrogel surface becomes insufficient to mediate cell attachment and cells have more affinity for each other than for the hydrogel resulting in clumping. As the cell clumps degrade the matrix, more RGD peptides are available and the cell affinity for the matrix increases, leading to cells spread over the entire hydrogel surface and dissolution of the cell aggregates. GFP gene expression was highest at 2-days and 25-days when the number of cells was the greatest and the cells were fully spread on the hydrogel surface, indicating that the amount of DNA/PEI polyplexes released is proportional to the number of cells, the area that the cells spread on the surface. For mMSCs cultured inside the hydrogel scaffold GFP gene expression was observed starting at 8 and 15-days for cells cultured homogeneously (Figure 3) and clustered (Figure 4) respectively. Gene transfer for the clustered cells is delayed because the cells must diffuse out of the cluster before they can be transfected. Extensive gene transfer at 15 and 16 days was observed for the mMSCs cultured as a cluster compared to homogeneously plated cells. For mMSCs cultured homogeneously, most rounded cells expressed GFP, which suggest that the production of GFP or the lack of cell-cell contacts may induce a rounded morphology. For the cells cultured as a cluster, however, the cells expressing GFP were spread, which indicate that cell-cell contacts are important for cell spreading inside these hydrogels.
Figure 2.
Gene transfer to cells seeded on top of the hydrogel scaffold (2D). mMSCs (8,000 cells) were plated on top of MMP degradable PEG hydrogels (6.0% PEG, 300μM RGD concentration, and 15μg DNA at N/P = 12). Inverted fluorescence microscopy was used to visualize the transfected cells. All fluorescent images were taken using a one second exposure time.
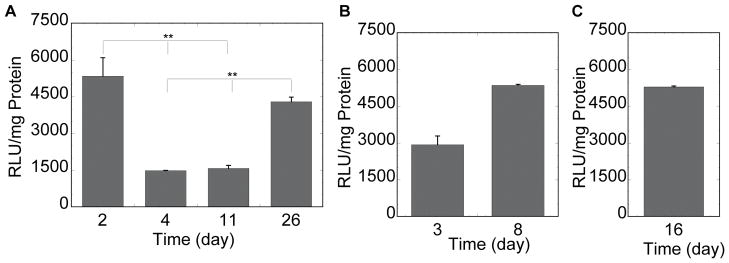
Figure 5.
Luciferase gene expression for cells seeded on and inside of DNA/PEI loaded hydrogels. mMSCs were plated on top (A) or inside (B, C) of MMP degradable PEG hydrogels. Initial cell densities were 8,000 cells for cells plated on top (A) of the gel, 150,000/100μL of gel for cells seeded inside the gel as single cells (B) and 5,000/cluster/30μL gel for cells seeded inside the gel as clusters (C). Luciferase activity was measured after hydrogel degradation at the time points shown. The data is plotted as a mean ± standard deviation. The symbol ** represents statistical significance to the level of p < 0.001 using the Tukey multiple comparisons test.
Figure 3.
Gene transfer to cells seeded inside of the hydrogel scaffold as single cells (3D homogeneous). mMSCs (150,000 cells) were seeded inside a 100μL MMP degradable PEG hydrogels (6.0% PEG, 300μM RGD concentration, and 15μg DNA at N/P = 12). Inverted fluorescence microscopy was used to visualize the transfected cells. All fluorescent images were taken using a one second exposure time.
Figure 4.
Gene transfer to cells seeded inside of the hydrogel scaffold as cluster (3D clustered). mMSCs (5000 cells) were clustered inside matrigel and seeded inside a 30μL MMP degradable PEG hydrogels (6.0% PEG, 300μM RGD concentration, and 15μg DNA/100μL gel at N/P = 12). Inverted fluorescence microscopy was used to visualize the transfected cells. All fluorescent images were taken using a one second exposure time.
To confirm that the observed fluorescence was indeed transgene expression, the hydrogels were degraded and cells were collected to measure luciferase activity at predetermined time points. For cells plated in 2D, luciferase gene expression was greatest at day 2 and day 26 with expression lowering at 4 and 11 days (Figure 5A), which correlates with the GFP expression profile for the later days of culture. Although the luciferase activity at day 2 was the same as that observed at 26 days, the GFP expression was not as visible at 2 days for the same exposure time. These results indicate that the amount of GFP expression per cells is lower at 2 days than it is at 26 days resulting in less visible GFP expression at 2 days and suggest that more cells are transfected at 2 days than 26 days with each cells producing less GFP and luciferase proteins. Although we originally anticipated that the amount of cells would affect the effectiveness of gene transfer from our hydrogel scaffolds, with more cells resulting in higher transgene expression, we found no correlation of cell number with transgene expression. Even though there are many more cells cultured inside the hydrogels than on top of the hydrogel (e.g. 30,000 versus 8,000 cells), the highest amount of luciferase transgene expression observed was comparable for all the seeding approaches (~4500 RLU/mg protein, Figure 5). Further, for the cells cultured in three dimensions the number of cells cultured in the cluster was much less than that in the homogeneous gel (5,000 versus 30,000 cells), but again the level of luciferase activity was similar (Figure 5B and C). These results suggest that the ability to internalize and process DNA/PEI polyplexes is limited for cells cultured in three dimensions. This result has been recently suggested also for cells cultured inside fibrin hydrogels loaded with lipid/DNA polyplexes [42]. Further, it suggests that when cells are seeded as a cluster, the cells are more efficient at degrading the gel, releasing the entrapped DNA and becoming transfected.
Unfortunately, it was difficult to determine the percent of cells transfected in these studies. In two dimensions, cell counting in clusters was not accurate and the low number of cells made flowcytometry analysis difficult. For the cells seeded inside the hydrogel, cell counting was even less accurate and flowcytometry analysis required the sacrifice of several hydrogels to get enough cells for accurate readings. We found that inverted fluorescence microscopy was the appropriate way to analyze GFP expression for cells seeded on or in hydrogel scaffolds. Although previously our laboratory [36] and others [29, 43] have reported green autofluorescence for cells culture in PEG hydrogels, we believe that the green fluorescence observed in this report is GFP expression because luciferase activity was also observed for these samples at similar time points. Further, the luciferase reading for untransfected cells is 50, which is at least one order of magnitude lower than that observed for the transfected samples. Although we were able to use traditional inverted fluorescence microscopy to image our hydrogel scaffolds, improvements to microscopy technology could greatly facilitate this type of research in the future. Microscopes capable of longer working distance objectives, without compromising brightness and speed would allow visualization deeper into the hydrogel scaffold. This type of technology would be of great interest to the tissue engineering community as a whole.
In conclusion, we have found that cells seeded on top of DNA/PEI-loaded enzymatically degradable hydrogel scaffolds are able to degrade the hydrogel and become transfected by the encapsulated polyplexes. Further, transgene expression was visible for up to 26 days. For cells seeded in three dimensions, the amount of transgene expression depended on the method of cell encapsulation. Cells seeded homogeneously resulted in less transgene expression than cells seeded in clusters, which suggest that matrix degradation and migration are key steps in the process of gene transfer for cell seeded inside hydrogel matrices. We believe that the ability to transfect cells slowly as the hydrogel matrix degrades could achieve the long term expression needed for the genetic modification of stem cells, without relying on the permanent insertion of the gene in the host chromosomes.
Acknowledgments
The authors would like to thank Talar Tokatlian (UCLA) for the helpful discussions and the Institute for Cell Mimetic Space Exploration (CMISE) at UCLA for the fluorescent microscope. We like to thank the National Institute of Health (NIH) and the National Institute for Bioimaging and Bioengineering (NIBIB) for funding (R21EB007730 (TS)).
References
- 1.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260(5110):920–6. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Suh H. Tissue restoration, tissue engineering and regenerative medicine. Yonsei Medical Journal. 2000;41(6):681–684. doi: 10.3349/ymj.2000.41.6.681. [DOI] [PubMed] [Google Scholar]
- 3.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 4.Ando S, et al. PLGA microspheres containing plasmid DNA: preservation of supercoiled DNA via cryopreparation and carbohydrate stabilization. J Pharm Sci. 1999;88(1):126–30. doi: 10.1021/js9801687. [DOI] [PubMed] [Google Scholar]
- 5.Bonadio J, et al. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5(7):753–9. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 6.Adami RC, et al. Stability of peptide-condensed plasmid DNA formulations. J Pharm Sci. 1998;87(6):678–83. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 7.Berry M, et al. Sustained effects of gene-activated matrices after CNS injury. Mol Cell Neurosci. 2001;17(4):706–16. doi: 10.1006/mcne.2001.0975. [DOI] [PubMed] [Google Scholar]
- 8.Segura T, Chung PH, Shea LD. DNA delivery from hyaluronic acid-collagen hydrogels via a substrate-mediated approach. Biomaterials. 2005;26(13):1575–84. doi: 10.1016/j.biomaterials.2004.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trentin D, et al. Peptide-matrix-mediated gene transfer of an oxygen-insensitive hypoxia-inducible factor-1 alpha variant for local induction of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2506–2511. doi: 10.1073/pnas.0505964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nature Reviews Genetics. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 11.Lungwitz U, et al. Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics. 2005;60(2):247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Boussif O, et al. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akinc A, et al. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. Journal of Gene Medicine. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 14.Abdallah B, et al. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Human Gene Therapy. 1996;7(16):1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, et al. Transgene expression in the brain stem effected by intramuscular injection of polyethylenimine/DNA complexes. Molecular Therapy. 2001;3(5):658–664. doi: 10.1006/mthe.2001.0324. [DOI] [PubMed] [Google Scholar]
- 16.Wiseman JW, et al. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Therapy. 2003;10(19):1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 17.Kichler A, et al. Intranasal gene delivery with a polyethylenimine-PEG conjugate. Journal of Controlled Release. 2002;81(3):379–388. doi: 10.1016/s0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 18.Rudolph C, et al. Nonviral gene delivery to the lung with copolymer-protected and transferrin-modified polyethylenimine. Biochimica Et Biophysica Acta-General Subjects. 2002;1573(1):75–83. doi: 10.1016/s0304-4165(02)00334-3. [DOI] [PubMed] [Google Scholar]
- 19.Gautam A, et al. Transgene expression in mouse airway epithelium by aerosol gene therapy with PEI-DNA complexes. Molecular Therapy. 2001;3(4):551–556. doi: 10.1006/mthe.2001.0300. [DOI] [PubMed] [Google Scholar]
- 20.Segura T, Schmokel H, Hubbell JA. RNA interference targeting hypoxia inducible factor 1alpha reduces post-operative adhesions in rats. J Surg Res. 2007;141(2):162–70. doi: 10.1016/j.jss.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 21.Iwai M, et al. Polyethylenimine-mediated suicide gene transfer induces a therapeutic effect for hepatocellular carcinoma in vivo by using an Epstein-Barr virus-based plasmid vector. Biochemical and Biophysical Research Communications. 2002;291(1):48–54. doi: 10.1006/bbrc.2002.6383. [DOI] [PubMed] [Google Scholar]
- 22.Aoki K, et al. Polyethylenimine-mediated gene transfer into pancreatic tumor dissemination in the murine peritoneal cavity. Gene Therapy. 2001;8(7):508–514. doi: 10.1038/sj.gt.3301435. [DOI] [PubMed] [Google Scholar]
- 23.Coll JL, et al. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy. 1999;10(10):1659–1666. doi: 10.1089/10430349950017662. [DOI] [PubMed] [Google Scholar]
- 24.Chun KW, et al. Controlled release of plasmid DNA from photo-cross-linked pluronic hydrogels. Biomaterials. 2005;26(16):3319–3326. doi: 10.1016/j.biomaterials.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 25.Quick DJ, Anseth KS. DNA delivery from photocrosslinked PEG hydrogels: encapsulation efficiency, release profiles, and DNA quality. Journal of Controlled Release. 2004;96(2):341–351. doi: 10.1016/j.jconrel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 26.Megeed Z, et al. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. Journal of Controlled Release. 2004;94(2–3):433–445. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Trentin D, Hubbell J, Hall H. Non-viral gene delivery for local and controlled DNA release. Journal of Controlled Release. 2005;102(1):263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 28.Wieland JA, Houchin-Ray TL, Shea LD. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. Journal of Controlled Release. 2007;120(3):233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, et al. Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration. Biomaterials. 2007;28(20):3163–3170. doi: 10.1016/j.biomaterials.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: A novel model system for proteolytically mediated cell migration. Biophysical Journal. 2005;89(2):1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutolf MP, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci U S A. 2003;100(9):5413–8. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutolf MP, et al. Repair of bone defects using synthetic mimetics of collagenous extracellular matrices. Nat Biotechnol. 2003;21(5):513–8. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- 33.Adeloew C, et al. The effect of enzymatically degradable poly(ethylene glycol) hydrogels on smooth muscle cell phenotype. Biomaterials. 2008;29(3):314–326. doi: 10.1016/j.biomaterials.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 34.Lutolf MP, Hubbell JA. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4(3):713–22. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 35.Ehrbar M, et al. Biomolecular hydrogels formed and degraded via site-specific enzymatic reactions. Biomacromolecules. 2007;8(10):3000–3007. doi: 10.1021/bm070228f. [DOI] [PubMed] [Google Scholar]
- 36.Lei Y, Segura T. DNA delivery from matrix metalloproteinase degradable poly(ethylene glycol) hydrogels to mouse cloned mesenchymal stem cells. Biomaterials. 2009;30(2):254–65. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Q, Wang GJ, Balian G. Steroid-induced adipogenesis in a pluripotential cell line from bone marrow. Journal of Bone and Joint Surgery-American Volume. 1997;79A(7):1054–1063. doi: 10.2106/00004623-199707000-00012. [DOI] [PubMed] [Google Scholar]
- 38.Cui QJ, et al. Alcohol-induced adipogenesis in a cloned bone-marrow stem cell. Journal of Bone and Joint Surgery-American volume. 2006;88A:148–154. doi: 10.2106/JBJS.F.00534. [DOI] [PubMed] [Google Scholar]
- 39.Shen FH, et al. Systemically administered mesenchymal stromal cells transduced with insulin-like growth factor-I localize to a fracture site and potentiate healing. Journal of Orthopaedic Trauma. 2002;16(9):651–659. doi: 10.1097/00005131-200210000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Hsiong SX, et al. Differentiation stage alters matrix control of stem cells. Journal of Biomedical Materials Research Part A. 2008;85A(1):145–156. doi: 10.1002/jbm.a.31521. [DOI] [PubMed] [Google Scholar]
- 41.Hsiong SX, et al. Integrin-adhesion ligand bond formation of preosteoblasts and stem cells in three-dimensional RGD presenting matrices. Biomacromolecules. 2008;9(7):1843–1851. doi: 10.1021/bm8000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei P, Padmashali RM, Andreadis ST. Cell-controlled and spatially arrayed gene delivery from fibrin hydrogels. Biomaterials. 2009;30(22):3790–9. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH, et al. Proteolytically degradable hydrogels with a fluorogenic substrate for studies of cellular proteolytic activity and migration. Biotechnology Progress. 2005;21(6):1736–1741. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]