Abstract
A phenotypic assay to determine coreceptor usage of HIV-1 has been developed for rapid testing of clinical samples. The assay is based on the synthesis of viral stock from full-length env amplicons isolated from patient’s plasma. Pseudoviral stock is generated rapidly by using an overlapping PCR method to assemble a CMV promoter to env, followed by co-transfection into producer cells with a HIV plasmid (pNL4-3.Luc.R−E−) containing a non-functional env. The coreceptor used by the viral quasispecies is tested by infection into U87.CD4.CCR5 and U87.CD4.CXCR4 cells. Viral entry is indicated by the expression of the luciferase gene in relative light units (RLU). The use of CXCR4 coreceptor by minor variants is confirmed with sufficient suppression of RLU by a CXCR4 inhibitor. Two statistical logistical tests are employed to confirm viral entry. This assay accurately assigned coreceptor usage of isolates of various subtypes and in the majority of samples of various viral loads. The sensitivity to detect minor species of CXCR4-using env is 1% at higher viral loads and 5% at less than 1000 copies/ml. This assay provides a sensitive, efficient and relatively low-cost approach suitable for use by research laboratories for assessing HIV-1 coreceptor usage of plasma samples.
Keywords: chemokine receptor, tropism, luciferase, pseudovirus, viral fitness, HIV, viral tropism, coreceptor usage, CCR5, CXCR4
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) infects host cells through an interaction of the viral envelope protein with the cell CD4 receptor and a chemokine coreceptor, usually CCR5 or CXCR4. Viruses that use CCR5 exclusively are termed R5-tropic viruses while those that use CXCR4 are termed X4-tropic viruses. Phenotypic methods of determining coreceptor usage on an uncloned viral population cannot differentiate dual-tropic viruses from mixtures of R5 and X4 variants, therefore viruses that use both coreceptors are termed dual-mixed (DM).
Accurate and efficient methods to test HIV-1 coreceptor tropism are needed both in the clinical care setting and in the research of disease pathogenesis and viral evolution. The recent introduction of coreceptor antagonists as a new class of antiretroviral drugs has made coreceptor testing essential for patients prior to starting such therapy and at the time of virological failure. A shift from an R5 to an X4-using virus or the expansion of an existing minor X4-tropic virus while being treated with a CCR5 antagonist could have significant consequences on HIV disease progression. Therefore, further study of viruses from HIV-infected patients in whom CCR5 antagonist therapy has failed is needed.
Currently, the Trofile assay (LabCorp [previously Monogram Biosciences], South San Francisco, CA) is the only commercially available phenotypic assay for determining coreceptor in clinical settings. This assay uses env sequences from patient plasma virus to construct pseudotyped viruses, which are then used to infect human cell lines that express CD4 together with either CXCR4 or CCR5 (Whitcomb et al. 2007). This method is well validated and can detect X4 virus when present at proportions as low as 0.3% of the virus population (Reeves et al. 2009).
An alternative rapid and efficient method of determining coreceptor usage of HIV-1 in plasma samples has been developed. This single-cycle assay is based on the generation of pseudotyped viruses using an overlap PCR to attach the CMV immediate enhancer/promoter to the 5’ end of a population of env amplicons without the need for ligation, bacterial transformation, plasmid amplification and isolation usually required for cloning env into expression vectors. This promoter-PCR (pPCR) has been used in combination with single genome amplification (SGA) to produce functional env clones for neutralization assays (Kirchherr et al. 2007). Here, the method of pPCR is employed on uncloned env amplicons to allow expression of a heterogeneous population of env genes and validated its use among reference HIV strains and clinical samples. The resulting assay provides a sensitive, efficient and relatively low-cost approach suitable for use by research laboratories for assessing coreceptor usage of HIV-1 in plasma samples.
2. Materials and Methods
2.1 Materials: Reference viruses, clinical samples and plasmids
Laboratory-adapted viruses and primary isolates with known coreceptor usage were used as controls and in validation studies for the assay. R5 viruses included JR-CSF, MJ4, YU2 and Q23; X4 viruses included LAI, NL4-3 and HXB2; dual-tropic viruses included SF2 and 89.6. All viruses were obtained from the AIDS Research and Reference Reagent Program (ARRRP, Rockville, MD). Viral RNA from low passage, patient-derived primary isolates were obtained for validation studies from 1) nine previously characterized isolates from the AIDS Clinical Trials Group (ACTG) protocol A5211, a phase 2b trial of the investigational CCR5 antagonist vicriviroc (Gulick et al. 2007; Hosoya et al. 2009), 2) five previously characterized subtype C samples from Botswana (Ndung'u et al. 2006), and 3) four subtype B isolates from patients with acute infections (Johnson et al. 1991; Rusconi et al. 1999; Tremblay et al. 2005; Tremblay et al. 2003). Additionally, 216 plasma samples from HIV-1 positive women enrolled in the Mashi study in Botswana with CD4 cell counts below 200/mm3 were tested to determine the success rate of env amplification at various plasma HIV-1 RNA levels. All subjects provided written informed consent and the study was approved and conducted accordingly to the Partners HealthCare Systems and Harvard School of Public Health institutional review boards and the Botswana Ministry of Health. The plasmid pNL43.LUC.-R-E-, which contains a full-length molecular clone of HIV-1 NL4-3 with the firefly luciferase gene inserted into nef and two frameshift mutations that result in nonfunctional env and vpr, was obtained from the ARRRP.
2.2 Amplification of HIV-1 env genes from viruses
Viruses derived from clinical samples were extracted from 140 μl of patient plasma or viral culture supernatant according to the QIAamp viral RNA Mini kit protocol (Qiagen; Valencia, CA), resuspended in a final volume of 60 μl and stored at −80°C for later use. For clinical samples the reverse transcription and first round PCR were performed using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen; Carlsbad, CA). In order to minimize possible resampling, the RT-PCR reactions were performed in triplicate for each sample in 30-μl reactions. In each reaction 4 μl of the extracted viral RNA was added as template to 15 μl of 2× buffer, 0.5ul of RNAse, 1.2 μl of SuperScript III RT/Platinum Taq HiFi mix, 0.6 μl of 20uM of reverse and forward primers: OFM19 5’-GCACTCAAGGCAAGCTTTATTGA GGCTTA-3’ and VIF1 5’-GGGTTTATTACAGGGACAGCAGCG-3’ and 8.1 μl of distilled water. For plasmids or DNA from reference viruses approximately 1 to 5 ng of template DNA was used in each PCR. All PCR reactions were limited to 20 or fewer cycles and extension times were increased to 1.5 minutes for amplification of each kilobase pair. The RT-PCR cycle conditions were as follows: one cycle of 55°C for 30 min followed by 94°C for 2 min and then 20 cycles of 94°C for 20 sec, 55°C for 30 sec and 68°C for 7 min followed by a final extension at 68°C for 5 min.
Triplicate reactions for each sample were combined and mixed well prior to their use as template for the second round PCR reaction, which was carried out in 3 separate 25-μl reactions. Primers for subsequent rounds of PCR were as described, with modifications in concentrations and reaction conditions (Kirchherr et al. 2007). Each reaction contained 0.01mol primers (env1Atopo 5’-CACCGGCTTAGGCATCTC CTATGGCAGGAAGAA-3’ and envN 5’-CTGCCAATCAGGGAAGTAGCCTTGTGT-3’) added to at least 1 μl of template DNA, 1 unit of Platinum Taq High Fidelity, 0.2 mM of dNTP mixture and 2 mM magnesium sulfate. For plasma samples known to have HIV-1 RNA levels less than 10,000 copies/ml, the amount of first-round product was increased to 2 to 4 μl. The 2nd round PCR cycle conditions were as followed: 94°C for 2 min, then 15 cycles of 94°C for 30 sec, 58°C for 30 sec, 68°C for 6 min followed by a final extension at 68°C for 5 min. The triplicate PCR reactions were combined and the presence of an approximately 3.5-kb amplicon was verified by agarose gel electrophoresis prior to the overlapping PCR step.
2.3 Overlapping PCR: Attachment of the CMV promoter to env genes
(i) Amplification of the CMV promoter
The CMV immediate enhance/promoter was amplified in 50-μl reactions with 1 unit of Platinum Taq polymerase HiFi, 100 pmol of pcDNA 3.3-TOPO cloning vector (Invitrogen; Carlsbad, CA) as template, 0.02 mol of each primer, 2mM magnesium sulfate and 0.2 mM dNTP mixture. Primers were: CMVenv 5’AGTAATCAATTACGGGGTCAT TAGTTCAT-3’ and CMVenv1A 5’-CATAGGAGATGCCCTAAGCCCGGTGGAGCTCTGC TTATATAGACCTC-3’ (Kirchherr et al. 2007). The following conditions were used: 94°C for 2 min, then 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 68°C for 1 min, followed by extension of 68°C for 5 min. The 0.8-kb amplicon was separated from the reaction mix by agarose gel electrophoresis and purified using QIAquick PCR purification columns (Qiagen; Valencia, CA). The concentration of the product was determined prior to its use in the promoter PCR reaction.
(ii) Promoter PCR (pPCR)
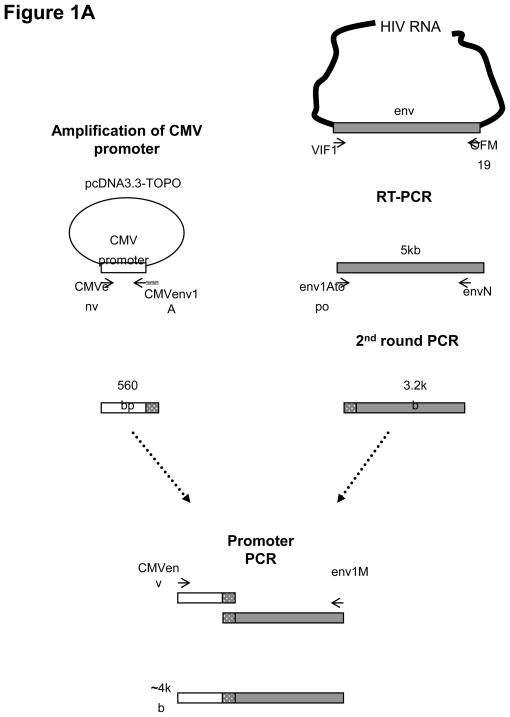
The CMV promoter was attached to the env amplicons by an overlapping PCR technique. For each sample, this 3rd-round of PCR was carried out in 3 or more separate 50-μl reactions; 1 to 2 μl of the 2nd-round product was used as template and added to 2 picomol of magnesium sulfate and 0.2 mM of dNTP mixture. The primers were CMVenv, described above, and env1M 5’-TAGCCCTTCCAGTCCCCCCTTTTCTT TTA-3’. The samples underwent 1 cycle of 94°C for 2 min, 15 cycles of 94°C for 30 sec, 55°C for 30 sec, 68°C for 6 min, followed by a final extension at 68°C for 10 min. The triplicate pPCRs for each patient sample were combined and the presence of the 4-kb PCR product was verified agarose gel electrophoresis. The pPCR amplicons were purified with the QIAQuick purification kits (Qiagen; Valencia, CA) (Figure 1A).
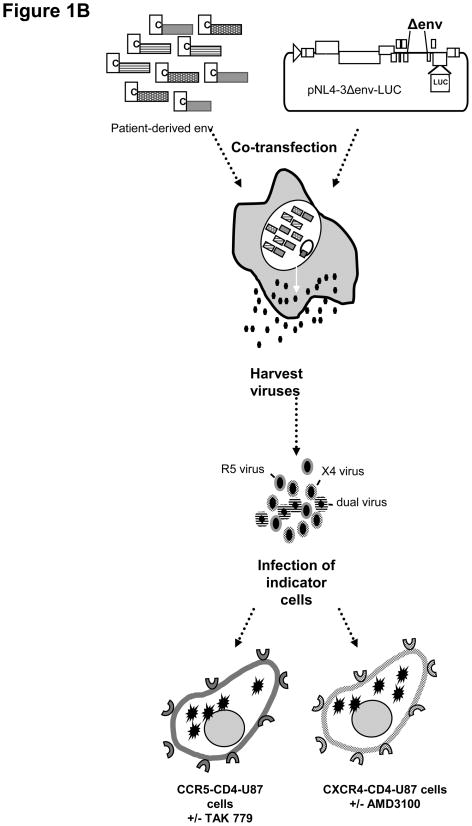
Figure 1. Schematic diagram of the methodology to determine HIV-1 coreceptor usage from clinical samples.
(A) Generation of promoter env PCR amplicons using overlapping PCR (Kirchherr et al, 2007). The env amplicon is generated from patient-derived viral RNA using RT-PCR followed by nested PCR to generate a 3.5 kb amplicon. The CMV promoter is amplified in a separate PCR and attached to the env amplicons by an overlapping PCR step. (B) Viral stock generation and viral infection. Env amplicons with attached CMV promoter are cotransfected into 293T cells with a plasmid containing a full-length HIV genome deleted in env and carrying a luciferase reporter gene in the nef region (pNL4-3.Luc.R-E-; (AIDS Research and Reference Reagent Program). The pseudotyped virons are harvested and used to infect two separate indicator cell lines, CCR5-CD4-U87 and CXCR4-CD4-U87 cells, in the absence and presence of inhibitors, TAK779 and AMD3100, respectively. Viral entry using the coreceptors is assessed by measuring the luciferase activity in RLU in each cell lines and is confirmed by greater than 50% inhibition of luciferase activity in the presence of the specific inhibitor. Two statistical tests are used to evaluate the raw RLU data to make an assignment of coreceptor usage.
2.4 Production of pseudotyped viruses
Pseudovirus stocks representing the env population from patient samples were produced by co-transfecting pPCR products together with pNL43.LUC.-R-E- into 293T cells. Six-well tissue culture plates were seeded with 0.9×106 cells per well the day prior to transfection (80% confluence) in a total volume of 1.5 ml of Dulbecco’s Modification of Eagle’s Medium (DMEM) with 4.5 g/L glucose, L-glutamine and sodium pyruvate with 10% fetal bovine serum (FBS). FuGENE6 (Roche Applied Science, Indianapolis, IN) was used for transfection and mixed with the total DNA in a ratio of 3 μl FuGENE6:1 μg of DNA. In each well of a 6-well plate, 6μl of FuGENE6 and 94 μl of reduced serum OPTImem media (GIBCO) were mixed and added to the DNA mixture containing 0.5 μg of pPCR product and 1.5 μg of pNL43.LUC.-R-E-. After incubation for 30 min at room temperature the mixture was added drop-by-drop onto the cells. Transfected cells were incubated for 48 hr at 37°C with 5% CO2 after which the pseudovirus stocks were passed through 0.45 μm filter prior to remove cellular debris prior to storage at −80°C. Supernatants of 293T cells transfected with pNL43.LUC.-R-E- without any pPCR product served as negative controls.
2.5 Single-cycle infection of indicator cell lines
The day prior to infection of harvested pseudoviruses U87.CD4.CCR5 and U87.CD4.CXCR4 cells were seeded in black 96-well microtiter plates at a density of 0.15×106 cells per well in 50 μl of DMEM supplemented with 15% FBS. The day of infection cells were at least 50% confluent and with a minimal number of large cell clumps, which prevented efficient infection. For wells designated to be treated with an inhibitor, either 2 μM AMD3100 (a CXCR4 antagonist) or 1 μM TAK779 (a CCR5 antagonist) was added to the cells prior to infection. Thawed stocks of pseudotyped virions (50 μl) were added to each well along with the addition of polybrene to a final concentration of 8μg/ml. The plates were centrifuged at 300g for 40 minutes at 37°C and then incubated in 37°C with 5% CO2 for 72 hours. All infections were performed in triplicate wells, and the results averaged. Since the majority of clinical isolates are capable of utilizing CCR5 as coreceptor, inhibition by TAK779 was used to verify infection via CCR5 only for selected samples that showed low luciferase activity on U87-CCR5 cells. Control infections were performed using pseudovirus stocks prepared with MJ4 (R5) and NL43 (X4) envelopes.
2.6 Luciferase assay
Levels of infection were measured by determining luciferase activity using the Luciferase assay system according to manufacturer’s protocol (Promega, Madison, WI). Briefly, the medium was removed and the cells rinsed with 300 μl of phosphate-buffered saline (PBS). Then 30 μl of Passive Lysis Buffer was added to each well and plates were either placed on a shaker for 20 min or underwent a single freeze-thaw cycle to permeabilize the cells. Once the plates returned to room temperature, 100 μl of the Luciferase Assay Reagent was added immediately prior to reading the plates in a luminometer. Luciferase activity was recorded as relative light units (RLU) detected over a 5-second period (Figure 1B).
2.7 Data analysis
Raw RLU data were transformed into a logarithmic scale to ensure constant error distribution across all signal intensities. Viruses were considered to use the CXCR4 coreceptor if they fulfilled two stringent criteria based on statistical tests: Criterion 1 determined if infection in U87-CXCR4 cells was above background; Criterion 2 determined if there was clear and sufficient suppression of infection in the presence of AMD3100. The logical tests applied were the following:
Criterion 1: A one-sided t-test compared mean luciferase activity of triplicate sample wells to wells infected with pseudovirus stocks prepared without envelope (background). Using a nominal level of significance of p<0.05, samples with RLU that were significantly above background were considered positive for infection of U87-CXCR4 cells and were then tested by criterion 2.
Criterion 2: A one-sided t-test compared mean luciferase activity in triplicate sample wells with and without AMD3100. Suppression of luciferase activity was considered significant when p-value <0.05 and the log fold-change in the presence of the inhibitor was >0.3 log10 (approximately 50% reduction in RLU).
For practical purposes all samples were initially tested on U87-CCR5 cells and in the absence and presence of AMD3100 in U87-CXCR4 cells. Samples that infected U87-CCR5 cells but did not fulfill both criteria for infection of U87-CXCR4 cells were considered to be R5 viruses. Samples that infected U87-CCR5 cells and fulfilled the two criteria for CXCR4-usage were considered DM viruses. Since pure X4 infections are rare cases, all samples were tested in U87-CCR5 cells alone without drug inhibition. When RLU levels were low (less than twice of background) then the sample was tested again in the presence of TAK779, a CCR5 inhibitor. In those cases the same two criteria were applied for infections in U87-CCR5 cells.
3. Results
3.1 Optimal conditions for PCR amplification of env from clinical samples
In order to characterize the plasma virus population as accurately as possible using bulk PCR, conditions were optimized to minimize recombination by increasing extension time, limiting cycle number, increasing the time of RT and use of proofreading Taq (Fang et al. 1998; Meyerhans et al. 1990). Infectivity of the pseudoviral stock generated was enhanced by the addition of 8 μg/ml polybrene during infection of indicator cells (U87) and spinoculation at 300g for 45 minutes. The use of spinoculation increased level of infectivity of the pseudoviral stock generated from pPCR without substantially increasing background luciferase activity (data not shown).
3.2 Pseudoviral stock generated from reference strains
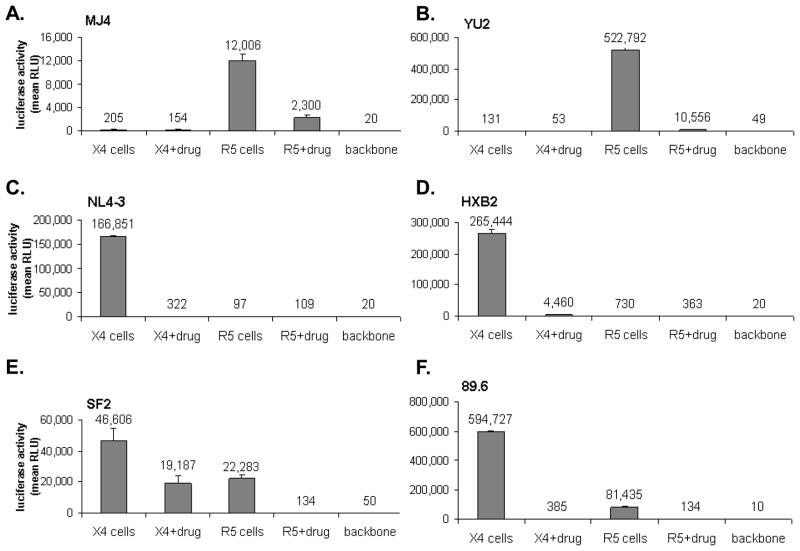
Pseudoviruses expressing env from the reference R5 strains YU2 and MJ4 were used to infect U87-CCR5 and U87-CXCR4 cell lines in the presence and absence of TAK227 or AMD3100, respectively. In the absence of inhibitors, the viruses produced high levels of luciferase activity in RLU in U87-CCR5 cells and background levels of RLU in U87-CXCR4 cells. In the presence of TAK227, infection of U87-CCR5 cells by YU2 was reduced by 98%, and infection by MJ4 by 81%; incubation with AMD3100 did not change the level of luciferase activity on U87-CXCR4 cells (Figure 2A-B). In contrast, pseudoviruses expressing env from the reference X4 strains NL4-3 and HXB2 exhibited high luciferase activity in U87-CXCR4 cells and low levels in U87-CCR5 cells. AMD3100 inhibited viral entry into U87-CXCR4 cells by 98–99%, whereas TAK227 had no effect on the already low RLU in the U87-CCR5 cells (Figure 2C–D). Pseudoviruses expressing env from the dual-tropic viruses SF2 and 89.6 infected both U87-CCR5 and U87-CXCR4 cell lines, producing moderate levels of luciferase activity in the absence of inhibitors. The RLU produced were approximately 2-fold higher on the U87-CXCR4 than on U87-CCR5 cells. The addition of TAK770 to U87-CCR5 cells or of AMD3100 to U87-CXCR4 cells, respectively, reduced the levels of luciferase activity by 99% in both cases, confirming the usage of both coreceptors by these two viral isolates (Figure 2E–F).
Figure 2.
pPCR tropism assay determination of previously characterized reference strains. U87-CXCR4 and -CCR5 cells were infected with pseudoviral stock from pPCR of env gene from R5-tropic MJ4 (A) and YU2 (B); X4-tropic NL4-3 (C) and HXB2 (D), and dual-tropic SF2 (E) and 89.6 (F). Infection was indicated by a mean RLU which was statistically above background (infection of pseudoviral stock generated by HIV-1 backbone; NL4-3delEnv-LUC plasmid with a non-functional env gene) and suppression of 50% or greater by an X4 or R5 inhibitor, AMD3100 or TAK 775, respectively.
3.3 Assay accuracy tested on reference clones and primary clinical isolates
Validation of the this pPCR tropism assay was performed by determining the coreceptor usage of a panel of previously characterized primary isolates and laboratory clones from subtypes A, B and C. The co-receptor usage of all nine HIV-1 laboratory isolates or cloned viruses (4 R5, 3 ×4 and 2 DM viruses) agreed in all cases with previously reported results (Table 1). Reproducibility was verified by retesting the majority of isolates in two or more independent assays, which gave consistent results. In the case of MJ4 (R5) and NL43 (X4), at least 20 different assay runs resulted in identical tropism assignments. In addition, the tropism of HIV-1 isolates previously characterized by independent laboratories was determined by the pPCR assay. Five subtype C primary isolates from Botswana that were previously characterized by p24 antigen production in U87-CD4 cells that expressed CCR5 or CXCR4 and by MT-2 cell assay were tested (Ndung'u et al. 2006). The pPCR tropism assay confirmed all the tropism assignments (Table 1). Coreceptor usage of the 9 subtype B samples previously tested by the Trofile assay and by a recombinant virus assay were also tested (Hosoya et al. 2009). Results of the pPCR assay were consistent with those obtained by Trofile assay. Lastly, four additional subtype B isolates previously characterized by MT-2 cell assay were tested with the current assay. Isolates DK and 14aPre, which produce syncytia on MT-2 cells, were both X4 using this assay, whereas RM and JC, which are both non-syncytium inducing on MT-2 cells, were R5 viruses in the pPCR assay (Kollmann et al. 2001; Tremblay et al. 2005) (Table 1).
TABLE 1.
Assay determination of viral tropism among isolates with known coreceptor usage
| Test samples | env subtypea | tropisma | no. replicates | no. fulfilled criterion 1b | no. fulfilled criterion 2c | log dec | assay result |
|---|---|---|---|---|---|---|---|
|
Reference virusesd | |||||||
| JR-CSF | B | R5 | 1 | 1 | 0 | R5 | |
| YU2 | B | R5 | 1 | 0 | NA | R5 | |
| Q23 | A | R5 | 2 | 2 | 0 | R5 | |
| MJ4 | C | R5 | 24 | 2 | 0 | R5 | |
| NL43 | B | X4 | 23 | 23 | 23 | 2.48 | X4 |
| HXB2 | B | X4 | 6 | 6 | 6 | 2.07 | X4 |
| LAI | B | X4 | 3 | 2 | 2 | 3.06 | X4 |
| SF2 | B | DM | 3 | 3 | 3 | 0.65 | DM |
| 89.6 | B | DM | 2 | 2 | 2 | 3.10 | DM |
|
Patient isolatese | |||||||
| 96BWMO1.5 | C | R5 | 2 | 2 | 0 | R5 | |
| 96BWMO3.2 | C | R5 | 2 | 2 | 0 | R5 | |
| 96BW.0502 | C | R5 | 3 | 1 | 1 | 1.21 | R5/DM |
| 96BW.1210 | C | R5 | 2 | 2 | 0 | R5 | |
| 96BW17A09 | C | DM | 2 | 2 | 2 | 3.27 | DM |
| RM-08 | B | R5 | 2 | 0 | NA | R5 | |
| JC-06 | B | R5 | 2 | 0 | NA | R5 | |
| DK | B | X4 | 2 | 2 | 2 | 2.55 | X4 |
| 14aPre | B | X4 | 1 | 1 | 1 | 0.30 | X4 |
| Subject 1 | B | DM | 1 | 1 | 1 | 1.56 | DM |
| Subject 4 | B | DM | 4 | 3 | 3 | 1.32 | DM |
| Subject 8 | B | DM | 2 | 2 | 2 | 0.86 | DM |
| Subject 9 | B | R5 | 3 | 0 | NA | R5 | |
| Subject 12 | B | R5 | 4 | 1 | 0 | R5 | |
| Subject 13 | B | R5 | 4 | 1 | 0 | R5 | |
| Subject 17 | B | R5 | 3 | 1 | 0 | R5 | |
| Subject 19 | B | R5 | 4 | 1 | 0 | R5 | |
| Subject 21 | B | R5 | 4 | 4 | 0 | R5 | |
Env subtype and tropism assignment based on published results from ARRRP and (Hosoya et al. 2009; Johnson et al. 1991; Ndung'u et al. 2006; Rusconi et al. 1999; Tremblay et al. 2005; Tremblay et al. 2003).
A one-sided t-test determines if infection in U87-CXCR4 cells is above background, using a nominal level of significance of p<0.05
A one-sided t-test determines if there is sufficient suppression of infection in the presence of AMD3100 with a log fold-change in the presence of the inhibitor is >0.3 log10 (approximately 50% reduction in RLU).
All of the reference viruses can be obtained from the AIDS Research Reagent and Reference Program (ARRRP), Rockville, MD.
Subtype C isolates were from Botswana (Ndung'u et al. 2006), 4 subtype B viruses (RM-08, JC-06, DK and 14aPre) were previously isolated from individuals with acute HIV infection (Johnson et al. 1991; Rusconi et al. 1999; Tremblay et al. 2005; Tremblay et al. 2003), and Subject 1–21 were isolates from ACTG study A5211 (Gulick et al. 2007; Hosoya et al. 2009) with tropism previously determined by the Trofile assay.
3.4 Sensitivity to detect minority populations
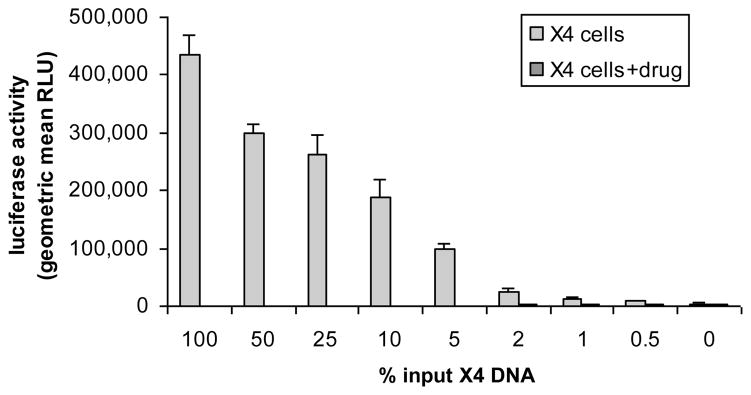
The sensitivity of the assay for detecting minority populations of X4 viruses in the background of predominantly R5 virus was tested using mixtures of plasmids carrying the HXB2 (X4) and MJ4 (R5) env at different ratios (Figure 3). The assay reproducibly detected X4-tropic virus present at a level of 1% (9 separate experiments). To determine the relationship between assay sensitivity and template input copy number, mixtures of HXB2 and MJ4 were diluted to contain a total of 1000 and 10,000 DNA copies/ml. The resulting X4 viruses were detected consistently in replicate experiments in which the env of the HXB2 plasmid was present at 5% in a total input of 1000 DNA copies/ml. At a total input of 10,000 copies/ml, CXCR4-using env could be detected approximately 67% of the time when HXB2 env constituted 0.5–1% of the input DNA. Overall, these dilution experiments verified that the threshold of this assay to detect a minority CXCR4-using variant is around 1% at higher total input copy numbers and at least 5% at lower copy numbers. This change in the sensitivity at lower percentage of minor CXCR4-using species results from failure to fulfill criterion 2 because of the overall low level of luciferase activity in U87-CXCR4 cells.
Figure 3.
Sensitivity to detect CXCR4-using variants within mixed viral populations. The results are the geometric mean RLU from multiple replicates with SE error bars. Minor species of X4-tropic env from HXB2 was detected consistently at 1% and approximately 50% of the times at 0.5%. Use of CXCR4 for entry was confirmed by ablation of the luciferase signal with the addition of AMD3100, which decreased the RLU to minimal levels. Luciferase activity in U87-CXCR4 cells decreased proportionally to the proportion of input DNA from X4 virus.
3.5 Success of pPCR depends on viral load input
Plasma samples from 203 HIV-1 subtype C-infected women in Botswana were tested using this assay. The pPCR was successful in 194 (95.6%) samples. The median virus load of the 9 samples that could not be amplified was 400 copies/ml as compared to 78,897 copies/ml for those samples with successful pPCR (p =0.048) (Table 2). Sample quality may have been a factor for the few samples with high virus loads that failed to amplify. An additional 12 samples with viral load of less than 50,000 initially failed to generate an amplicon by pPCR. With repeat RNA extraction after ultracentrifugation for 1 hour at 300g to concentrate the viral particles and an increase of 1–2 folds of input RNA for the RT-PCR reaction resulted in successful env amplification. Infectious pseudotyped viral stocks were obtained from all samples that underwent successful PCR amplification of env and the final pPCR step.
TABLE 2.
Amplification of env with varying viral loads
| viral load (copies/ml) | no. tested | no. failed |
|---|---|---|
| <400 | 6 | 4 |
| 401–1000 | 5 | 0 |
| 1001–5000 | 18 | 0 |
| 5001–10,000 | 9 | 0 |
| 10,001–90,000 | 55 | 2 |
| 90,001–200,000 | 35 | 0 |
| 200,001–750,000 | 42 | 1 |
| >750,000 | 5 | 0 |
| not available | 28 | 2 |
| TOTAL | 203 | 9 |
4. Discussion
The described method to determine HIV-1 coreceptor usage is a rapid, sensitive, relatively low cost assay suitable for use by research laboratories for determining co-receptor usage of HIV-1 from clinical samples. The assay consistently detects CXCR4-using variants present at proportions as low as 5% of the population in samples containing at least 1000 copies/mL, with a threshold of detection of 0.5–1.0% in samples containing equal or greater than 10,000 copies/mL of viral template. This level of sensitivity is comparable to that reported by other phenotypic assays, but not as sensitive as the enhanced Trofile assay which can detect minority CXCR4-using variants to 0.3% of the population (Trouplin et al. 2001; Van Baelen et al. 2007; Whitcomb et al. 2007). Actual sensitivity in clinical samples depends not only on the input virus load but is also a function of infectivity of the envelopes being assayed. In addition, inefficiencies due to RNA extraction and reverse transcription of the template viral RNA also affect sensitivity of the assay (Braun and Wiesmann 2007). Therefore, as with other tropism assays the sensitivity predicted by the reconstruction experiments using mixtures of HXB2 and MJ4 env-encoding plasmids most likely gives an upper bound of the actual level of sensitivity of the assay to detect minor CXCR4-using variants.
Much of the initial work demonstrating an association between HIV-1 tropism and disease progression relied on the MT-2 cell assay, which tested the ability of HIV-1 to replicate and induce syncytia in MT-2 cells (Japour et al. 1994; Koot et al. 1993; Schuitemaker et al. 1992). Contemporary assays that test coreceptor usage directly using recombinant viruses or pseudoviruses that express HIV-1 env from patient virus correlate well with results of the MT-2 cell assay (Coakley et al. 2009; Hosoya et al. 2009). Assays using pseudoviruses have a theoretical advantage over recombinant virus assays or assays requiring virus isolation because they avoid the labor-intensive and potentially prolonged process of in vitro virus isolation and amplification, thereby eliminating potential bias introduced by selection during in vitro passage. The potential for in vitro passage to bias results is particularly concerning given the results of some studies that suggest as few as two mutations in the V3 loop can confer the ability to use CXCR4 (Cocchi et al. 1996; Fouchier et al. 1992; Hwang et al. 1991).
In the described experiments a majority of clinical samples tested was successfully amplified and produced viral stock which generated a phenotypic read-out. As expected difficulty with PCR amplification occurred more often in samples with low viral load, but PCR amplification was unsuccessful in a few samples with high viral loads. Poor sample quality likely was a contributing factor, as other samples from the same patients obtained at different time points were successfully amplified, suggesting that primer mismatch did not explain the initial failure to amplify. The high success rate of pPCR in samples from chronically infected patients, in whom HIV-1 diversity is expected to be high, suggests that this method could easily be applied to the study of large patient cohorts with a wide range of viral loads. Although the plasma samples tested in these experiments were all from Botswana, where HIV-1C is the most prevalent subtype (Novitsky et al. 2002), the PCR primers used were equally successful in amplifying env from subtypes A and B (Table 1).
Lastly, results of the assay were fully concordant with results of the Trofile assay in samples from nine subjects tested by both methods. Although full validation would require comparison in a larger number of samples, these results suggest that the pseudoviruses generated by the pPCR together with the statistical approach used to analyze the luciferase data generated from infection of the U87.CD4.CCR5 and U87.CD4.CXCR4 cells accurately determines coreceptor usage of plasma virus. Pseudoviruses produced using pPCR express levels of env that are approximately 2-fold lower than those produced by cloning env into CMV expression vectors, such as those used in the Trofile assay (Kirchherr et al. 2007). Conditions were optimized through systematic testing to compensate for the decreased infectious titer associated with the pPCR approach. The addition of polybrene and a centrifugation step during infection (“spinoculation”) increase the effective infectious titer of the pseudovirus by increasing virus-cell contact (O'Doherty et al. 2000; Thomas et al. 2007).
This pPCR tropism assay is well-suited to the study of viral tropism in large cohort studies because of its efficiency in amplifying the HIV-1 viral env gene from plasma HIV-1 RNA and the rapid generation of viral stocks. A pseudotyped viral stock representing the circulating viral species could be obtained after two days, after which infection of indicator cell lines occurs in another two days; total time needed to complete the assay is approximately 7 days. The methodology described is tailored for rapid high-throughput testing, optimized to test multiple samples at the same time in 98-well cell culture plates and the use of an automated injector and plate reader. Although the assay is optimized for the testing of plasma samples, it can be adapted to determine coreceptor usage of virus isolated from proviral DNA samples and other compartments, such as specimens of CSF, breast milk and semen.
Acknowledgments
to the contributions of Drs. Myron Essex, Vladimir Novitsky, Shahin Lockman and Joseph Makhema for the subtype C clinical samples from Botswana; the ACTG A5211 protocol team for the use of clinical isolates and their corresponding Trofile assay data; and Drs. Martin Hirsch and Cecil Trembley for the subtype B acute infection isolates are gratefully acknowledged. Dr. Manish Sagar and Dr. Zixin Hu provided valuable scientific advice throughout the study. The following reagents were obtained from the NIH AIDS Research and Reference Reagent Program: U87-CD4-CCR5 and U87-CD4-CXCR4 cells from H. Deng and D.R. Littman, and AMD3100. The generous donation of TAK779 by Takeda Pharmaceuticals is also acknowledged. This work was supported by NIH grants RR016482 and U01 AI068636 (an ACTG Virology Support Laboratory contract) (DRK), R01 HD037793 (ME), R01 HD044391 (SL), and the Harvard Center for AIDS Research Grant and Burroughs-Wellcome-ASTMH Tropical Research Fellowship (NHL).
Footnotes
Presented in part at the 17th Conference on Retroviruses and Opportunistic Infections, 16–19 February, 2010, San Francisco, California, abstract 278.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nina H. Lin, Email: nhlin@partners.org.
Daniel M. Negusse, Email: dnegusse@hsph.harvard.edu.
Rameen Beroukhim, Email: rameen@broad.mit.edu.
Francoise Giguel, Email: fgiguel@partners.org.
Shahin Lockman, Email: slockman@hsph.harvard.edu.
Myron Essex, Email: messex@hsph.harvard.edu.
Daniel R. Kuritzkes, Email: dkuritzkes@partners.org.
References
- Braun P, Wiesmann F. Phenotypic assays for the determination of coreceptor tropism in HIV-1 infected individuals. Eur J Med Res. 2007;12:463–72. [PubMed] [Google Scholar]
- Coakley E, Reeves JD, Huang W, Mangas-Ruiz M, Maurer I, Harskamp AM, Gupta S, Lie Y, Petropoulos CJ, Schuitemaker H, van 't Wout AB. Comparison of human immunodeficiency virus type 1 tropism profiles in clinical samples by the Trofile and MT-2 assays. Antimicrob Agents Chemother. 2009;53:4686–93. doi: 10.1128/AAC.00229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–7. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- Fang G, Zhu G, Burger H, Keithly JS, Weiser B. Minimizing DNA recombination during long RT-PCR. J Virol Methods. 1998;76:139–48. doi: 10.1016/s0166-0934(98)00133-5. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Groenink M, Kootstra NA, Tersmette M, Huisman HG, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–7. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick RM, Su Z, Flexner C, Hughes MD, Skolnik PR, Wilkin TJ, Gross R, Krambrink A, Coakley E, Greaves WL, Zolopa A, Reichman R, Godfrey C, Hirsch M, Kuritzkes DR. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-Infected, treatment-experienced patients: AIDS clinical trials group 5211. J Infect Dis. 2007;196:304–12. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- Hosoya N, Su Z, Wilkin T, Gulick RM, Flexner C, Hughes MD, Skolnik PR, Giguel F, Greaves WL, Coakley E, Kuritzkes DR. Assessing HIV-1 tropism: A comparison of assays using replication-competent virus versus plasma-derived pseudotyped virions. J Clin Microbiol. 2009a doi: 10.1128/JCM.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya N, Su Z, Wilkin T, Gulick RM, Flexner C, Hughes MD, Skolnik PR, Giguel F, Greaves WL, Coakley E, Kuritzkes DR. Assessing human immunodeficiency virus type 1 tropism: Comparison of assays using replication-competent virus versus plasma-derived pseudotyped virions. J Clin Microbiol. 2009b;47:2604–6. doi: 10.1128/JCM.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–4. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- Japour AJ, Fiscus SA, Arduino JM, Mayers DL, Reichelderfer PS, Kuritzkes DR. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–4. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VA, Merrill DP, Videler JA, Chou TC, Byington RE, Eron JJ, D'Aquila RT, Hirsch MS. Two-drug combinations of zidovudine, didanosine, and recombinant interferon-alpha A inhibit replication of zidovudine-resistant human immunodeficiency virus type 1 synergistically in vitro. J Infect Dis. 1991;164:646–55. doi: 10.1093/infdis/164.4.646. [DOI] [PubMed] [Google Scholar]
- Kirchherr JL, Lu X, Kasongo W, Chalwe V, Mwananyanda L, Musonda RM, Xia SM, Scearce RM, Liao HX, Montefiori DC, Haynes BF, Gao F. High throughput functional analysis of HIV-1 env genes without cloning. J Virol Methods. 2007;143:104–11. doi: 10.1016/j.jviromet.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann C, Tremblay C, Giguel F, Chou TC, Hirsch MS. In vitro anti-HIV-1 synergy between non-nucleoside reverse transcriptase inhibitors nevirapine and efavirenz. Antivir Ther. 2001;6:143–4. [PubMed] [Google Scholar]
- Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–8. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–91. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndung'u T, Sepako E, McLane MF, Chand F, Bedi K, Gaseitsiwe S, Doualla-Bell F, Peter T, Thior I, Moyo SM, Gilbert PB, Novitsky VA, Essex M. HIV-1 subtype C in vitro growth and coreceptor utilization. Virology. 2006;347:247–60. doi: 10.1016/j.virol.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, Ndung'u T, Klein I, Chang SY, Peter T, Thior I, Foley BT, Gaolekwe S, Rybak N, Gaseitsiwe S, Vannberg F, Marlink R, Lee TH, Essex M. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–51. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–80. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Coakley E, Petropoulos CJ, Whitcomb JM. An enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Journal of Viral Entry. 2009:3. [Google Scholar]
- Rusconi S, Merrill DP, La Seta-Catamancio S, Citterio P, Offord RE, Hirsch MS. Effective inhibition of HIV-1 isolated from patients with acute primary HIV-1 infection by aminooxypentane-RANTES. Aids. 1999;13:1144–5. doi: 10.1097/00002030-199906180-00022. [DOI] [PubMed] [Google Scholar]
- Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–60. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Ott DE, Gorelick RJ. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J Virol. 2007;81:4367–70. doi: 10.1128/JVI.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay CL, Giguel F, Chou TC, Dong H, Takashima K, Hirsch MS. TAK-652, a novel CCR5 inhibitor, has favourable drug interactions with other antiretrovirals in vitro. Antivir Ther. 2005;10:967–8. [PubMed] [Google Scholar]
- Tremblay CL, Poulin DL, Hicks JL, Selliah S, Chamberland A, Giguel F, Kollmann CS, Chou TC, Dong H, Hirsch MS. Favorable interactions between enfuvirtide and 1-beta-D-2,6-diaminopurine dioxolane in vitro. Antimicrob Agents Chemother. 2003;47:3644–6. doi: 10.1128/AAC.47.11.3644-3646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouplin V, Salvatori F, Cappello F, Obry V, Brelot A, Heveker N, Alizon M, Scarlatti G, Clavel F, Mammano F. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J Virol. 2001;75:251–9. doi: 10.1128/JVI.75.1.251-259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays. J Virol Methods. 2007;146:61–73. doi: 10.1016/j.jviromet.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, Chappey C, Kiss LD, Paxinos EE, Petropoulos CJ. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–75. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]