1. Introduction
A diverse range of non-viral gene delivery carriers has been developed and studied as alternatives to viral gene delivery carriers. The advantages of non-viral gene delivery carriers include: non-immunogenicity, flexibility in DNA cargo capacity, convenience in handling and manufacture, and effectiveness in therapeutic gene incorporation [1–3]. There are also potential drawbacks to non-viral gene delivery carriers including: lower in vivo transfection efficiency, cytotoxicity, and lack of cell-specific targeting [4, 5]. These potential drawbacks have previously limited the adoption of non-viral gene delivery carriers for in vivo gene therapy. To address these potential drawbacks, a new generation of polymeric gene carriers has been developed. Biodegradable linkages and cell-penetrating peptide modifications have been introduced to non-viral gene carriers to improve transfection efficiency and cell viability [6–15]. These modifications, however, do nothing to improve cell-targeting specificity. This lack of cell-targeting specificity has been a major obstacle to the application of polymeric gene carriers to the treatment of cell-type specific diseases, including central nervous system and cardiovascular diseases. Previous studies have suggested that it may be possible to develop specific ligand-modified gene delivery carriers which would offer some degree of cell selectivity [16, 17].
PCM is an isolated phage that displays a 20 amino acid peptide (WLSEAGPVVTVRALRGTGSW) and binds primary cardiomyocytes 180 times more avidly than control phages [18, 19]. For this study, the PCM peptide sequence was used as a specific cardiomyocyte targeting ligand for the delivery of genes for the potential treatment of cardiovascular disease. Cardiovascular disease is the leading cause of death in the United States. Cardiomyocyte apoptosis is a central pathophysiologic mechanism which underlies the development of cardiovascular disease [20]. The Fas gene has been identified as an inducer of cardiomyocyte apoptosis, and many studies using Fas siRNA have demonstrated that inhibition of Fas in turn inhibits cardiomyocyte apoptosis without immune stimulation [21–25]. In this study, we describe the ability of our newly synthesized PCM-modified bioreducible polymer carrier containing Fas siRNA to down regulate Fas gene expression and inhibit cardiomyocyte apoptosis.
Recently, we demonstrated that disulfide-linked bioreducible polymers are more efficient than PEI for the intracellular delivery of siRNA [26, 27]. This increased efficiency is likely due to enhanced decomplexation of the siRNA/bioreducible polymer polyplexes as a result of degradation of the disulfide bonds in the reductive environment of the cytoplasm. In this study, the synthesis of PCM-modified bioreducible poly(CBA-DAH), (PCD) and its characterization were reported. The specific cardiomyocyte targeting of the PCM modification was evaluated as compared to NIH 3T3 fibroblasts, and the higher cellular binding and uptake toward cardiomyocytes was examined using competition assay with free PCM peptide. To investigate Fas siRNA delivery into cardiomyocyte, gene silencing and the anti-apoptotic effects were measured by real-time RT PCR and FACS analysis.
2. Materials and Methods
2.1. Materials
Hyperbranched poly(ethylenimine) (bPEI, Mw = 25 kDa), tert-butyl-N-(6-aminohexyl) carbamate (N-Boc-1,6-diaminohexane, N-Boc-DAH), trifluoroacetic acid (TFA), triisobutylsilane (TIS), and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). N, N′-Cystaminebisacrylamide (CBA) was purchased from PolySciences, Inc. (Warrington, PA). The plasmid pCMV-Luc, containing a firefly luciferase reporter gene, which was inserted into a pCI plasmid vector with the CMV promoter (Promega, Madison, WI), was amplified in E. coli DH5R and isolated using a standard Maxiprep kit (Invitrogen, Carlsbad, CA). The Luciferase assay system and reporter lysis buffer were purchased from Promega (Madison, WI). All cell culture products including fetal bovine serum (FBS), Dulbecco’s phosphate buffered saline (PBS), antibiotics, trypsin-EDTA and Dulbecco’s modified Eagle’s medium (DMEM) were obtained from Invitrogen (Gibco BRL, Carlsbad, CA). For FACS analysis, YOYO-1 iodide (1 mM solution in DMSO) and PI staining solution (1.0 mg mL) were also purchased from Invitrogen. Cysteine-terminated PCM peptide (NH2CWLSEAGPVVTVRALRGTGSW-CONH2, Mw ~2,245 Da) was synthesized and purified by HPLC by the Health Sciences Center core research facilities at the University of Utah (Salt Lake City, UT). siRNAs were chemically synthesized and supplied by Dharmacon Co. (Lafayette. CO). The target sequence used for rat Fas siRNA was ACA CGG ACA GGA AAC ACT A. Green fluorescent protein (GFP) siRNA (target sequence: GCA CGA CTT CTT CAA GTC C) was used as the control siRNA without sequence homology to the Fas gene.
2.2. Synthesis of the PCM-Conjugated Polymer, PCM-poly (CBA-DAH) (PCM-PCD)
Poly(CBA-DAH), (PCD) was synthesized as described previously [28]. Briefly, PCD was synthesized by Michael reaction of equivalent moles of N-Boc-DAH and CBA in aqueous MeOH (9:1, v/v) at 60 °C for 4 days. The reaction was terminated with 10% mole excess of N-Boc-DAH and further reacted for one day. After precipitation with diethyl ether, the Boc groups were de-protected by the reagent solution (TFA:triisobutylsilane: H2O= 95: 2.5: 2.5, v/v) in an ice bath for 30 min. The crude product was then dialyzed against ultrapure water with dialysis membrane (MWCO=1,000 Da), followed by lyophilization. The synthesized PCD had an average molecular weight of 6,250 Da (polydispersity index, 1.13), which was determined by size exclusion chromatography (SEC, Superose 12 column) with standard poly[N-(2-hydroxypropyl) methacrylamide].
PCD was activated with 2.5 molar equivalents of the hetero-bifunctional linker, SM (PEG)2 (Pierce, Rockford, IL), per 10 DAH groups of the PCD in anhydrous DMF for 4 h at room temperature and purified with a desalting column. Cysteine-terminated PCM (-SH) was added to the activated polymer and the reaction mixture was stirred overnight at room temperature. The crude product was purified with precipitation and dialysis (MWCO=3,000 Da). After lyophilization, the synthesis of PCM-PCD was confirmed by 1H NMR (400 MHz, D2O), SEC and UV/Vis spectrophotometery.
2.3. Polyplex Formation and Characterization
The plasmid DNA or siRNA condensing ability of PCM-PCD was examined by agarose gel electrophoresis. Polyplexes were prepared in HEPES Buffered Saline (HBS, 10 mM HEPES, 1 mM NaCl, pH 7.4) at various weight ratios. An agarose gel (1.0%, w/v) containing a SYBR gel staining solution was prepared in TAE (10 mM Tris/HCl, 1% (v/v) acetic acid, 1mM EDTA) buffer. After 30 min of incubation at room temperature, the samples were electrophoresed at 100 V for 30 min, and the migration of DNA bands was visualized with a UV illuminator using a Gel Documentation System (Bio-Rad, Hercules, CA).
For particle size measurement, a fixed amount of DNA (5 μg) and siRNA (3 μg) were complexed with different amounts of polymers in 0.4 mL HBS buffer. After 30 min incubation at room temperature, the polyplex solution was diluted 10-fold with deionized water. Zeta-potential values and average sizes of the polyplexes were examined using a Zetasizer 3000HS (Malvern Instruments, USA) with a He–Ne Laser beam, 633 nm fixed scattering angle of 90° at 25 °C.
2.4. Cytotoxicity
The cytotoxicity of the polyplexes was measured by MTT assay. The cells, NIH 3T3 and H9C2, were seeded in 24-well plates at a density of 5.0 × 104 cells/well and incubated for 24 h in DMEM medium containing 10% FBS at 37 °C. Polyplexes were formed with plasmid DNA (0.5 μg/well) at different weight ratios and incubated for 30 min. The cells were treated with the formed polyplexes for 4 h in serum-free DMEM, followed by 24 h further incubation in DMEM medium with 10% FBS. The MTT solution (50 μL, 2 mg/mL) was then added and cells were further incubated for 2 h at 37 °C. After the medium was removed, DMSO (300 μL) was added to each well. The absorption was measured at 570 nm using a microplate reader (model 680, Bio-Rad Laboratory, Hercules, CA). The cell viability was determined as a percentage relative to untreated control cells. All experiments were performed in triplicate.
2.5. In Vitro Transfection Study
2.5.1. Luciferase Assay
For the Luciferase assay, the NIH 3T3 and H9C2 cells were seeded in 24-well plates at a density of 5.0 × 104 cells/well and incubated for 24 h in DMEM medium containing 10% FBS at 37 °C. Plasmid DNA (0.5 μg/well) was complexed with polymer at different weight ratios and incubated for 30 min. The cells were treated with the polyplexes for 4 h in serum-free media and then further incubated for 2 days in DMEM medium containing 10% FBS. The cells were rinsed with DPBS and treated with 200 μL of reporter lysis buffer, followed by shaking for 30 min at room temperature. Luciferase activity was measured by using 100 μL of luciferase assay reagent on a luminometer (Dynex Technologies Inc., Chantilly, CA). All experiments were performed in triplicate.
2.5.2. Fas Gene Silencing and Apoptosis Assay
H9C2 cells were seeded in a 6-well plate at a density of 1.0 × 105 cells/well and maintained for 24 h in DMEM medium containing 10% FBS. Fas siRNA (1.0 μg) was complexed with polymer at different weight ratios and incubated for 30 min. The cells were treated with polymer:Fas siRNA polyplexes for 4 h in serum-free media and then further incubated for 24 h in fresh DMEM medium containing 10% FBS. For hypoxic conditions, the cells transfected with Fas siRNA polyplexes were incubated in a hypoxia chamber (BD GasPak EZ system, San Diego, CA), which catalytically reduces oxygen to undetectable levels within 150 min [29]. The media was also changed to a hypoxic DMEM medium with low glucose that had been placed in the GasPak chamber 24 h before the experiments. Then, the cells were stimulated with 1 μg anti-Fas antibody/ml (MBL International Corporation, Watertown, MA) to induce Fas-mediated apoptosis. The cells were incubated under hypoxic conditions for 48 h at 37 °C. For surface Fas detection, the cells were collected and washed twice with washing buffer (PBS containing 2% FBS) and then incubated with anti-Fas antibody (MBL International Corporation) at 4 °C for 1 h and washed with washing buffer. FITC-labeled anti-mouse IgG (MBL International Corporation) was added to the cells for 30 min at 4 °C. The extent of Fas expression by the cells was detected by flow cytometry on the FL1 channel (FACS Caliber, Becton-Dickinson, Mountain View, CA) using Cell Quest software. A total of 10,000 events were acquired for each analysis.
2.5.3. Quantitative Real-Time PCR
Total RNA was isolated from the transfected H9C2 cells using TRIzol® Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. A reverse transcription reaction was performed with 1 μg pure total RNA using SuperScript™ III (Invitrogen, Carlsbad, CA), and quantitative realtime PCR was performed using a 7500 Fast Real-Time PCR System(Applied Biosystems, Foster City, CA, USA). 2μl of cDNA and Gene expression PCR Master Mix (Applied Biosystems) was used for PCR. Quantification of Fas gene expression in H9C2 cells was performed with Taq-Man Gene Expression Assays® (Applied Biosystems). All reactions were done in a 20 μl reaction volume following the manufacturer’s instructions. The PCR amplification reaction was carried out at 50°C for 2 min, at 95°C for 10 min, followed by 40 cycles of 95 °C for 15 s and at 60 °C for 1 min. The level of target gene Fas expression was determined by the comparative Ct method, whereby relative amounts of Fas mRNA were normalized to GAPDH mRNA.
To measure apoptotic cells, the apoptotic cell fraction (sub-G0/1) was measured by flow cytometry using PI staining after 48 h incubation. The cells were collected, washed twice with cold PBS and fixed with a 70% EtOH solution for 30 min at −20 °C. The cells were stored in PI staining solution (Invitrogen) for 4 h at 4 °C in the dark according to the manufacturer’s protocol. The fluorescence profiles of the PI-stained cells were collected on the FL2 channel by flow cytometry.
2.6. Cellular Binding and Uptake and Competition Assay using Flow Cytometry
NIH 3T3 and H9C2 cells were seeded at a density of 2.0 × 105 cells/well in a 6-well plate in DMEM medium containing 10% FBS and grown for 24 h. pDNA was labeled with YOYO-1 iodide (1 molecule of the dye per 20 base pairs of nucleotide) 30 min before use. The polyplexes were prepared with YOYO-1 labeled plasmid DNA (pDNA 1.0 μg) and polymers. After 30 min incubation, the cells were treated with polyplex for 3 h at 37 °C in serum-free medium. For the competition assay, the polyplexes were co-incubated with free PCM peptide. After removing the medium, the cells were washed with PBS, trypsinized and collected by centrifugation. The collected cells were suspended in 1 mL PBS, and cellular uptake was examined using a BD FACScan analyzer.
2.7. Statistical analysis
Results are expressed as mean values ± standard deviation (SD). Differences between groups were assessed by one-way analysis of variance (ANOVA) using SPSS 12.0 software (SPSS Inc., Chicago, IL, USA). One-way ANOVA followed by Tukey post hoc analysis was used to identify significance between groups.
3. Results and Discussion
3.1. Development and Evaluation of Peptide-Modified Polymers
The sequence of PCM, (a 20 amino acid peptide, WLSEAGPVVTVRALRGTGSW) selected by in vitro bio-panning, showed a high selectivity for cardiomyocytes. PCM modification was performed by conjugating a cysteine-terminated PCM to an activated poly(CBA-DAH) using a SM(PEG)2 cross-linker. PCM conjugation to PCD was confirmed and determined by UV/Vis and 1H-NMR spectrophotometry. The presence of the PCM peptide and PEG spacers on PCD was confirmed by the NMR peaks of tryptophan (W, aromatic ring, 5.9–6.9 ppm) and PEG (CH2CH2O, 3.6 ppm). The molar ratio of PCM peptide conjugated to PCD was determined by quantifying absorbance at 280 nm using UV/Vis spectrophotometry. The conjugation ratio through NMR and UV/Vis spectrophotometry was found to be, on average, 1 PCM per 6 DAH amines of PCD. The average molecular weight of PCD-PCM was also measured by an FPLC-SEC system (SEC, Superose 12 column). The molecular weight of PCM-PCD was estimated to be 15 kDa and its polydispersity index was 1.19.
3.2. Complex Formation and Characterization of Polyplexes
Cationic polymers formed complexes with polyanionic DNA (or siRNA) by self-assembly. Complex formation with PCM-modified PCD was examined by agarose gel electrophoresis (Figure 1). The polyanionic genes were condensed with the positively charged polymer at increasing weight ratios and studied for electrophoretic mobility in a gel retardation assay. PCM-PCD alone was not able to complex genes and demonstrated the native plasmid DNA (or siRNA) band in the gel retardation assay migrating uninhibited up to a weight ratio of 10. For more precise analysis and confirmation of complex formation, DLS was used to assess polyplex sizes. PCM-PCD/DNA alone was not able to form stable and compact nanoparticles, with nanoparticle sizes greater than 400 nm for all weight ratios, too large for efficient gene delivery [30]. The inability of PCM-PCD to form compact polyplexes may be due to the loss of cationic charge necessary for interaction with polyanionic genes, caused by the conjugation of PCM to the polymer via amine functional groups.
Figure 1.
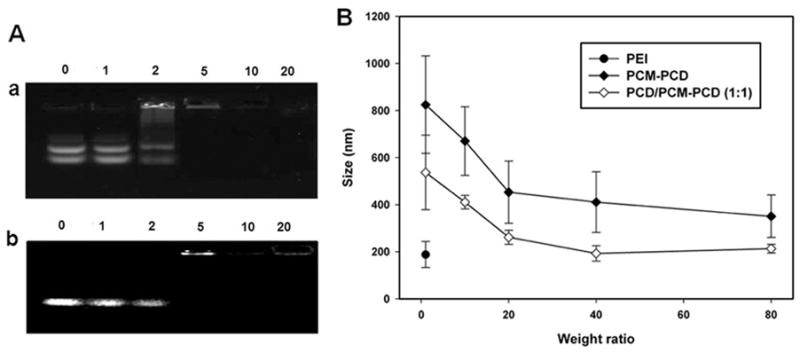
Polyplex formation assay. (A) Agarose gel electrophoresis retardation assay of plasmid DNA (a) and siRNA (b) by PCM-PCD/PCD (1:1). Plasmid DNA or siRNA only (0. 5 μg, lane 1), Weight ratios of polymer/DNA = 1, 2, 5, 10 and 20 (lanes 2, 3, 4, 5 and 6, respectively). (B) Polyplex size measurement with plasmid DNA at various weight ratios (n=3).
The overall percentage of conjugation of PCM to the primary amines of PCD was 20%. PCM modification of PCD results in a loss of positively charged primary amines and increased rigidity of PCD due to the presence of the additional PCM peptides. The decrease in cationic charge of the PCD with the addition of the PCM was detected by the potential values of the polyplex, for which the zeta potentials of the PCM-PCD/plasmid DNA polyplexes were significantly lower (+23.3 mV) than the measured potential of the unmodified PCD polyplexes (+38.1 mV) at the same weight ratio. To overcome this problem, PCM-PCD was mixed with unmodified PCD. By mixing DNA and siRNA with both PCM-conjugated PCD and unconjugated PCD, stable polyplexes could be formed. As shown in Figure 1, the extra cationic character added by unconjugated PCD enabled the PCD/PCM-PCD (1:1, w/w) mixture to completely condense plasmid DNA into particles with diameters less than 200 nm at a weight ratio of over 40:1. For siRNA, polypex formation followed a similar pattern (data not shown). These results show that the PCM modified bioreducible polymer can be formulated to complex with polyanionic genes efficiently, and the size of that complex is appropriate for effective gene delivery.
3.3. Cell Selectivity and Specific Binding to Cardiomyocytes
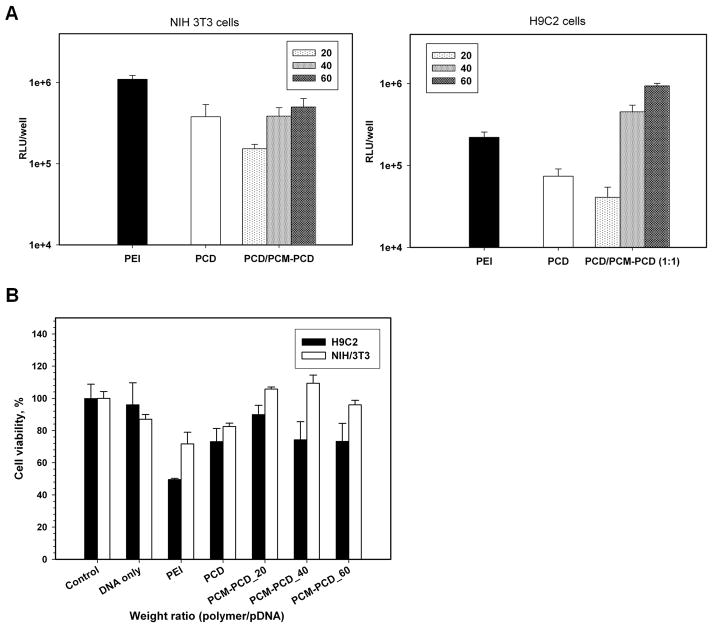
Luciferase assay and flow cytometry was used to quantify the specificity of PCM-modified polymer/pDNA (or siRNA) for gene delivery to cardiomyocytes. The transfection experiments with PCM-modified PCD were performed in H9C2 and NIH 3T3 cells. The PCM-PCD showed significantly increased reporter gene expression compared to the expression observed following transfection with unmodified PCD at weight ratios of greater than 40:1 in H9C2 cells, but there was no significant difference between the two polymers when transfecting NIH 3T3 cells (Figure 2). In addition, in vitro cytotoxicity by MTT assay showed good cell viability after transfection using PCM-PCD polyplexes, similar to that seen with nontoxic bioreducible PCD alone (Figure 2).
Figure 2.
Transfection efficiency and cytotoxicity of polymers/pDNA in H9C2 and NIH 3T3 cells. Each data point represents the mean ± standard deviation (n = 3). (A) For luciferase activity, cells were differentiated for 2 days. Results were expressed as RLU/well at various weight ratios. (B) Relative cell viability at various weight ratios of polyplexes was evaluated by MTT assay.
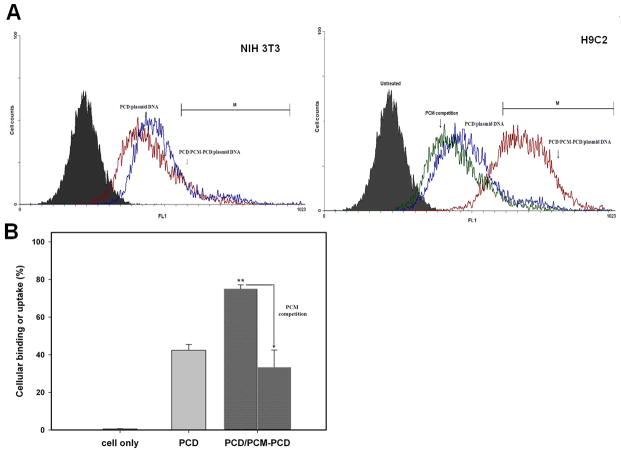
The binding and uptake by H9C2 cells of PCM-PCD nanoparticles mixed with PCD was investigated by FACS analysis using YOYO-1 labeled DNA. There was no difference in cellular uptake in non-cardiomyocyte NIH 3T3 cells. However, PCM-modified nanoparticles displayed significantly greater cellular binding and uptake in H9C2 cells than in NIH 3T3 cells. The PCM-PCD nanoparticles also showed greater binding and uptake than PCD in H9C2 cells (Figure 3). These results indicate that a bioreducible polymer conjugated with PCM residues has the ability to differentiate between cardiomyocyte and non-cardiomyocyte cell types. Specific cellular binding of PCM peptide was further confirmed by competition experiments through co-incubation with an excess of free PCM ligand. As shown by FACS analysis (Figure 3), the presence of the excess free PCM peptide blocked the uptake of the PCD modified with PCM peptide. Binding and cellular uptake was decreased from 75% to 33% in the presence of the PCM competitive ligand. These transfection experiments and cellular uptake studies indicate that the PCM-conjugated polymer is able to bind specifically to H9C2 cells, demonstrating the specific cardiomyocyte targeting ability which can be achieved with the addition of PCM to PCD.
Figure 3.
Cellular binding or uptake assay and competition experiment by flow cytometry. (A) Fluorescence histogram intensity of averaged cellular binding and uptake with YOYO-1-labeled pDNA in NIH 3T3 cells and H9C2 cells, and plotted as fluorescence intensity vs. count plot. For competition assays with free ligand to H9C2 cells, excess free ligand was incubated with the cells before treatment with nanoparticles. (B) Cellular binding % gated in M1 region in H9C2 cells. Each data point represents the mean ± standard deviation (n = 3). ** = P < 0.01
3.4. Fas siRNA Delivery to H9C2 Cells Using Peptide-Modified Carriers
3.4.1. Fas Gene Silencing Evaluation
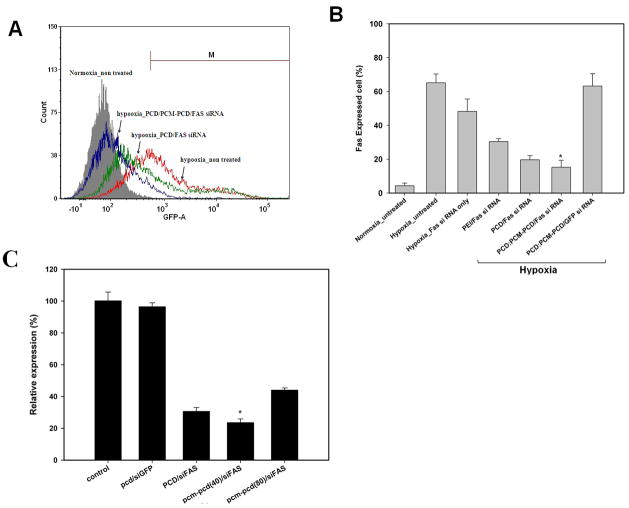
The cardiomyocyte-targeting PCM-conjugated polymer was evaluated for Fas siRNA delivery to cardiomyocytes for prevention of cardiomyocyte apoptosis. The ability of Fas gene silencing by PCM-polymer/Fas siRNA polyplexes to inhibit apoptosis under hypoxic conditions was verified in H9C2 cells by flow cytometry [31]. FACS demonstrated that Fas was expressed in 65% of all cells under hypoxic conditions, as compared to being expressed in only 10% of cells under normoxic conditions. Fas expression was decreased by Fas siRNA delivered by the bioreducible polymers PCD and PCD/PCM-PCD. We have previously reported efficient siRNA delivery using bioreducible polymers [27]. There was an improvement in Fas gene silencing with the PCM-modified PCD carrier compared to the unmodified PCD carrier (P<0.05) (Figure 4).
Figure 4.
Cardiomyocyte-targeted Fas gene silencing of PCM modified bioreducible polymer. (A) The representative histogram of Fas expression in H9C2 cells transfected by Fas siRNA formulations under normoxic or hypoxic conditions. The Fas expressed cells were measured by flow cytometry. (B) The graph indicates the results of Fas expressed H9C2 cells gated by M region. * = P < 0.05 vs all other groups. (C) The relative FAS expression level under hypoxia was normalized to that in non-treated H9C2 cells (control, 100%). * = P < 0.05 vs all other groups
Real-time-PCR was performed to confirm Fas gene silencing in H9C2 cells (Figure 4C). The mRNA expression of Fas was significantly reduced in H9C2 cells transfected with Fas siRNA using PCD and PCM-PCD as compared to GFP siRNA-transfected or non-transfected cardiomyocytes. Quantitative real-time PCR analysis demonstrated a 77% reduction in Fas gene expression in H9C2 cells treated with Fas siRNA delivered using the PCM-PCD/PCD carrier compared to non-transfected control cells. This Fas gene silencing as measured by FACS analysis and real-time PCR appears to be due to the targeted delivery of PCM-PCD/Fas siRNA polyplexes into cardiomyocytes.
3.4.2. Anti-Apoptotic Effect
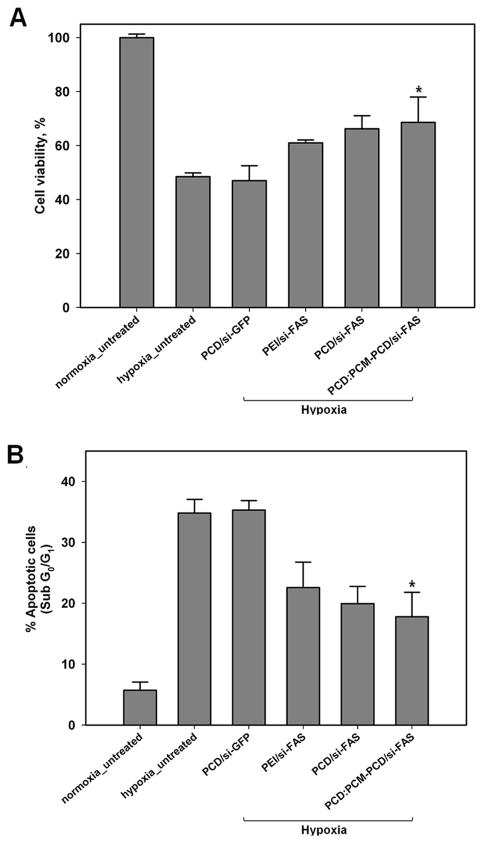
The cardiomyocyte-targeted anti-apoptotic effects of PCM-conjugated polymer with Fas siRNA were evaluated in H9C2 cells after exposure to hypoxia. Propidium iodide (PI) flow cytometric assays and MTT assays were performed after transfection with Fas siRNA and exposure to hypoxic conditions (Figure 5). Under normoxic conditions, cells were healthy and few were in the sub-G0/1 phase (apoptotic and necrotic cells, 5.72 ± 1.3%), whereas the percentage of apoptotic cells under hypoxic conditions was 34.8 ± 2.3%. Incubation with PCD/PCM-PCD Fas siRNA under hypoxic conditions displayed significantly higher cell viability (69.6 ± 2.9%, P<0.05) for transfected cells than non-transfected cells. The sub-G0/1 population was also reduced significantly in transfected cells, to 17.8 ± 3.9%, compared to the non-transfected cells under hypoxic conditions. There was no significant difference in apoptotic populations between cell groups under normoxic conditions. These results suggest that the PCM-modified bioreducible polymer/Fas siRNA polyplexes facilitate Fas gene silencing and inhibit apoptosis in cardiomyocytes under hypoxic conditions.
Figure 5.
Anti-apoptotic effect of PCM modified bioreducible polymer H9C2 cells. (A) Cell viability was determined by MTT assay. (B) The graph indicates the sub-G1 cell fractions of apoptotic cells after PI staining after 48 h. All experiments were performed in triplicate. The data are expressed as the mean ± SD. * = P < 0.05 compared to untreated and scrambled si-RNA (GFP si-RNA) under hypoxia.
4. Conclusion
In this study, we report the development of a cardiomyocyte-targeting polycationic polymer for siRNA delivery to silence Fas gene expression and inhibit cardiomyocyte apoptosis. Cardiomyocyte apoptosis is a pathophysiologic mechanism underlying a number of forms of heart disease. The synthetic PCM peptide is an especially promising moiety for targeting cardiomyocytes. PCM modified polymers can efficiently and specifically bind cardiomyocytes, improving transfection efficiency without significant cytotoxicity. We believe that PCM modification is a promising approach for cardiomyocyte-targeted siRNA delivery to inhibit cardiomyocyte apoptosis.
Acknowledgments
This work was financially supported by NIH grants HL 065447 (SWK) and HL071541 (DAB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu F, Huang L. Development of non-viral vectors for systemic gene delivery. J Control Release. 2002;78:259–266. doi: 10.1016/s0168-3659(01)00494-1. [DOI] [PubMed] [Google Scholar]
- 2.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 3.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, et al. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J Am Chem Soc. 2004;126:2355–2361. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 5.Wright MJ, Rosenthal E, Stewart L, Wightman LML, Miller AD, Latchman DS, et al. Beta-galactosidase staining following intracoronary infusion of cationic liposomes in the in vivo rabbit heart is produced by microinfarction rather than effective gene transfer: a cautionary tale. Gene Ther. 1998;5:301–308. doi: 10.1038/sj.gt.3300590. [DOI] [PubMed] [Google Scholar]
- 6.Christensen LV, Chang C-W, Kim WJ, Kim SW, Zhong Z, Lin C, et al. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 7.Kim T-i, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nam HY, Hahn HJ, Nam K, Choi W-H, Jeong Y, Kim D-E, et al. Evaluation of generations 2, 3 and 4 arginine modified PAMAM dendrimers for gene delivery. Int J Pharm. 2008;363:199–205. doi: 10.1016/j.ijpharm.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Nam HY, Nam K, Hahn HJ, Kim BH, Lim HJ, Kim HJ, et al. Biodegradable PAMAM ester for enhanced transfection efficiency with low cytotoxicity. Biomaterials. 2009;30:665–673. doi: 10.1016/j.biomaterials.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Koo H, Jin G-w, Kang H, Lee Y, Nam HY, Jang H-s, et al. A new biodegradable crosslinked polyethylene oxide sulfide (PEOS) hydrogel for controlled drug release. Int J Pharm. 2009;374:58–65. doi: 10.1016/j.ijpharm.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Choi JS, Nam K, Park J-y, Kim J-b, Lee J-K, Park J-s. Enhanced transfection efficiency of PAMAM dendrimer by surface modification with L-arginine. J Control Release. 2004;99:445–456. doi: 10.1016/j.jconrel.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Lundberg P, Langel U. A brief introduction to cell-penetrating peptides. J Mol Recognit. 2003;16:227–233. doi: 10.1002/jmr.630. [DOI] [PubMed] [Google Scholar]
- 13.Lim Y-b, Kim C-h, Kim K, Kim SW, Park J-s. Development of a safe gene delivery system using biodegradable polymer, Poly[a-(4-aminobutyl)-l-glycolic acid] J Am Chem Soc. 2000;122:6524–6525. [Google Scholar]
- 14.Jarver P, Langel U. Cell-penetrating peptides--a brief introduction. Biochim Biophys Acta. 2006;1758:260–263. doi: 10.1016/j.bbamem.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Kim I-D, Lim C-M, Kim J-B, Nam HY, Nam K, Kim S-W, et al. Neuroprotection by biodegradable PAMAM ester (e-PAM-R)-mediated HMGB1 siRNA delivery in primary cortical cultures and in the postischemic brain. J Control Release. 2010;142:422–430. doi: 10.1016/j.jconrel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami S, Sato A, Nishikawa M, Yamashita F, Hashida M. Mannose receptor-mediated gene transfer into macrophages using novel mannosylated cationic liposomes. Gene Ther. 2000;7:292–299. doi: 10.1038/sj.gt.3301089. [DOI] [PubMed] [Google Scholar]
- 17.Park I-K, Lasiene J, Chou S-H, Horner PJ, Pun SH. Neuron-specific delivery of nucleic acids mediated by Tet1-modified poly(ethylenimine) J Gene Med. 2007;9:691–702. doi: 10.1002/jgm.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire MJ, Samli KN, Johnston SA, Brown KC. In vitro Selection of a peptide with high selectivity for cardiomyocytes in vivo. J Mol Biol. 2004;342:171–182. doi: 10.1016/j.jmb.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 19.Barry MA, Dower WJ, Johnston SA. Toward cell-targeting gene therapy vectors: selection of cell-binding peptides from random peptide-presenting phage libraries. Nat Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 20.Nabel EG. Cardiovascular Disease. N Engl J Med. 2003;349:60–72. doi: 10.1056/NEJMra035098. [DOI] [PubMed] [Google Scholar]
- 21.Hamar P, Song E, Kökény G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephanou A, Scarabelli TM, Brar BK, Nakanishi Y, Matsumura M, Knight RA, et al. Induction of apoptosis and Fas receptor/Fas ligand expression by ischemia/reperfusion in cardiac myocytes requires serine 727 of the STAT-1 transcription factor but not tyrosine 701. J Biol Chem. 2001;276:28340–28347. doi: 10.1074/jbc.M101177200. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Li W, Min J, Ou Q, Chen J. Fas siRNA reduces apoptotic cell death of allogeneic-transplanted hepatocytes in mouse spleen. Transplant Proc. 2003;35:1594–1595. doi: 10.1016/s0041-1345(03)00438-x. [DOI] [PubMed] [Google Scholar]
- 24.Wesche-Soldato DE, Chung CS, Lomas-Neira J, Doughty LA, Gregory SH, Ayala A. In vivo delivery of caspase-8 or Fas siRNA improves the survival of septic mice. Blood. 2005;106:2295–2301. doi: 10.1182/blood-2004-10-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue T-L, Ma X-L, Wang X, Romanic AM, Liu G-l, Louden C, et al. Possible involvement of stress-activated protein kinase signaling pathway and Fas receptor expression in prevention of ischemia/reperfusion-induced cardiomyocyte apoptosis by carvedilol. Circ Res. 1998;82:166–174. doi: 10.1161/01.res.82.2.166. [DOI] [PubMed] [Google Scholar]
- 26.Kim SH, Jeong JH, Ou M, Yockman JW, Kim SW, Bull DA. Cardiomyocyte-targeted siRNA delivery by prostaglandin E2-Fas siRNA polyplexes formulated with reducible poly(amido amine) for preventing cardiomyocyte apoptosis. Biomaterials. 2008;29:4439–4446. doi: 10.1016/j.biomaterials.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SH, Jeong JH, Kim T-i, Kim SW, Bull DA. VEGF siRNA delivery system using arginine-grafted bioreducible poly(disulfide amine) Mol Pharm. 2008;6:718–726. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou M, Wang X-L, Xu R, Chang C-W, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjug Chem. 2008;19:626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imhof A, Heinzer I. Continuous monitoring of oxygen concentrations in several systems for cultivation of anaerobic bacteria. J Clin Microbiol. 1996;34:1646–1648. doi: 10.1128/jcm.34.7.1646-1648.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zauner W, Ogris M, Wagner E. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv Drug Deliv Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 31.Wanka H, Kessler N, Ellmer J, Endlich N, Peters BS, Clausmeyer S, et al. Cytosolic renin is targeted to mitochondria and inducesapoptosis in H9c2 rat cardiomyoblasts. J Cell and Mol Med. 2009;13:2926–2937. doi: 10.1111/j.1582-4934.2008.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]