Abstract
Ribonucleic acid interference (RNAi) is a powerful molecular tool that has potential to revolutionize the treatment of cancer. One major challenge of applying this technology for clinical application is the lack of site-specific carriers that can effectively deliver short interfering RNA (siRNA) to cancer cells. Here we report the development and assessment of a cancer-cell specific magnetic nanovector construct for efficient siRNA delivery and non-invasive monitoring through magnetic resonance imaging (MRI). The base of the nanovector construct is comprised of a superparamagnetic iron oxide nanoparticle core coated with polyethylene glycol (PEG)-grafted chitosan, and polyethylenimine (PEI). The construct was then further functionalized with siRNA and a tumor-targeting peptide, chlorotoxin (CTX), to improve tumor specificity and potency. Flow cytometry, quantitative RT-PCR, and fluorescence microscopy analyses confirmed receptor-mediated cellular internalization of nanovectors and enhanced gene knockdown through targeted siRNA delivery. The ability of this nanovector construct to generate specific contrast enhancement of brain tumor cells was demonstrated through MR imaging. These findings suggest that this CTX enabled nanoparticle carrier may be well suited for delivery of RNAi therapeutics to cancer cells.
Keywords: Nanoparticle, Gene Therapy, Cancer, Bioconjugation, MRI, siRNA
1. Introduction
RNA interference (RNAi) is a mechanism by which living cells regulate gene expression using small RNA molecules. Short interfering RNA (siRNA) is double stranded, non-coding RNA, which binds to complementary mRNA to direct gene silencing through argonaute, an endonuclease within the RNA-induced silencing complex (RISC) [1,2]. RNAi is a rapidly developing frontier in the field of gene therapy that shows tremendous potential as a therapeutic approach for the treatment of various diseases caused by aberrant gene expression [3], and cancer is a prime target where this technology can be applied [4,5]. By silencing the genes that contribute to uncontrolled cell growth, siRNA treatments can curb pathogenesis and potentially induce cancer cell death. However, unlike conventional chemotherapeutics, RNA molecules are anionic, hydrophilic, and cannot be internalized by cells through passive diffusion [5]. Furthermore, upon internalization they are ineffectively trafficked by cancer cells, hindering their potency [6]. Therefore, there is considerable interest in the development of safe and effective carriers to facilitate both the delivery and intracellular trafficking of siRNA.
A broad spectrum of siRNA delivery constructs, including viral vectors, liposomes, cationic polymers and dendrimers, cell-penetrating peptides, semiconductor quantum dots, and gold and magnetic nanoparticles have been investigated [6-12]. Non-viral methods are preferred over viral approaches because they present a lower risk of immunogenicity, and do not produce oncologic side effects [6]. Thus, in recent years a great deal of attention has been paid towards the development of non-viral carriers for siRNA delivery [13]. A common characteristic among these constructs is their net positive charge, which contributes to both complex formation with the anionic siRNA, and interaction with the negatively charged cell membrane [14-16]. From these carriers, several formulations (e.g. liposomes, and cationic polymers) have emerged and shown great potential for clinical implementation for cancer therapy [6,17]. For example a lipoplex formulation developed by Alnylam inc. is currently under Phase I clinical investigation for the treatment of liver cancer [18]. Also, Silence Therapeutics has developed a liposome formulation for the treatment of solid tumors [19].
Although significant advances have been made in developing gene delivery vehicles, poor site specificity, low efficacy in gene silencing, and lack of non-invasive delivery monitoring remain major hurdles towards clinical advancement [20]. We have recently developed a magnetic nanoparticle platform consisting of a superparamagnetic iron oxide (Fe3O4) core coated with a cationic copolymer of chitosan-grafted-polyethelyne glycol (PEG) and polyethylenimine (PEI) for non-viral DNA delivery [21]. In this system, the superparamagnetic core enables non-invasive monitoring of DNA delivery in real-time by magnetic resonance imaging (MRI). The combination of chitosan and PEG form a PEGylated chitosan coating on the iron oxide core, stabilizing the nanoparticle from agglomeration [22]. Cationic PEI was incorporated into the coating to complex negatively charged DNA by electrostatic interaction, and to enable proper intracellular trafficking. This unique formulation has shown efficacy to safely deliver plasmid DNA in vivo and transfect brain tumor cells [21].
In present study we aim to develop a new multifunctional magnetic nanovector based on our previously developed nanoparticle platform, which would enable cancer-specific targeting, effectively deliver sufficient dose of siRNA to target cells to induce gene silencing, while providing the capability of carrier monitoring through MRI. In designing this nanovector, siRNA is covalently attached to nanoparticles to prevent its degradation by extracellular or intracellular enzymes, and thus improving the efficacy in gene silencing. To enable site specificity, the targeting peptide chlorotoxin (CTX) was covalently attached to the nanoparticles. CTX is a 4-kDa cationic peptide originally purified from the venom of the Leiurus quinquestriatus scorpion, and it was chosen because of its high affinity to the vast majority of brain tumors (74 of 79 World Health Organization classifications of brain tumors) as well as prostate, skin, and colorectal cancers [23,24]. It is postulated that the target of CTX on cancer cells is associated with the membrane-bound matrix metalloproteinase-2 (MMP-2) protein complex, which is up-regulated on brain tumors and other invasive cancer [24,25]. Here, we present our nanovector design and examine its ability to deliver siRNA to brain tumor cells through receptor-mediated endocytosis, and to specifically knockdown the transgene expression of green fluorescence protein (GFP) in C6/GFP+ glioma cells.
2. Materials and methods
2.1. Materials
All reagents were purchased from Sigma Aldrich (St. Louis, MO) unless otherwise specified.
2.2. Nanoparticle synthesis
Iron oxide (Fe3O4) nanoparticles coated with chitosan, PEG, and PEI were prepared as previously described [21]. Briefly, iron oxide nanoparticles were co-precipitated in the presence of chitosan-grafted PEG polymer (chitosan-g-PEG). 1.2-kDa PEI was then modified with succinimidyl iodoacetate (SIA; Molecular Biosciences, Boulder, CO) at a 1.2:1 molar ratio in thiolation buffer (0.1M sodium bicarbonate, 5mM EDTA, pH 8.0) through N-hydroxy succinimide ester chemistry. Concurrently, nanoparticles coated with chitosan-g-PEG were modified with 2-iminothiolane (2IT; Molecular Biosciences, Boulder, CO) by adding 30 mg of 2IT to 3 ml of nanoparticles (1 mg of Fe/ml). Both reactions were shielded from light and preceded under gentle shaking for one hour. After the one-hour incubation, excess 2IT was removed by gel permeation chromatography using a PD-10 desalting column (GE Healthcare, Piscataway, NJ) equilibrated with thiolation buffer. The modified PEI was added to the purified nanoparticle (216 mg PEI/g Fe3O4) and reacted in the dark for >1 hr under gentle shaking. The resulting nanoparticle/PEI complex was stored at 4°C and allowed to react overnight before removing excess PEI using S-200 Sephacryl resin (GE Healthcare, Piscataway, NJ) equilibrated with HEPES buffer (20mM, pH 7.4). The formed iron oxide nanoparticles coated with chitosan-g-PEG and subsequently modified with PEI modified are referred herein as NP.
2.3. Coating polymer characterization
The degree of chitosan PEGylation was determined using a UV spectroscopy method as reported previously [26]. Briefly, a dried sample of PEGylated chitosan was weighed, and subsequently dissolved in 0.1 N HCl solution. The absorbance of solutions at a wavelength of 201 nm was then measured using UV spectroscopy and compared to a standard of pure chitosan to determine chitosan content. The following formula was then applied to determine the degree of pegylation: PEG weight in solution = Total sample weight – measured chitosan weight. Using this method we determined the molar ratio of chitosan:PEG to be 2.2.
2.4. siRNA preparation
siRNA sequences designed to knockdown GFP expression, and modified with thiol (a set without thiol modification were also acquired) and Dy547 were purchased from Dharmacon, Lafayette, CO. The specific sequences were 5′GCAAGCUGACCCUGAAGUUCUU3′–antisense, 5′GAACUUCAGGGUCAGCUUGCUU3′–sense. These sequences were acquired with protected-thiol modifications on the 5′ end of the sense strand and with Dy547 modification on the 5′ end of the antisense strand. siRNA sequences were received as single strands and were annealed to their complementary strand in annealing buffer (12 mM potassium chloride, 1.2 mM HEPES, 0.04 mM magnesium chloride, pH 7.5) by incubating at 95°C for five minutes, then 37°C for 1 hr, and then stored at −20°C.
2.5. Nanoparticle-siRNA complex (NP-siRNA) formation
A suspension of NP in thiolation buffer was prepared at a concentration of 1 mg of Fe/ml. Amine groups on the surface of NP were then modified with SIA by addition of 200 μg of SIA dissolved in DMSO. Concurrently, the annealed siRNA with protected-thiol was deprotected by adding 57.3 mg/mL Tris(2-Carboxyethyl) phosphine Hydrochloride (TCEP·HCl; Molecular Biosciences, Boulder, CO) at a 1:1 volume ratio. Both reactions proceeded in the dark with gentle rocking for 1 hr. After 1 hr, both the SIA-modified NP (NP-SIA) and the deprotected siRNA were purified using Zeba™ Micro Spin Desalting Columns (Thermo Fisher Scientific, Waltham, MA) equilibrated with thiolation buffer supplemented with 150mM NaCl. NP-SIA and deprotected siRNA were then mixed in PBS (5 mM EDTA, pH 7.4) at concentrations corresponding to the wt:wt (Fe mass of NP : siRNA mass) ratios tested (0:1, 0.1:1, 0.5:1, 1:1, 5:1, 10:1, 20:1) and immediately vortexed. The solutions were incubated overnight at 4°C to allow the formation of NP-siRNA complexes.
2.6. Nanoparticle-siRNA-chlorotoxin complex (NP-siRNA-CTX) formation
A 1 mg/mL solution of chlorotoxin (CTX; Alamone Labs, Jerusalem, Israel) was prepared in thiolation buffer and reacted with 2IT at a 1.2:1 molar ratio of 2IT:CTX for 1 hour in the dark. Concurrently, NP-siRNA complexes were reacted with SM(PEG)12 (Thermo Fisher Scientific, Waltham, MA) at 216 μg of SM(PEG)12/mg Fe2O3 in the dark with gentle rocking for 30 minutes. NP-SM(PEG)12 was then reacted with CTX-2IT at 1 μg CTX per 4.5 μg Fe for one hour in the dark. The resultant NP-siRNA-CTX was purified using Zeba™ Micro Spin Desalting Columns equilibrated with PBS, and stored at 4°C.
2.7. siRNA binding assay
A 4% low melting point agarose gel was prepared with 0.05 μg/mL ethidium bromide. While maintaining a uniform concentration of siRNA, samples of NP :siRNA complexes were prepared at NP:siRNA weight ratios (Fe mass of NP: siRNA mass) ranging from 0:1 to 20:1. siRNA binding was analyzed by gel electrophoresis at 55 V for 90 min. Images were acquired on a Gel Doc XR (Bio-Rad Laboratories, Hercules, CA).
2.8. siRNA release assay
NP-siRNA complexes were prepared at the optimal ratios determined from the binding assay (10:1, w/w), and reacted in 100 mM glutathione for 90 min at 37°C. Samples were then treated with heparin (1,000 units/ml, 10 μL heparin/μg siRNA) and incubated for 30 min at room temperature to block the electrostatic interaction between the NP and siRNA. siRNA release was analyzed on a 4% low melting point agarose gel containing 0.05 μg/mL ethidium bromide, running at 55 V for 90 min. Images were obtained on a Gel Doc XR. siRNA release was quantified using the Quantity One software package (Bio-Rad Laboratories, Hercules, CA).
2.9. CTX binding assay
To quantify the degree of CTX attachment to nanoparticles, NP-siRNA-CTX were prepared as described above without purification of unbound CTX through S-200 sephacryl resin. Free, unreacted CTX was separated from the CTX conjugated to NPs through SDS-PAGE and quantified using the Quantity One software package and a standard curve of known concentrations of CTX. CTX conjugated to NPs was calculated by subtracting the amount of free CTX from the total amount of CTX in the reaction.
2.10. Nanoparticle characterization
Nanoparticle samples were prepared in 20 mM HEPES buffer (pH 7.4) to a concentration of 100 μg/mL, and then analyzed for hydrodynamic size and zeta potential using a DTS Zetasizer Nano (Malvern Instruments, Worcestershire, UK). For stability studies, nanoparticle samples were diluted to a concentration of 100 μg/mL in the indicated solution, then analyzed for hydrodynamic size using a DTS Zetasizer Nano, and imaged using a digital camera.
2.11. Cell culture
C6 rat glioma cells (ATCC, Manassas, VA) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Atlanta Biological, Lawrenceville, GA) and 1% antibiotic-antimycotic (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2. Enhanced Green Fluorescent Protein (EGFP) fluorescing C6 cells (GFP+ C6) were produced by stably transfecting C6 cells with the pEGFP-N1 vector using the Effectene transfection reagent (Qiagen, Valencia, CA) following the manufacturer’s protocol. 48 hrs post-transfection, cells were sorted using a FACS Vantage and maintained in fully supplemented DMEM with 1 mg/ml G-418.
2.12. Cell transfection
The day before transfection, cells were plated at 200,000 cells per well in 12-well plates. For transfection of cells with NP-siRNA and NP-siRNA-CTX formulations, cells were treated with 2 μg/mL of siRNA for 2 hrs under normal cell culture conditions. After the 2-hour incubation the media was replaced and cells were incubated for an additional 48 hrs before analysis. For trasnfection of cells with free siRNA, cells were treated with siRNA at 2 μg/ml for 48 hrs under normal cell culture conditions. For transfection of cells with PEI/siRNA, siRNA (20 μg/ml, serum free DMEM) and 25-kDa hyperbranched PEI (52 μg/ml, serum free DMEM) were mixed and allowed to form the PEI/siRNA complex for 15 min, and cells were then treated with the complexes (2 μg of siRNA/ml in serum free DMEM) for 24 hrs. After the 24-hour incubation cell culture media was replaced with serum contain DMEM, and cells were assayed after an additional 24 hours of growth. Transfections of cells with siRNA using Dharmafect 4 (Lafayette, CO) and Lipofectamine 2000 (Invitrogen, Carlsbad, CA) were performed according to the manufacturer’s instructions.
2.13. Alamar blue cell viability analysis
Cells were grown to confluence on 12-well plates, and treated as described for cell transfection experiments. After treatment, samples were washed with PBS three times, and incubated for 1 hr with 10% Alamar blue (Invitrogen) in phenol-free DMEM (supplemented with 10% FBS and 1% antibiotic–antimycotic). The percent reduction of Alamar blue was determined following the manufacturer’s protocol. Cell counts of treated and untreated samples were compared to determine percent viability of treated samples (untreated cells assumed to represent 100% viability).
2.14. Confocal fluorescence microscopy
50,000 treated cells were plated on each of 24 mm glass cover slips and allowed to attach for 24 hrs. Cells were then washed with PBS and fixed in 4% formaldehyde (Polysciences Inc., Warrington, PA) for 30 min. Cells were then washed 3 times with PBS, and stained with membrane stain WGA-AF647 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Cover slips were then mounted on two microscope slides using Prolong Gold antifade solution (Invitrogen, Carlsbad, CA) containing DAPI for cell nuclei staining. Images were acquired on a LSM 510 Meta confocal fluorescence microscope (Carl Zeiss Inc., Peabody, MA) with the appropriate filters.
2.15. Endosomal escape assay
C6 cells (200,000 cells/ cover slip) were plated on 24 mm glass cover slips and allowed to attach for 24 hrs under normal cell culture conditions. Cells were then treated with the membrane impermeable dye Calcein (0.25 mM, Invitrogen), physical mixtures of calcein and NP-siRNA-CTX, or calcein and NP/siRNA. After 2 hrs, excess dye was washed off, and samples were prepared for confocal fluorescence microscopy as described above. Fluorescence imaging was used to evaluate endosome escape (indicated by diffuse cytoplasmic calcein fluorescence).
2.16. Flow cytometry
Cells treated with a transfection reagent were washed with PBS, and detached using TrypLE Express (Invitrogen, Carlsbad, CA), and resuspended in PBS containing 2% FBS. At least 10,000 cells were then analyzed using a BD FACSCanto flow cytometer (Beckton Dickinson, Franklin Lakes, NJ) and data analyses were performed using the FlowJo software package (Tree Star, Ashland, OR).
2.17. Real-time PCR
For gene expression analysis, cells were removed from culture 48 hrs after transfection and RNA was extracted using the Qiagen RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. mRNA reverse transcription (RT) was performed with a BioRad iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). DNA transcripts were then probed using BioRad iQ SYBR Green Supermix with Qiagen QuantiTect Primer Assays for GFP and rat β-actin. Thermocycling was performed with a BioRad CFX96 Real-Time Detection System under the following conditions: 95°C for 15 min, 45 cycles of denaturation (15 s, 94°C), annealing (30 s, 55°C), and extension (30 s, 72°C). The relative expression of GFP was compared to the expression of β-actin and normalized to the untreated cells as a control.
2.18. In vitro MRI
Cell samples (1 million cells) were suspended in 50 μL of 1% agarose. For nanovector samples, 25 μL nanoparticles dispersed in PBS were mixed with 25 μL of 1% agarose. T2 relaxation measurements were performed on a 4.7-T Bruker magnet (Bruker Medical Systems, Karlsruhe, Germany) equipped with Varian Inova spectrometer (Varian, Inc., Palo Alto, CA). A 5 cm volume coil and spin-echo imaging sequence were used to acquire T2-weighted images. Images were acquired using a repetition time (TR) of 3000 ms and echo times (TE) of 13.6, 20.0, 40.0, 60.0, 90.0 and 120.0 ms. The spatial resolution parameters were: acquisition matrix of 256 × 128, field-of-view of 35 × 35 mm, section thickness of 1 mm, and two averages. The T2 map was generated by NIH ImageJ (Bethesda, MD) based on the equation, SI = A*exp(−TE/T2) + B, where SI is the signal intensity, TE is the echo time, A is the amplitude, and B is the offset. R2 maps were generated by taking the reciprocal of T2 maps.
3. Results and discussion
3.1. Nanovector synthesis
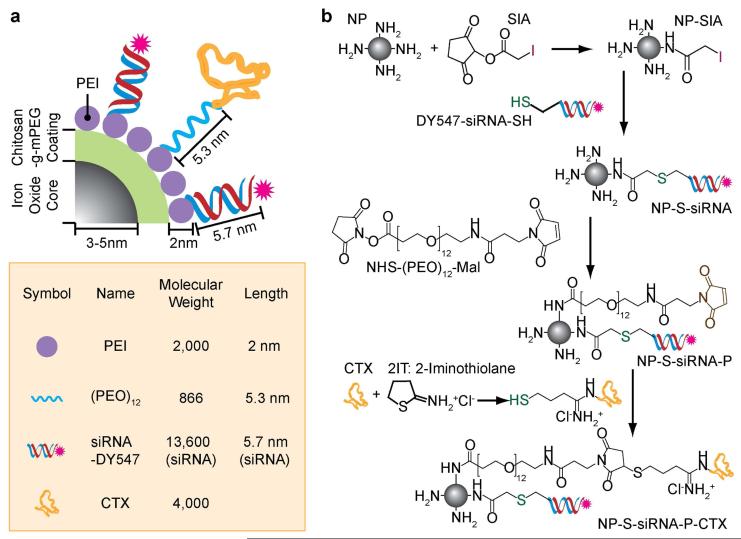
The chemical components and assembly of the cancer targeting magnetic nanovector (NP-siRNA-CTX) developed in this study are shown in Figure 1. As shown in Figure 1a, the nanovector contains a superparamagnetic iron oxide nanoparticle core (6–10 nm diameter) coated with chitosan-g-PEG (2.2 molar ratio of chitosan:PEG, see Experimental Section). PEI is subsequently attached to the chitosan-g-PEG coating to enable proper intracellular trafficking [21]. PEI is known for its ability to facilitate endosome escape via the “proton sponge effect” wherein the influx of protons and counter-ions into the endosome increases the osmotic pressure, leading to swelling and rupture of endosomes and release of its contents [27]. The base nanoparticle (NP), i.e., the iron oxide nanoparticles coated with chitosan-g-PEG copolymer and PEI, was synthesized following a process reported in our previous study [22].
Figure 1.
Schematic representation for development of a cancer targeting magnetic siRNA nanovector (NP-siRNA-CTX). (a) Chemical illustration of NP-siRNA-CTX. (b) Chemical reaction scheme for the conjugation of NHS-PEO12-malemide, siRNA, and CTX to NP.
Outlined in Figure 1b are the details of our chemical scheme used to assemble siRNA, PEO linker, and CTX on the NP surface. Fluorescently modified siRNA (Dy547-siRNA with 21 nucleotide and 5.7 nm in length), designed to knockdown the transgene expression of green fluorescence protein (GFP), was covalently attached to the PEI on the nanoparticle by reacting its amine groups with thiol-reactive iodoacetyl groups of SIA on NP. A heterobifunctional PEO linker, N-hydroxysuccinimide (NHS)-polyethylene oxide (PEO)12-maleimide of 5.3 nm in length was also attached to the PEI to serve as an anchor for further attachment of the targeting ligand CTX, which could allow CTX to be exposed outwardly and effectively interact with target cells. CTX was first thiolated through reaction with 2-iminoothiolane (2IT), and then conjugated to the PEO linker through the reaction of its thiol group with the malemide group of the PEO, to form the nanovector (NP-siRNA-CTX). A nanovector without CTX (i.e., NP-siRNA) was also prepared as a control in our study. Previously we have demonstrated that CTX peptides anchored to short chains of PEO can assemble on nanoparticle surfaces as multivalent displays prompting enhanced affinity to cancer cells compared to the free peptide [28]. This concept was integrated in the design of our current system to ensure proper molecular assembly of the peptide on the NP surface.
3.2. Characterization of siRNA loading and stability of the nanovector
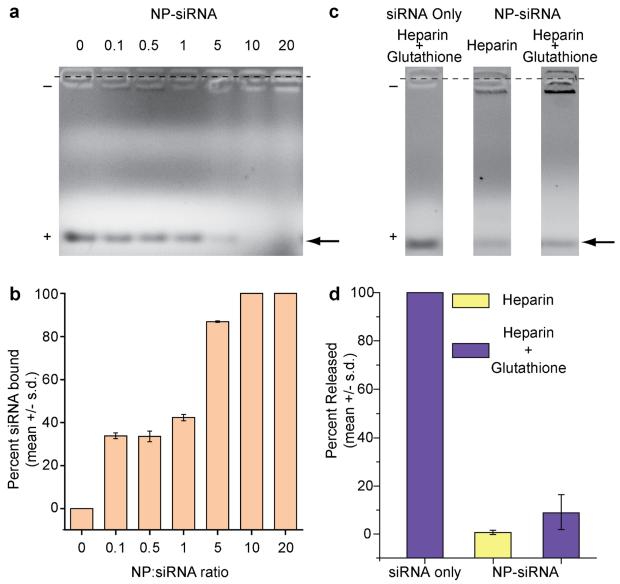
The siRNA loading efficiency of the nanovector was assessed using gel retardation assays (Figure 2). The NP-siRNA-CTX was prepared at varying NP:siRNA weight ratios (Fe mass of NP : siRNA mass) and the reaction time was optimized to allow for the completion of the reaction for each ratio. The reaction products were then loaded in agarose gel wells without purification (Figure 2a). siRNA bound to NP remained in wells, while unbound siRNA migrated towards the positive electrode (bottom). Figure 2b shows the corresponding quantitative volume measurements for the intensities of migrating siRNA bands. As shown, at a ratio of 10, 100% of the siRNA was completely bound to NP, and thus this ratio was used for subsequent experiments in this study.
Figure 2.
Assessment and optimization of siRNA attachment to NP and the subsequent verification of linkage chemistry. (a) Gel retardation assay analyzing siRNA attachment onto base NPs at different weight ratios of NP to siRNA (the arrow points to the band of interest). (b) Corresponding quantitative analysis (gel shown in a) of degree of siRNA binding. (c) Gel retardation assay analyzing siRNA release profiles of NP-siRNA complex through treatment with Heparin (electrostatic disrupting agent) and Glutathione (reducing agent) (the arrow points to the band of interest). (d) Corresponding quantitative analysis (gel shown in c) of percent siRNA released from NPs.
To verify that a stable covalent thioether linkage was formed between NP and siRNA, we studied the release properties of the nanovector after treating with heparin, or heparin+glutathione mixture. For these assays, the strongly anionic molecule, heparin, and glutathione, were used as an electrostatic disrupting agent and a reducing agent, respectively, to examine the stability of covalent linkage between siRNA and NP in the presence of exterior competitive molecules and in reductive environments. Naked siRNA was provided as a reference of unbound siRNA. Figure 2c shows the images of gels analyzed in these assays, and Figure 2d shows the corresponding quantitative results in term of percent siRNA released, as determined by the volume-based intensity measurements of siRNA bands. The results of these assays showed that the NP-siRNA complex exhibited negligible siRNA release in response to either heparin or glutathione treatment confirming the formation of a robust non-cleavable covalent linkage between the NP and siRNA. Covalent attachment of siRNA to a nanocarrier is desirable for in vivo applications because it ensures that the siRNA and carrier will remain intact during blood circulation. In this study we used a thioether linkage to facilitate covalent conjugation of siRNA to the nanoparticles. To favor thioether bond formation over electrostatic binding of the negatively charged siRNA with cationic NP the reaction was performed in a high ionic strength buffer (see Materials and methods section).
The physicochemical properties of nanoparticles, particularly the hydrodynamic size and zeta potential, are known to greatly influence their behavior both in vitro and in vivo, and internalization by cells [29]. The hydrodynamic size and zeta potential of the nanovector were measured by dynamic light scattering (DLS) and these results are summarized in Table 1. The NP-siRNA nanovectors (NP:siRNA ratio of 10:1) have an average core size of 7.5 nm, a hydrodynamic size of 111.9 ± 52.4 nm, and a cationic zeta potential of 19.6 ± 9.7 mV. The number of siRNA and CTX molecules per nanoparticle were calculated to be 3.8, and 5 respectively, as determined by gel retardation assays. The magnetic relaxivity of each NP-siRNA, which determines the detectability of the magnetic nanovectors by MRI, was evaluated by R2 measurements, and was found to be 214.07 S−1 mM−1. The hydrodynamic size of NP-siRNA falls within the acceptable size range (5 < d < 200 nm) to ensure in vivo success and evasion of sequestration by macrophages [30].
Table 1.
Primary physicochemical properties of NP-siRNA-CTX
| Core Size (nm) |
Hydrodynamic Size (nm) |
Zeta Potential (mV) |
siRNA Molecules / NP |
CTX Molecules / NP |
Magnetic Relaxivity R2 (S−1.mM−1) |
|---|---|---|---|---|---|
| 7.5 | 111.9 ± 52.4 | + 19.6 ± 5.7 | 3.8 | 5 | 214.07 |
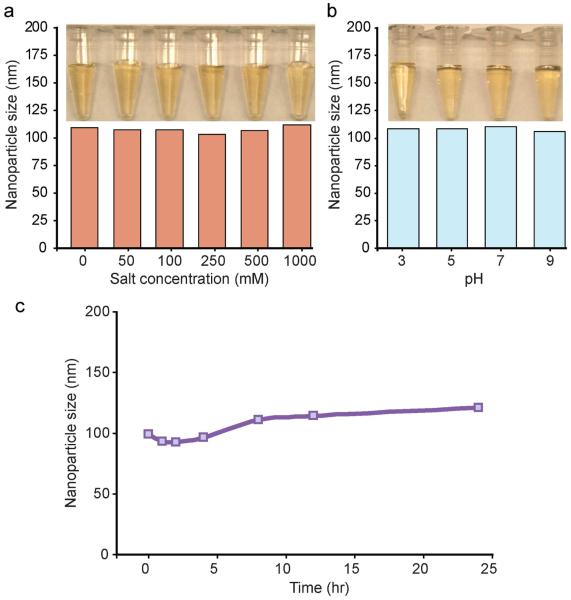
The stability of the nanovector in biological media, which is critical to their clinical success, was evaluated by monitoring their hydrodynamic size changes in solutions over a range of pH and salt concentrations, and in DMEM cell culture media containing 10% FBS at 37°C over a 24-hour period (Figure 3). Our results showed that no appreciable change in the hydrodynamic size of nanovectors was observed in response to increased NaCl concentrations over 0–1000 mM (Figure 3a) or to pH changes from 3 to 9 (Figure 3b). This can also be visualized by photographs shown in Figures 3a-b, which shows transparent nanovector suspensions with no sign of color change, agglomeration, nor precipitation. Similarly, the hydrodynamic sizes of nanovectors in the cell culture media remained approximately constant over the 24-hour period (Figure 3c). Combined, these colloidal stability tests confirm that the NP-siRNA construct is stable under biologically relevant conditions.
Figure 3.
Colloidal-stability of NP-siRNA in various media assessed at 37°C over a period of 24 hours. (a) Hydrodynamic size of NP-siRNA in solutions of various NaCl concentrations, and corresponding photo images of the tested samples. (b) Hydrodynamic size of NP-siRNA in solutions of varing pH, and corresponding photo images of the tested samples. (c) Hydrodynamic size of NP-siRNA in DMEM/10%FBS cell culture medium measured over time.
The colloidal stability in biological environments can be a challenging issue in clinical use of any nanoparticle-based constructs due to the large surface area to volume aspect ratio of nanoscale materials. Charged cationic nanoparticles, in particular, often display poor stability in cell culture conditions, as they tend to adsorb proteins from the biological environment through electrostatic interaction [31,32], causing fouling or precipitation. Previous studies have shown that iron oxide nanoparticles coated with PEI or other cationic polymers are greatly susceptible to aggregation in cell culture environments [32]. In the design of our nanovectors, we have integrated PEG to prevent non-specific absorption of proteins. Hydrophilic PEG coatings have been shown to resist protein absorption and provide steric hindrance preventing nanoparticles from aggregation [33]. Our nanovectors have PEG incorporated in both the initial shell coating and the outer layer exposed to solution environment. These combined measures contribute to the enhanced stability of the nanovector construct, even over the NaCl and pH ranges tested here.
3.3. Receptor mediated internalization of nanovectors by cancer cells
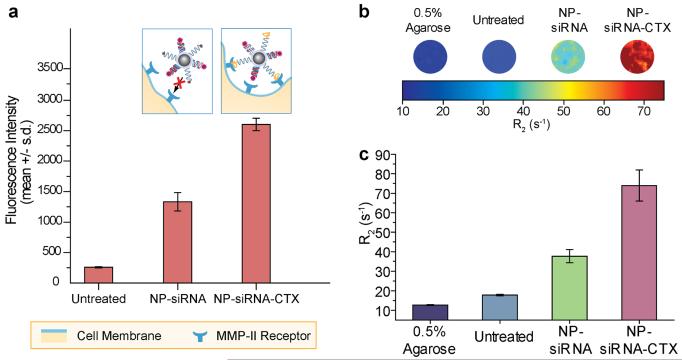
To evaluate the enhanced affinity of our CTX targeted nanovectors to tumor cells, C6 cells were treated with either NP-siRNA-CTX or NP-siRNA (control), and their internalization by the cells were assessed with flow cytometry and MRI. For these experiments, C6/GFP+ cells were treated with 20 μg of Fe/ml (2 μg of siRNA/ml) of NP-siRNA or NP-siRNA-CTX for two hrs prior to analysis. Figure 4a shows the results of the flow cytometry assay, which monitored Dy547 fluorochrome attached to the siRNA. As shown, NP-siRNA-CTX was internalized by cells 2-fold more than NP-siRNA. As a reference, the mean intensity of untreated cells is also plotted. The cartoon diagrams above the graph in Figure 4a comparatively illustrate how the surface chemistry of each nanovector regulates the internalization of the nanoparticles. NP-siRNA does not bind MMP-2 receptors expressed by the cancer cells (shown as blue claws), while NP-siRNA-CTX readily binds to glioma cells through the affinity of CTX to MMP-2.
Figure 4.
Receptor-mediated internalization of siRNA by target cells monitored by flow cytometry and MRI. (a) Flow cytometry analysis of siRNA internalization by C6/GFP+ cells 2 hrs post treatment with either NP-siRNA or NP-siRNA-CTX; also shown is the result for cells receiving no nanovector treatment (Untreated) as a reference. The cartoon diagram above the bar charts conceptually illustrates the interaction of the non-targeting NP-siRNA and targeting NP-siRNA-CTX with cells. (b) R2 relaxivity maps of C6/GFP+ cells treated with either NP-siRNA or NP-siRNA-CTX acquired 2 hrs post treatment; also shown are the images for untreated cells and agarose as references. (c) Corresponding R2 values for each condition shown in b.
This preferential cell binding for NP-siRNA-CTX was further demonstrated by MRI phantom imaging through the inherent magnetic properties exhibited by the iron oxide core of the nanovector. Figure 4b shows colorized R2 maps that highlight the degree of contrast enhancement produced due to the internalization of nanovectors by cells, and Figure 4c shows the corresponding R2 values measured from each sample. The cells treated with NP-siRNA-CTX produced the highest signal enhancement, which was 4.2 fold higher than those of untreated cells, and 2 fold higher than those of cells treated with NP-siRNA. Both the flow cytometry assay and MRI experiments indicated that CTX modification of nanovector promoted receptor-mediated internalization yielding improved delivery of siRNA to cancer cells. These findings suggest that while some of nanoparticles are able to enter cells through adsorptive mediated endocytosis, it is evident that a substantial enhancement in the cellular internalization of nanovectors and MRI contrast can be achieved through targeting with CTX. The results also suggested that the magnetic properties possessed by the nanovector would potentially allow for clinical treatment monitoring through MRI detection.
3.4. Nanovector escape from endosome compartments
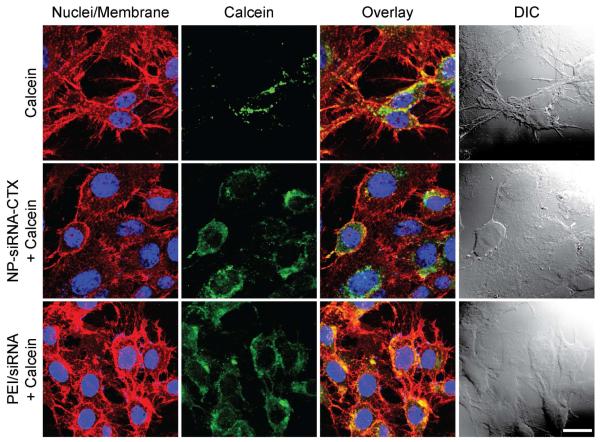
Upon internalization of siRNA carriers by cancer cells, the nanocarrier, along with its siRNA cargo, becomes localized in endosomal compartments and have no access to the cytoplasm to initiate the RNAi pathway. We incorporated PEI in our nanovector design to enable the nanovector to escape the endosome through the proton sponge effect [10,27]. Here, we utilized an endosomal integrity assay [34] to assess the ability of nanovectors to escape endosomal compartments. In this assay, a membrane impermeable fluorescent dye, Calcein (0.25 mM), was delivered alone or co-delivered along with NP-siRNA-CTX or PEI/siRNA complexes (commercial transfection reagent) to C6 cells, and the treated cells were maintained for 2 hrs under normal cell culture conditions. The intracellular localization of calcein was then visualized using fluorescence microscopy with DAPI (cell nuclei) and WGA-647 (cell membranes) counterstaining (Figure 5). As shown in cells treated with calcein alone, the membrane impermeable calcein did not spread throughout cells indicating the integrity of these endosomes. Conversely, in cells co-treated with calcein and NP-siRNA-CTX or PEI/siRNA nanovectors, calcein was distributed throughout the cells, which indicates the disruption of the endosomes and release of calcein into the cytoplasm of the cells. This visual observance verifies that NP-siRNA-CTX is able to promote endosomal escape similarly to PEI/siRNA.
Figure 5.
Confocal fluorescence images of cells for assessment of endosomal escape. C6 cells were treated with either calcein alone (negative control, first row) or co-treated with calcein + NP-siRNA-CTX (test sample, second row), or calcein + PEI/siRNA (positive control, third row). The cell nuclei and membranes are stained in blue and red, respectively, and calcein is shown in green. The DIC images are provided to colocalize the cells in fluorescence images. The scale bar represents 20 μm.
3.5. Receptor mediated gene knockdown and monitoring of toxicity
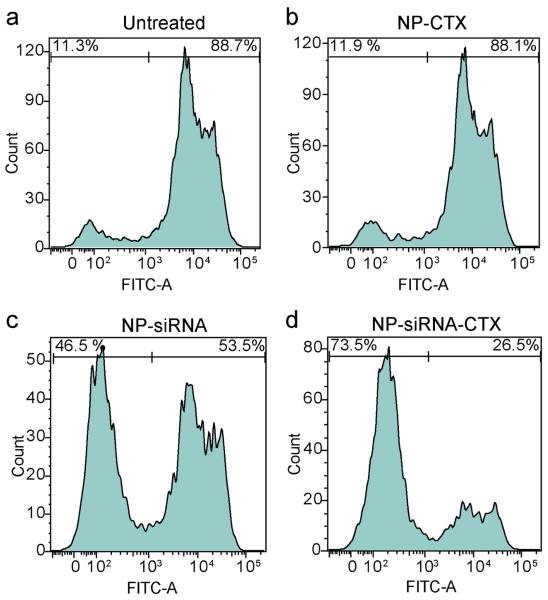
After confirming that the nanovector was able to promote endosomal escape, we evaluated the ability of the nanovector to knockdown the transgene expression of GFP in C6/GFP+ cells after the cells were treated with either NP-siRNA or NP-siRNA-CTX. The study also included a treatment of cells with NP-CTX as negative control nanoparticles (i.e., targeting nanoparticles that carry no siRNA) to confirm that NP and CTX do not have an effect on GFP expression. C6/GFP+ cells were treated with each nanovector at a concentration of 2 μg/ml of siRNA for two hrs, then analyzed using flow cytometry 48 hrs post treatment. Figure 6 shows the percentage of GFP expressing cells post treatment. The GFP expression profile of untreated C6/GFP+ cells, serving as a baseline reference, indicates that 88.7% of the cells were GFP positive (Figure 6a). Cells treated with NP-CTX (Figure 6b) showed no appreciable change in GFP expression compared to untreated cells (88.1% versus 88.7%), which confirms that NP and CTX did not affect GFP expression, as expected. Cells treated with NP-siRNA (Figure 6c) exhibited a 35% reduction in GFP expression (53.5% versus 88.7%). Significantly, cells treated with NP-siRNA-CTX treatment (Figure 6d) showed a 62% reduction in GFP expression compared to untreated cells (26.5% versus 88.7%). These results suggest that through the receptor-mediated cellular internalization of NP-siRNA-CTX, we can achieve enhanced knockdown of GFP expression in C6/GFP+ cells.
Figure 6.
Receptor-mediated GFP knockdown characterized at the protein leval by flow cytometry. (a) Untreated C6GFP+ cells. (b) Cells treated with NP-CTX (20 μg/ml of Fe; iron equivalence to siRNA containing samples). (c) Cells treated with NP-siRNA (2 μg/ml siRNA). (d) Cells treated with NP-siRNA-CTX (2 μg/ml siRNA).
It should be noted that the non-targeting but cationic vectors can be internalized by cells non-specifically through adsorptive mediated endocytosis [33], as we also observed here. However, prior studies have also shown this mechanism of cellular internalization is not as effective as targeted delivery and yet can only be used to treat a certain percentage of the cellular population [35]. Alternatively, as demonstrated here, targeted delivery can ensure that a greater percentage of the cell receives an effective dose of siRNA.
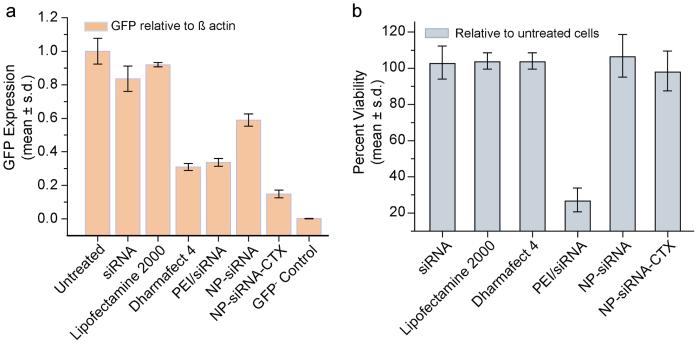
We further demonstrated the targeted siRNA delivery and enhanced gene knockdown by our NP-siRNA-CTX nanovector through quantitative RT-PCR analysis (Figure 7). In these experiments C6/GFP+ cells were treated with nanovectors as described above. For comparison, we also evaluated the efficacies of the following additional gene transfection reagents: naked siRNA, NP-siRNA, PEI/siRNA, Dharmafect 4 and Lipofectamine 2000. Here, PEI/siRNA is a commonly used cationic complex with high gene transfection efficiency [21,36], and Dharmafect 4 and Lipofectamine 2000 are two of the most widely used commercial gene transfection reagents. Cells were treated with one of these reagents and analyzed 48 hrs after treatment. As negative control, non-transfected C6 cells (GFP−) were also assayed to obtain a reference for cells with zero GFP expression.
Figure 7.
Receptor-mediated GFP knockdowns analyzed at the RNA level by qRT-PCR and cell viability analyized using Alamar blue. (a) GFP expression of C6/GFP+ analyzed as untreated, or 48 hrs post treatment with one of the following reagents: siRNA (2 μg/ml), Lipofectamine 2000, Dharmafect 4, PEI/siRNA, NP-siRNA, and NP-siRNA-CTX (all at 2 μg/ml of siRNA). As a reference, cells not expressing GFP (i.e. GFP−) were also analyzed (GFP– Control). Expression is normalized to B-actin levels. (b) Viability of C6/GFP+ glioma cells 48 hrs post treatment with one of above transfection reagents, determined by percent reduction of Alamar Blue and normalized to untreated cells.
Figure 7a shows the GFP mRNA transcripts of cells for each treatment relative to the housekeeping gene β-actin. NP-siRNA treatment reduced GFP mRNA transcripts to 0.58 relative to those in untreated cells, representing a 1.7 fold knockdown in gene expression. NP-siRNA-CTX treatment reduced GFP mRNA levels to 0.23 relative to those in untreated cells, a 4.3 fold knockdown in gene expression. These results are consistent with those obtained from the flow cytometry experiments. By comparing the transfection efficiencies of these two nanovectors, we observed a 2.5-fold enhancement in gene knockdown as a result of the targeted delivery by incorporation of CTX in the nanovector. Cells treated with naked siRNA or Lipofectamine 2000 did not show significant GFP expression knockdown, and PEI/siRNA and Dharmafect 4 treatments were able to knockdown GFP expression by ~3 folds. NP-siRNA-CTX nanovector was most efficient among all transfection reagents evaluated here, highlighting its advantage as a targeted delivery system.
Figure 7b shows the cell viability after cells were treated with each transfection reagent. It is seen that only PEI/siRNA treatment generated significant cytotoxicity. The high cytotoxicity exhibited by PEI/siRNA can be attributed to the presence of unmodified PEI whose toxicity is well documented in the literature [37]. The toxic effect of PEI in NP-siRNA-CTX (NP-siRNA as well) was modulated by inclusion of PEG in our nanovector design. This modification shields the cationic nature of PEI and thus reduces (or eliminates) toxicity associated with PEI, as is also shown in other studies [21,36].
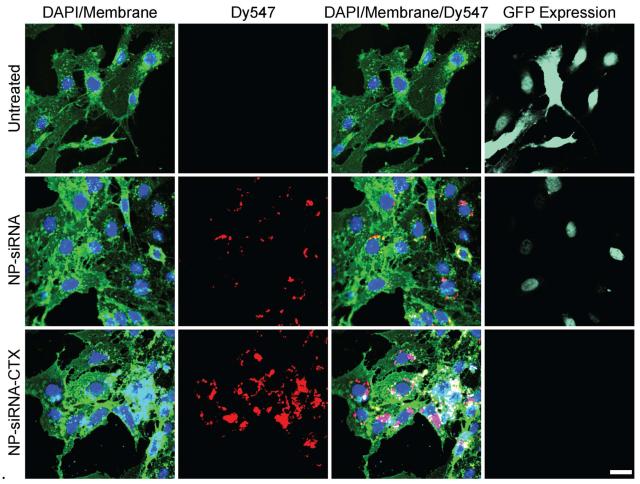
To visualize the targeted delivery of siRNA and enhanced knockdown of gene expression by NP-siRNA-P-CTX, cells treated with either NP-siRNA-CTX or NP-siRNA were examined by confocal fluorescence microscopy (Figure 8). In these fluorescence images, cell nuclei were stained with DAPI (blue) and membranes with WGA-647 (green). The images of untreated cells are provided as a reference. Treatments with NP-siRNA and NP-siRNA-CTX were administered as described above. Figure 8 shows that the relative amount of siRNA (red, Dy547, second column) internalized by cells treated with NP-siRNA-CTX is significantly higher than in cells treated with NP-siRNA. The overlay images (third column) reveal that the delivered siRNA molecules are predominantly localized in the perinuclear region of cells, where siRNA molecules are recognized by the RISC complex [6]. This observation confirms the proper trafficking of siRNA within cells. The GFP expression analysis (light green, fourth column) showed that the NP-siRNA treatment reduced the GFP expression of cells compared to the untreated cells, but GFP+ cells were still noticeably present. On the other hand, for cells treated with NP-siRNA-CTX, little to no GFP expression was observed. This fluorescence analysis confirmed the trends demonstrated by flow cytometry and qRT-PCR analyses.
Figure 8.
Confocal fluorescence images of C6GFP+cells illustrating targeted siRNA delivery and enhanced knockdown of gene expression. Cells were imaged 48 hrs post treatment with NP-siRNA (second row) or NP-siRNA-CTX (third row), with untreated cells (first row) as a reference. Cell nuclei are shown in blue, cell membranes in green, siRNA in red, and GFP expression levels in light green (fourth column). The scale bar corresponds to 20 μm.
4. Conclusions
Efficient and specific delivery of siRNA to target cancer cells continues to be the major hurdle to application of siRNA for cancer therapy. Nanovectors, when properly engineered, can be used to overcome these hurdles. Here, we have developed a targeted magnetic siRNA nanovector that demonstrates both increased siRNA internalization by target tumor cells and intracellular trafficking towards enhanced knockdown of targeted gene expression. Furthermore, we demonstrated that this same nanovector could enhance MRI contrast in vitro, potentially enabling monitoring of treatment in vivo through MRI. The elevated specificity and potency of this nanovector system make it a potential candidate of gene therapy for treatment of malignant tumors.
Acknowlegments
This work is supported by NIH grants (R01CA134213 and R01EB006043). O. Veiseh F. Kievit acknowledge support through NCI training grant (T32CA138312). C. Fang acknowledges the support through an NCI/NSF IGERT fellowship. We acknowledge lab assistance from Katie Huebner and Christine Epperson and the use of resources at the Department of Immunology’s cell analysis facility, Keck Microscopy Imaging Facility, and the Diagnostic Imaging Science Center (DISC) at the University of Washington.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–6. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- [2].Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003;21:1457–65. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- [3].Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- [4].Dykxhoorn DM, Chowdhury D, Lieberman J. RNA interference and cancer: endogenous pathways and therapeutic approaches. Adv Exp Med Biol. 2008;615:299–329. doi: 10.1007/978-1-4020-6554-5_14. [DOI] [PubMed] [Google Scholar]
- [5].Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–62. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- [6].Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61:721–31. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61:710–20. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- [9].Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nat Mater. 2010;9:272–8. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- [10].Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J Am Chem Soc. 2008;130:9006–12. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9:2402–6. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–3. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharm. 2009;6:651–8. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–84. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- [15].Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Akhtar S, Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: potential impact on siRNA-mediated gene silencing activity and specificity. Adv Drug Deliv Rev. 2007;59:164–82. doi: 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- [17].Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–9. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68:9788–98. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- [20].Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- [21].Kievit FM, Veiseh O, Bhattarai N, Fang C, Gunn JW, Lee D, et al. PEI-PEG-chitosan copolymer coated iron oxide nanoparticles for safe gene delivery: synthesis, complexation, and transfection. Adv Funct Mater. 2009;19:2244–51. doi: 10.1002/adfm.200801844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, et al. Specific targeting of brain tumors with an optical/magnetic resonance imaging nanoprobe across the blood-brain barrier. Cancer Res. 2009;69:6200–7. doi: 10.1158/0008-5472.CAN-09-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lyons SA, O’Neal J, Sontheimer H. Chlorotoxin, a scorpion-derived peptide, specifically binds to gliomas and tumors of neuroectodermal origin. Glia. 2002;39:162–73. doi: 10.1002/glia.10083. [DOI] [PubMed] [Google Scholar]
- [24].Veiseh M, Gabikian P, Bahrami SB, Veiseh O, Zhang M, Hackman RC, et al. Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007;67:6882–8. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- [25].Sontheimer H. An unexpected role for ion channels in brain tumor metastasis. Exp Biol Med (Maywood) 2008;233:779–91. doi: 10.3181/0711-MR-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu D, Wei Y, Yao P, Jiang L. Determination of the degree of acetylation of chitosan by UV spectrophotometry using dual standards. Carbohydr Res. 2006;341:782–5. doi: 10.1016/j.carres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- [27].Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60:149–60. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- [28].Veiseh O, Gunn JW, Kievit FM, Sun C, Fang C, Lee JS, et al. Inhibition of tumor-cell invasion with chlorotoxin-bound superparamagnetic nanoparticles. Small. 2009;5:256–64. doi: 10.1002/smll.200800646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sun C, Lee JSH, Zhang MQ. Magnetic nanoparticles in MR imaging and drug delivery. Adv Drug Deliv Rev. 2008;60:1252–65. doi: 10.1016/j.addr.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine. 2008;3:703–17. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fang C, Bhattarai N, Sun C, Zhang M. Functionalized nanoparticles with long-term stability in biological media. Small. 2009;5:1637–41. doi: 10.1002/smll.200801647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Petri-Fink A, Steitz B, Finka A, Salaklang J, Hofmann H. Effect of cell media on polymer coated superparamagnetic iron oxide nanoparticles (SPIONs): colloidal stability, cytotoxicity, and cellular uptake studies. Eur J Pharm Biopharm. 2008;68:129–37. doi: 10.1016/j.ejpb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- [33].Veiseh O, Gunn JW, Zhang M. Design and fabrication of magnetic nanoparticles for targeted drug delivery and imaging. Adv Drug Deliv Rev. 2010;62:284–304. doi: 10.1016/j.addr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Agrawal A, Min DH, Singh N, Zhu H, Birjiniuk A, von Maltzahn G, et al. Functional delivery of siRNA in mice using dendriworms. Acs Nano. 2009;3:2495–504. doi: 10.1021/nn900201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:15549–54. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Veiseh O, Kievit FM, Gunn JW, Ratner BD, Zhang M. A ligand-mediated nanovector for targeted gene delivery and transfection in cancer cells. Biomaterials. 2009;30:649–57. doi: 10.1016/j.biomaterials.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58:1523–31. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]