Abstract
This paper presents a novel statistical framework for human cortical folding pattern analysis that relies on a rich multivariate descriptor of folding patterns in a region of interest (ROI). The ROI-based approach avoids problems faced by spatial-normalization-based approaches stemming from the deficiency of homologous features between typical human cerebral cortices. Unlike typical ROI-based methods that summarize folding by a single number, the proposed descriptor unifies multiple characteristics of surface geometry in a high-dimensional space (hundreds/thousands of dimensions). In this way, the proposed framework couples the reliability of ROI-based analysis with the richness of the novel cortical folding pattern descriptor. This paper presents new mathematical insights into the relationship of cortical complexity with intra-cranial volume (ICV). It shows that conventional complexity descriptors implicitly handle ICV Differences in Different ways, thereby lending Different meanings to “complexity”. The paper proposes a new application of a non-parametric permutation-based approach for rigorous statistical hypothesis testing with multivariate cortical descriptors. The paper presents two cross-sectional studies applying the proposed framework to study folding Differences between genders and in neonates with complex congenital heart disease. Both studies lead to novel interesting results.
Keywords: cerebral cortical folding, statistical shape analysis, gender differences, neonatal development
1. Introduction
Cerebral cortical folding [Armstrong et al. (1995); Ono et al. (1990); Van-Essen (1997)] forms an underpinning for the cognitive skills and behavioral traits in humans. It is one of the major maturational processes of the human brain that occurs rapidly throughout fetal and early postnatal life and is a major factor contributing to human intelligence. For the last few decades, magnetic resonance (MR) imaging has enabled in vivo studies of human cortical folding patterns.
One class of approaches to folding analysis rely on spatial normalization [Nordahl et al. (2007); Yeo et al. (2008); Yu et al. (2007)] and subsequently perform statistical hypothesis testing at every voxel or surface element in the normalized space. However, the difficulty in finding a large number of homologous features [Lyttelton et al. (2007); Mangin et al. (2004); Van-Essen and Dierker (2007)] may directly affect the normalization and, thereby, the reliability of findings in the clinical study. Furthermore, the phenomenon of cortical folding has an inherent large-scale or non-local character that may be difficult to capture with point-based descriptors.
A second class of approaches propose region-based folding descriptors [Batchelor et al. (2002); Pienaar et al. (2008); Rodriguez-Carranza et al. (2008); Van-Essen and Drury (1997)], which avoid the challenges associated with normalization by reducing spatial sensitivity from a voxel to a region of interest (ROI) that can be reliably defined in each individual based on observed homologous features. Examples of such ROIs can be brain lobes, regions around major sulci/gyri, etc. Some descriptors quantify surface complexity alone: fractal dimension (FD) [Griffin (1994); Majumdar and Prasad (1988); Thompson et al. (1996)] captures the rate of increase in surface area over multiscale representations of the surface; gyrification index (GI) [Zilles et al. (1988)] is the ratio of the length of a planar/2D curve to the length of its convex hull/envelope; convexity ratio (CR) [Batchelor et al. (2002)] is the ratio of the area of the surface to the area of the convex hull of the surface; isoperimetric ratio (IPR) [Batchelor et al. (2002); Im et al. (2008); Toro et al. (2008)] is the ratio of the surface area to the two-third power of the volume enclosed by the surface; average curvedness (AC) [Awate et al. (2008)] measures the deviation of the surface from a plane; another measure is the 2D centroid of the 1D histogram of curvature (HC) [Pienaar et al. (2008)]. Some folding descriptors capture partial folding characteristics by integrating specific measures for all surface patches: intrinsic curvature index (ICI) [Van-Essen and Drury (1997)] integrates degrees of hemisphericity; mean curvature norm (MCN) [Batchelor et al. (2002)] integrates degrees of hemisphericity and cylindricity; Gaussian curvature norm (GCN) [Batchelor et al. (2002)] integrates degrees of hemisphericity and saddle-likeness; average shape index (AS) [Awate et al. (2008)] integrates shape indices. Thus, typical ROI-based approaches produce scalar or low-dimensional summary statistics for the entire ROI, risking information loss.
This paper presents a new ROI-based statistical framework for folding analysis relying on a rich multivariate non-local descriptor that captures multiple aspects of surface geometry [Awate et al. (2009a,b)]. The proposed descriptor is a joint probability density function (PDF) of two complementary/independent variables, one capturing surface curvedness and the other capturing surface shape index. The resulting surface folding descriptor lies in a space having hundreds or thousands of dimensions. The paper proposes a new application of a nonparametric permutation-based approach for statistical hypothesis testing with multivariate cortical descriptors. In these ways, the proposed framework couples the reliability of ROI-based analysis with the richness of the proposed descriptor. This paper validates the proposed framework, on simulated as well as clinical data, and presents applications on two neuroimaging studies, in adults and neonates.
While several folding studies concern neurodevelopmental disorders, studies on gender Differences, in the normal population, have received very little attention. Moreover, while one study [Luders et al. (2004)] using the FD measure reported higher complexity in adult females, two recent studies [Im et al. (2008); Toro et al. (2008)] using the IPR measure report higher complexity in larger adult brains (i.e. males). The study in this paper elucidates these seemingly-conflicting findings. This paper provides new theoretical insights into relationships between folding measures with intra-cranial volume (ICV), pinning them down to the fundamental issues of scale and replication. It shows that standard folding measures in the literature imbibe Different meanings of “complexity”. This paper proposes two systematic methods for handling ICV changes in folding studies using the proposed descriptor and shows that while the findings using one method are consistent with [Luders et al. (2004)], those using the other method are consistent with [Im et al. (2008); Toro et al. (2008)].
The second study in this paper deals with cortical folding in the operculum in neonates with complex congenital heart disease (CHD). The operculum includes language areas and the sensory motor cortex for the mouth, tongue, and throat. There is growing evidence of immature features or delayed maturation of the brains of full-term infants with complex CHD [Licht et al. (2009); Miller et al. (2007)]. This immaturity probably gives rise to unexpected vulnerability to a white-matter injury termed periventricular leukomalacia, previously seen only in premature infants. Abnormally low fetal blood oxygenation and blood flow in the brain are likely causes of this maturational delay. While direct evidence is lacking, there are Differences in the circulatory patterns in fetuses with Different forms of complex CHD. This paper quantitatively evaluates cortical folding in the operculum in two key subtypes of CHD, namely hypoplastic left heart syndrome (HLHS) and transposition of the great arteries (TGA). This paper reports Differences in not only the complexity of folding patterns, but other geometrical aspects as well.
2. Methods and Materials
2.1. A Novel Multivariate High-Dimensional Folding Descriptor
This section describes a novel high-dimensional multivariate surface descriptor that captures multiple aspects of surface geometry.
For surface , at every point , the principal curvatures κmin(m) and κmax(m) describe the local geometry [DoCarmo (1976)] (upto second order and upto a translation and rotation). The space < κmin, κmax > can be reparameterized, by a polar transformation, into the orthogonal bases of curvedness C and shape index S that meaningfully separate notions of bending and “shape” [Koenderink and van Doorn (1992); Koenderink (1990)].
Novel cortical folding descriptor
We propose the joint PDF as the multivariate high-dimensional descriptor of cerebral cortical folding patterns for surface . Figure 1 provides a visual overview of these concepts.
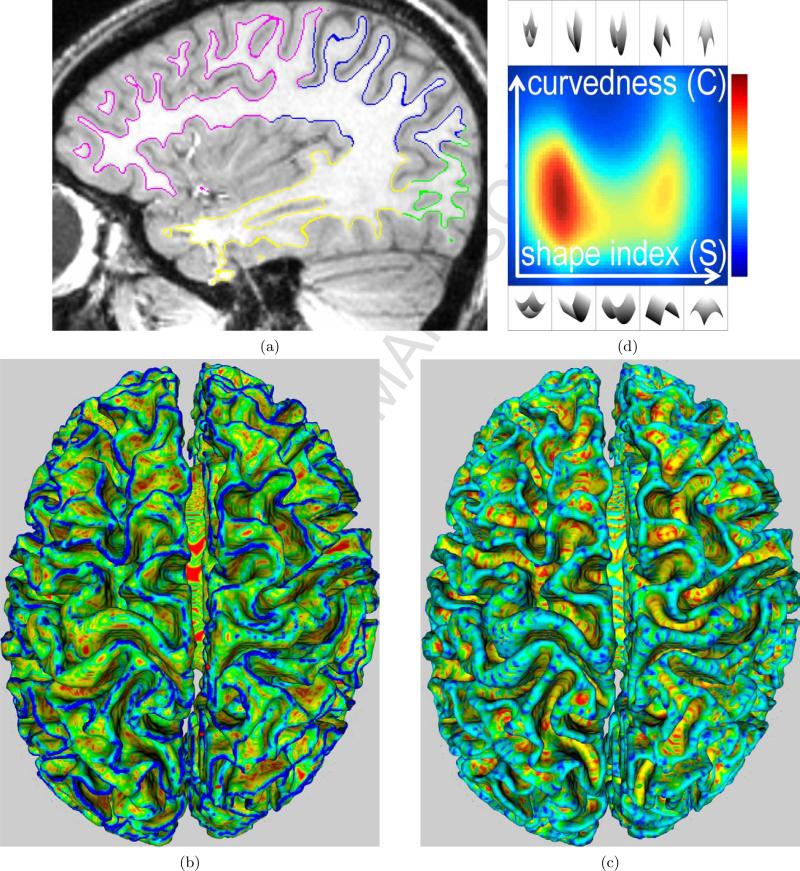
Figure 1.
(a) A sagittal slice of a head MR image overlapped with the cortical surface . (b) Curvedness values C(m) painted on (red→blue ≡ zero→high). In this figure, red areas are almost flat and blue areas are highly curved. (c) Shape-index values S(m) ∈ [−1, 1] painted on (red→blue ≡ −1 → 1). In this figure, red/yellow areas are concave, blue/cyan areas are convex, and green areas are saddles. (d) Proposed descriptor (blue→red ≡ 0 → 1 probability; colormap shown on right). For all plots of in this paper: horizontal axis ≡ S, vertical axis ≡ C. The illustrations of local surface patches, at the top and bottom of this figure, depict patches for the entire range of possible S values; corresponding patches at the top and bottom have the same S value; all patches at the top have the same C value that is higher than the C value for all the patches at the bottom; the left half of the domain comprises concave patches, while the right half comprises convex patches.
In a way, subsumes scalar descriptors such as ICI, MCN, GCN, AC, AS, and HC, because these descriptors can be computed using a subset of the values/information in the proposed descriptor . The next paragraph provides a brief sketch of the underlying theoretical modeling framework. The paragraphs following that provide details on the practical aspects in estimating from data.
Modeling cortical surfaces
We employ the following generative model for the proposed studies of human cerebral cortical surfaces. We assume cortical surfaces, in ROIs, to be smooth 2-D Riemannian manifolds [Davatzikos and Prince (1995); Joshi et al. (1995)] that possesses a smooth 2-parameter planar parameterization . The smooth parameterization also implies a smooth spatial variation of curvedness and shape-index values on the surface. Let us consider C̃ : Ω → [0, ∞) and S̃ : → [−1, 1] as continuous random fields describing the curvedness and shape index, respectively, at each point on the surface. Let us also consider the joint PDF that captures the dependencies between C̃(ω) and S̃(ω), ∀ω ∈ Ω, for a specific class of surfaces. Thus, human cortical surfaces, in a study group, can be considered as independent instances of a single field of random vectors (C̃, S̃). Consider a finite collection of points {f(ω1), . . . , f(ωT)} = {m1, . . . , mT } uniformly distributed over the surface . Then, an MR image yields a discretization (in Ω) {(C̃(ω1), S̃(ω1)), . . . , (C̃(ωT), S̃(ωT))} of an instantiation of the field of random vectors (C̃, S̃). For making analysis tractable, we assume that each observation (C̃(ω), S̃(ω)), ∀ω ∈ Ω, is randomly drawn from a single PDF , i.e. the random field is stationary [Papoulis and Pillai (2001)]. The complexity and variability in cortical folding in the human population suggests that dependencies between random vectors (C̃(ω1), S̃(ω1)), (C̃(ω2), S̃(ω2)) decrease at a fast rate with increasing geodesic distance between surface locations f(ω1) and f(ω2) or, equivalently, ω1 and ω2, i.e. the random field is mixing [Papoulis and Pillai (2001)].
Estimating the proposed cortical descriptor
For a given surface , we propose to estimate the folding pattern descriptor from the sample {(C(mt), S(MTi)) : t = 1, . . . , T}. A consistent nonparametric estimate for the folding descriptor is the Parzen-window kernel density estimate [Parzen (1962),Lu and Chen (2004)]: , where G((μ1, μ2), ∑) is a 2D Gaussian kernel with mean (μ1, μ2) and covariance ∑. Consistency requires an optimal choice of ∑t, dependent on T , and we employ a penalized maximum-likelihood scheme [Chow et al. (1983)] to estimate ∑t; the literature provides many schemes [Wand and Jones (1995)].
The aforementioned estimation scheme for relies on the sample {m1, . . . , mT } being uniformly distributed over the surface . We obtain a reasonably uniform distribution of points by obtaining an implicit surface parameterization as a level set of a distance transform on a Cartesian grid [Osher and Paragios (2003); Sethian (1999)]; more sophisticated approaches exist in the literature e.g. [Peyre and Cohen (2006)]. We employ the time-e cient sparse-field formulation [Whitaker (1998)] for the level-set fitting procedure. This paper represents the level set in a grid of isotropic voxels of size ν3 mm3 with ν = 0.4 mm. To reduce effects of noise, level-set fitting incorporates smoothing. The level-set fitting, performed at sub-pixel accuracy, is driven by two forces resulting from: (i) the data-consistency term that enforces the level set to align with the (discrete) boundary between white matter (WM) and gray matter (GM) given by the tissue probabilities, at voxels, after segmentation [Awate et al. (2006)] and (ii) the mean-curvature term that enforces smoothness of the level set and counteracts some of the “noise”/errors in the segmentation as well as the limited resolution (discrete nature) of MRI data. Figures 1(b) and (c) show the fitted level-set surface where every point is colored by the values of C and S, respectively.
Empirically, we find that values of C virtually never exceed cmax = (15ν)−1 mm−1 equivalent to a minimum radius of curvature of 15ν = 6 mm. Figures 1(b) and (c) indicate that this degree of smoothing continues to capture essential folding pattern information in typical cortical surfaces. The lower limit of 6 mm is more conservative than the limit of 3 mm in [Pienaar et al. (2008)]. Imaging limitations, on voxel sizes and signal-to-noise ratios, risk fidelity in capturing sharper surface features.
Discretizing on an I × J grid leads to an IJ-dimensional descriptor. In this paper, we choose I = J = 64, which yields a 4096-dimensional folding descriptor for each cortical surface. The trade-offs involved in choosing the grid size, i.e. I and J, are very similar to those involved in standard voxel-based morphometry in the neuroimaging literature regarding smoothing of the data and the scale(s) at which statistically-significant effects are searched for in the data [Jones et al. (2005); Worsley et al. (1996)]; this analysis relies on the matched-filter theorem in linear filtering in signal processing. For instance, coarser grids result in a higher degree of smoothing. In addition to such trade-offs, the trade-offs in our case are also related to the amount of available data, i.e. the sample size T underlying the density estimation. For example, a larger sample size (resulting from a larger anatomical region of interest in the study) can allow a higher-resolution probing of the data to reliably search for subtle effects in . On the other hand, larger IJ increases the computation time. Lastly, the choice of the grid size is also determined by the relationship between the clinical hypotheses of interest and the associated size of the region(s) in the < C, S > domain.
Figure 1(d) shows a typical , which is multimodal and far from standard parametric PDFs, thus justifying nonparametric PDF estimation for reliability of the clinical study. In practice, typical ROIs yield sample sizes T in the range of thousands or tens of thousands, producing (i) very narrow kernel bandwidth estimates (∑t has small eigen values) such that is desirably close to zero and (ii) robust estimations. Moreover, the PDF mass very close to the c = 0 axis (shape index is undefined for a plane) is also negligible: for sufficiently small cmin and δ. This paper sets cmin = 0.001.
2.2. A Novel Testing Scheme for Multivariate Density Analysis
This section proposes a new application of a known nonparametric permutation-based approach, i.e. the statistical nonparametric mapping (SnPM) algorithm [Nichols and Holmes (2002)], for statistical hypothesis testing with N multivariate cortical descriptors in a clinical study, i.e. ,
Typical hypothesis tests, e.g. cross-sectional, longitudinal, regression, etc., are subsumed in the framework of general linear models (GLM). The GLM framework has been applied extensively for voxel-based neuroimaging studies of brain function and structure, via the statistical parametric mapping algorithm (SPM) [Ashburner and Friston (2000); Friston et al. (1995)]. SPM entails running parametric GLM tests at each voxel in the image followed by corrections for multiple comparisons via, for instance, Gaussian field theory. However, such parametric approaches make strong assumptions on the data concerning the parametric distributions of the values at each point in the domain and the dependencies within neighborhoods [Nichols and Holmes (2002)]. Such strategies can be prone to spurious results when the underlying assumptions become invalid.
Permutation tests [Fisher (1935)], on the other hand, are nonparametric and rely on the less inclusive assumption of exchangeability: under the permutation-test null hypothesis, i.e. both groups of surfaces being generated by one distribution, the independent and identically-distributed observations are exchangeable. Compared to parametric tests, permutation tests are more stringent (stronger control over Type-1 error) and more robust to random noise in the imaging measurements and random inaccuracies in the post-processing of image data. In this way, permutation tests can lead to more reliable inferences from clinical studies. A rigorous hypothesis-testing scheme based on nonparametric permutation testing for voxel-based studies is the SnPM algorithm [Nichols and Holmes (2002)].
The proposed strategy for testing di ers from conventional strategies in two ways: (i) unlike typical usage of the SnPM algorithm involving functions on the image domain [Nichols and Holmes (2002)] or surface domain [Styner and Gerig (2003)], both of which necessitate spatial normalization, we propose to apply the SnPM algorithm to discretized cortical descriptors ; (ii) unlike conventional multivariate hypothesis testing (e.g. using Hotelling T2 statistic), the SnPM algorithm provides the locations (pixels and clusters), in the domain < C, S >, for significant differences/effects.
The proposed multivariate hypothesis-testing algorithm is as follows:
Empirically select thresholds cmin > 0 and cmax > cmin for curvedness values and a very small ∈ such that, ∀n = 1, . . . , N: .
For the domain [cmin, cmax] × [−1, 1], construct a regular rectangular tessellation of the desired resolution. Denote the resulting I × J rectangular bins by {bij : i = 1, . . . , I; j = 1, . . . , J}.
For all surfaces n = 1, . . . , N and all bins {bij}, compute the probabilities .
Use the N 2D images of probability values, P1, . . . , PN, as input for permutation testing via the SnPM algorithm [Nichols and Holmes (2002)]. The SnPM algorithm indicates (i) a set of locations {(i, j)} and (ii) a set of clusters exhibiting statistical significance for the underlying GLM experiment. Figure 3 shows a validation study that is explained in detail later in Section 3.2.
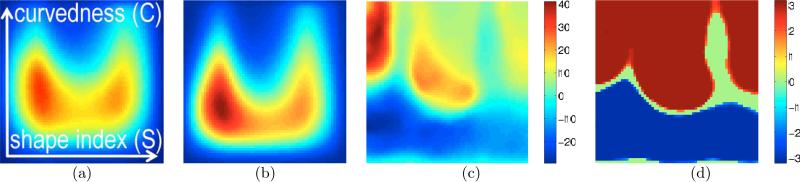
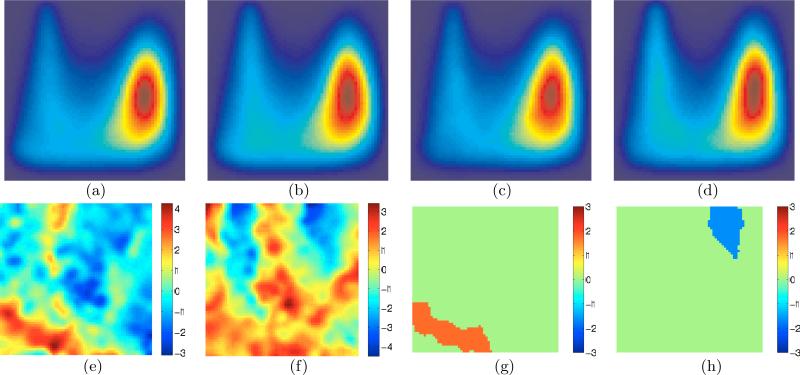
Figure 3.
Validation : a simulated cross-sectional study between the group of cortical surfaces (interface between GM and WM) of 20 BrainWeb [Aubert-Broche et al. (2006)] images and a second group created by moderately smoothing the 20 BrainWeb surfaces. For all plots in this paper, horizontal axis ≡ S, vertical axis ≡ C, coordinates for the bottom left corner : (C, S) = (cmin, −1); bottom right corner : (C, S) = (cmin, 1); top left corner : (C, S) = (cmax, −1). (a)-(b) Mean of the multivariate surface descriptors for the n = 1, . . . , 20 original and smoothed surfaces, respectively, as proposed in Section 2.1; red≡high and blue≡low values. (c) The t-statistic map for the original and smoothed surfaces; t > 0 ⇒ Poriginal > Psmoothed. Expectedly, the map indicates that the original surfaces have larger mass (red/yellow) in high-curvedness regions as compared to the smoothed surfaces. (d) The significant locations (p < 0.05) produced via permutation testing [Fisher (1935); Nichols and Holmes (2002)]. For all plots in this paper, p values (corrected; permutation testing) for significant locations/clusters are visualized by coloring them by the associated z score [Papoulis and Pillai (2001)], e.g. z(p = 0.05) = 1.65, z(p = 0.005) = 2.58.
2.3. Complexity and Volume Relationships : New Insights
This section presents new theoretical insights into (i) relationships between folding and ICV and (ii) different meanings of “complexity” underlying descriptors.
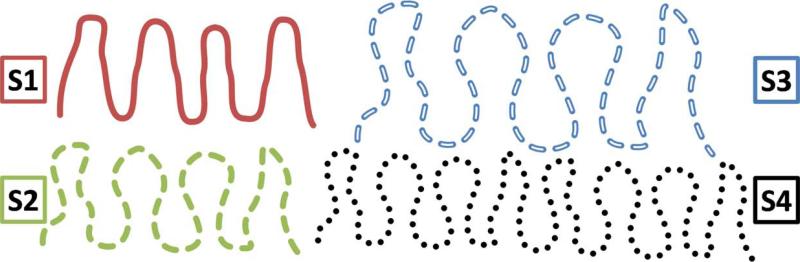
In Figure 2, S1 and S2 occupy equal volumes (i.e. the volume of their convex hulls) but S2 has a larger number of finer-scale features than S1. Desirably so, typical complexity measures, e.g. aforementioned measures in Section 1, inform that S2 is more complex than S1.
Figure 2.
What does “complexity” mean when volumes differ: issues of scale and replication. S1 and S2 occupy the same volume (i.e. volume of their convex hulls); S2 is more complex than S1. S3 and S4 occupy larger volumes than S2. S3 enlarges/scales the folds in S2. S4 replicates the folds in S2. How do we compare the complexities of (i) S3 and S2 and (ii) S4 and S2 ?
Now consider surfaces occupying different volumes. ICV increase can be associated with two kinds of effects on cortical surfaces: (i) folds are scaled up/enlarged, e.g. comparing S2 and S3, or (ii) folds are replicated, e.g. comparing S2 and S4. This section shows that the meaning of “complexity” imbibed in folding descriptors reflects how the descriptors handle scaling and replication.
One class of measures is invariant to the aforementioned processes of scaling and replication; this includes GI and CR, which are both normalized by the convex-hull surface area. Thus, GI and CR inform that S2, S3, and S4 have equal complexity.
A second class of measures is designed to be invariant to scale, but not replication; this includes IPR, ICI, MCN, GCN, and AC [Awate et al. (2008); Batchelor et al. (2002); Im et al. (2008); Toro et al. (2008)], which are normalized by surface-patch area or ICV2/3. Thus, these measures inform that S3 and S2 have equal complexity, but S4 is more complex than S2.
A third class of measures is invariant to replication, but not scale; this includes FD [Luders et al. (2004)], HC [Pienaar et al. (2008)], and the proposed in Section 2.1. Unlike the first two classes, these measures are not normalized via area or ICV2/3. Thus, they inform that S4 and S2 are equally complex, but S3 is less complex than S2.
We now propose a new scale-invariant descriptor. Isotropically enlarging or rescaling a volume by a factor β3, where β > 1, reduces curvedness values of every point on the surface, lying within that volume, by a factor of β. Indeed, unit-area patches in enlarged surfaces appear more planar, as dictated by Taylor's theorem. Thus, a scale-invariant version of is , where is the ratio of the mean group ICV to the ICV for cortical surface . Similar to the second class of measures described in this section, informs that S3 and S2 have equal complexity, but S4 is more complex than S2.
Subsequent sections denote and .
2.4. Clinical Cohorts and Imaging
This paper applies the proposed framework for two clinical studies using the following cohorts:
Healthy adults: The clinical cohort comprised T1-weighted MR images (1 mm3 isotropic voxels) of 30 females (mean age 34.8 years, standard deviation 9.6 years) and 27 males (mean age 36 years, standard deviation 11 years), obtained after enforcing quality-assurance checks on every image in the designed brain database described in [Mortamet et al. (2005)].
Neonates with complex congenital heart disease: The clinical cohort [Licht et al. (2009)] comprised 42 neonates with complex CHD (29 with HLHS, 13 with TGA) between 1 − 2 weeks of age, before undergoing corrective heart surgery. Neonates with independent risk factors (e.g. shock, end-organ injury, intra-uterine growth retardation) for abnormal brain development were excluded. MR images were acquired on a 3T scanner with voxel sizes around 0.88×0.88×1.5 mm3 using T1-weighted, T2-weighted, and FLAIR schemes. The HLHS and TGA groups were well matched by age and brain volume. The left and right operculums in every image were parcellated semi-automatically with expert supervision.
3. Validation of the Proposed Framework
3.1. Cortical Folding Analysis Pipeline
The studies in this paper employed the following processing sequence: (i) brain extraction [Smith (2002); Yushkevich et al. (2006)], ROI parcellation (atlas registration [Avants and Gee (2004)] on healthy adults), denoising, inhomogeneity correction [Vovk et al. (2006)], and (for neonatal images only) contrast enhancement via adaptive histogram equalization; (ii) automatic intensity-based probabilistic tissue segmentation [Awate and Gee (2007); Awate et al. (2006); Song et al. (2007)]; (iii) resample the segmentation to an isotropic voxel size of 0.4 × 0.4 × 0.4 mm3; (iv) define to be the cortical GM-WM interface corresponding to a cortical-WM membership of 0.5; the GM-WM interface is estimated much more reliably than interface between GM and cerebrospinal fluid, especially in neonatal/pediatric populations; (v) represent cortical surface in the ROI, to subvoxel accuracy, as a level set of a distance transform [Osher and Paragios (2003)] (vi) compute curvedness C(m) and shape-index S(m) values at every voxel m on the zero crossing of the level set , (vii) estimate the proposed multivariate folding pattern descriptor in the ROI as described in Section 2.1 and discretize it on an I × J = 64 × 64 Cartesian grid, (viii) perform multivariate statistical hypothesis testing as described in Section 2.2 using the SnPM algorithm. The implementation and visualization in this paper relied on the Insight Toolkit [ITK (2010)], Matlab, ITK-SNAP [Yushkevich et al. (2006)], and the Visualization Toolkit [VTK (2010)].
3.2. Validation using Simulated MRI (BrainWeb) of Healthy Adults
We validated the proposed framework for folding pattern analysis using 20 simulated images from the BrainWeb [Aubert-Broche et al. (2006)] repository having ground-truth segmentations. We simulated a cross-sectional study between (i) the group of GM-WM surfaces in the BrainWeb images and (ii) another group of surfaces obtained after slightly smoothing the surfaces in the first group (mean curvature flow, time step 0.24, iterations 4). Figure 3(a) and (b) show the means of the surface descriptors in the two groups. As expected, the smoothed surfaces have the mean PDF, shown in Figure 3(b), shifted slightly downwards, i.e. towards regions of low curvature, relative to Figure 3(a).
The t-statistic map (Figure 3(c)) clearly reveals the difference between the two groups. The blue (or red) significant locations (Figure 3(d)), at level of significance α = 0.05, correspond to those locations whose t statistics were less than (or greater than) the lowest (or greatest) 100α-th percentile of the permutation distribution of the smallest (or largest) t statistic over the < C, S > domain. Similarly, significant clusters (in all figures in this paper) are shown in blue (or red) when the sizes of the clusters formed by thresholding the negative (or positive) t statistics are larger than the 100α-th percentile of the permutation distribution of the maximum cluster size obtained after thresholding the negative (or positive) t statistics.
Figures 3(c) and (d) indicate that (i) the first (BrainWeb) group has a larger mass in the top of the plot, while the second (smoothed surfaces) group has more mass on the bottom part of the plot; and (ii) there is a high degree of (left-right) symmetry in the plots with respect to the vertical centerline. Indeed, the low degree of smoothing of the cortical surfaces is expected to have a greater effect in reducing the complexity/curvedness of the surfaces by eliminating fine-scale variations as compared to causing significant changes in the distribution of concave/convex patches. In this way, that the proposed framework indicates that the differences between the groups are mainly in cortical complexity.
3.3. Validation using Clinical MRI of Healthy Adults
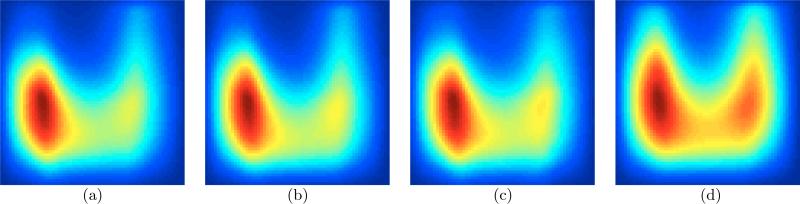
This validation experiment used 50 MR images of healthy adults and measured the average fraction of cortical surface area comprising concave surface patches that belong predominantly to sulci. The left halves, i.e. S < 0, of the plots concern concave patches (mainly associated with sulci), while the right halves, i.e. S > 0, concerns convex patches (mainly associated with gyri). The mass in the left half (c ≥ 0, s < 0) of the average PDF descriptor is a robust estimate of the fraction of the surface area of the cortical surface buried in sulci. We estimated this fraction to be 0.58 for the frontal, parietal, and temporal lobes in adults, which comes very close to the published value of 0.61 for GM-WM interfaces of entire brains [Van-Essen and Drury (1997); Zilles et al. (1988)]. For the occipital lobes, however, our estimate of this fraction was lower, i.e. 0.54. Figure 4 shows this phenomenon clearly, i.e. the PDF in Figure 4(d) has more mass (red) on the right (convex gyral regions) relative to the PDFs in Figure 4(a)-(c). These experiments demonstrate that the proposed descriptor is sensitive to changes in cortical shape and can provide clinically-relevant estimates consistent with the literature on cortical anatomy.
Figure 4.
Validation : measuring the fraction of surface area of the cortical GM-WM surface comprising concave surface patches (predominantly sulci) in 50 healthy adults. (a)-(d) Proposed surface descriptors for the frontal, parietal, temporal, and occipital lobes, respectively. The asymmetry between the left and right halves, i.e. concave and convex patches, is consistent with published clinical studies [Van-Essen and Drury (1997); Zilles et al. (1988)]. Note the reduced left-right asymmetry in (d) relative to (a)-(c).
4. Results and Discussion
4.1. Gender Differences in Cortical Complexity and Shape in Adults
This section describes the results of applying the proposed framework to study gender differences in adults. A discussion on these results is continued in Section 5.
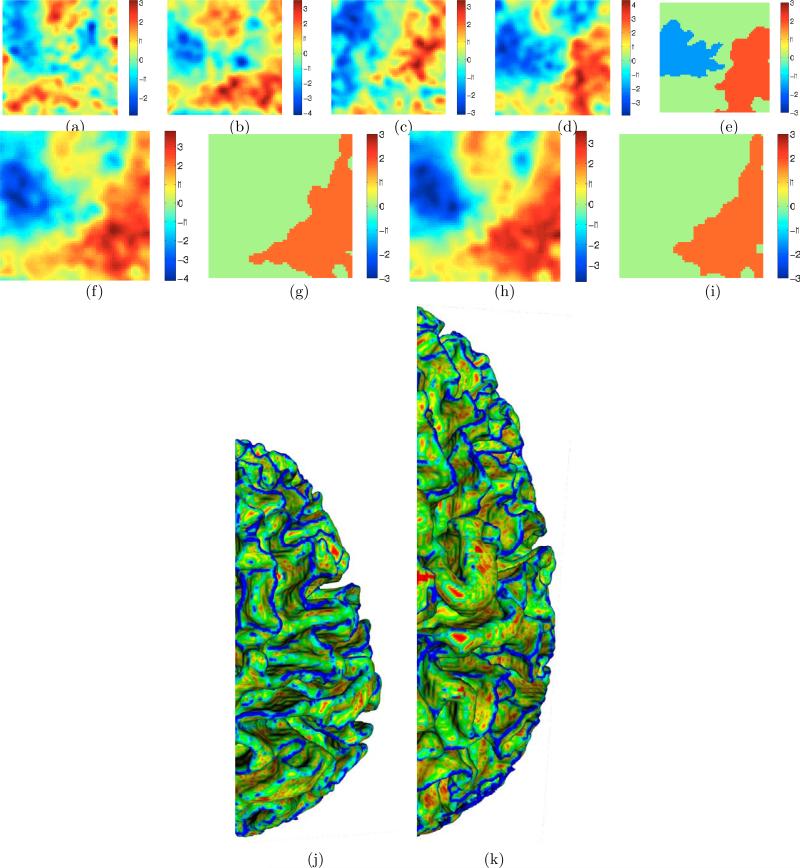
The proposed replication-invariant folding descriptor Preplication, described in Section 2.1, produces t maps (Figure 5(a)-(d)) indicating larger mass (red) for males in low-curvedness regions (bottom half) and larger mass (red) for males in convex regions (right half). The SnPM algorithm indicates significant clusters for the occipital lobes (Figure 5(e)), but not for other lobes. Nevertheless, when the ROI size is increased to a hemisphere, these e ects get significantly strengthened; evident in t maps (Figure 5(f),(h)) and significant clusters (Figure 5(g),(i)). These results show that female cortical surfaces are more “complex” based on the meaning of “complexity” ingrained in Preplication (consistent with [Luders et al. (2004)]) and significantly less convex than those of males. Figures 5(j)-(k) visualize the (accentuated) complexity differences by displaying colored cortical surfaces on a chosen larger-than-average male brain and a chosen smaller-than-average female brain in the cohort. Referring back to Figure 2, these results rule out analogizing female and male cortical surfaces using S2 and S4, respectively, because that would make female cortical surfaces less “complex” than what the results indicate.
Figure 5.
(a)-(d) Student's t statistics for a cross-sectional study with Preplication (positive t ≡ larger value in males) for frontal, parietal, temporal, and occipital lobes, respectively, in the left hemisphere. Similar patterns exist for lobes in right hemisphere. (e) Significant clusters (via permutation testing) for left occipital lobe. (f ),(h) t statistics and (g),(i) significant clusters (via permutation testing) for Preplication for left hemisphere (4 lobes) and whole brain (8 lobes), respectively. A similar pattern exists for the right hemisphere. Note the larger mass (red) for males in the low-curvedness regions (bottom half) and larger mass (red) for males in the convex regions (right half). (j)-(k) Selected smaller-than-average female and larger-than-average male brains, respectively, painted by C values (red→blue ≡ zero→high) to enable/accentuate visualization of the differences. Thus, based on the meaning of complexity ingrained in Preplication, the female cortex appears more “complex”, i.e. more blue/cyan. Compare this with Figure 6(e-f).
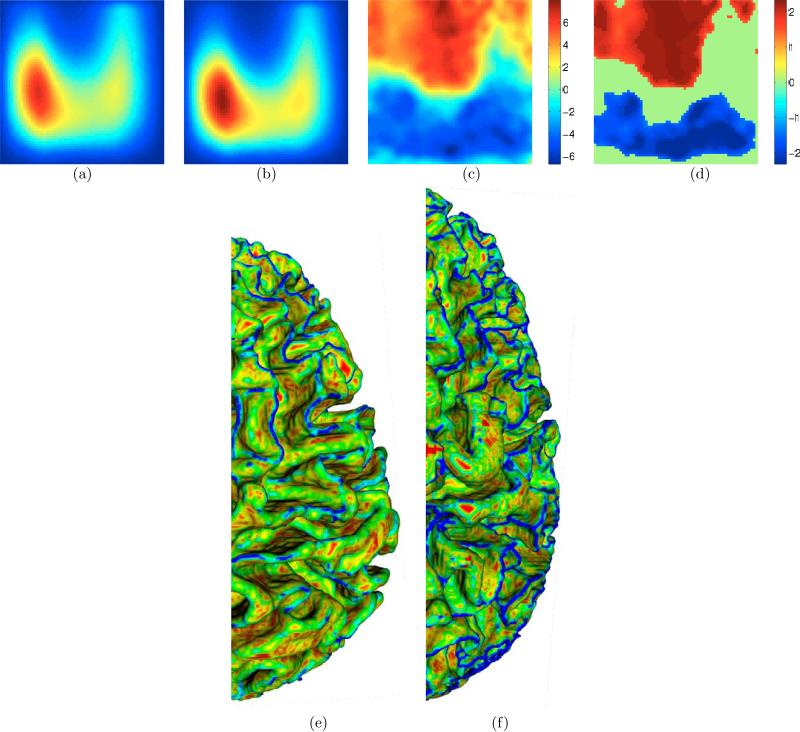
The proposed scale-invariant folding descriptor Pscale, described in Section 2.3, produces t maps (Figure 6(c)) indicating larger mass (red) for males in high-curvedness regions (top half) for all lobes. Complexity differences are very strong, producing significant locations (Figure 6(d)) and clusters for all lobes, and overwhelm shape-index differences. These results show that, when ICV differences have been accounted for via rescaling, male cortical surfaces are more “complex” than those of females based on the meaning of “complexity” ingrained in Pscale. This interpretation is consistent with [Im et al. (2008); Toro et al. (2008)] that analyzed brains, in the same way as the scale-invariant IPR measure, via the ratios of their surface areas to the two-thirds power of their volumes. Figures 6(e)-(f) help visualize the (accentuated) complexity differences by displaying colored cortical surfaces on a large male brain and a small female brain that is rescaled so that these two brains have equal volume. Referring back to Figure 2, these results rule out analogizing female and male cortical surfaces using S2 and S3, respectively, because that would make male cortical surfaces less “complex” than what the results indicate.
Figure 6.
(a)-(b) Average of Pscale for males and females, respectively, for the left frontal lobe. (c)-(d) t statistics (positive t ≡ larger value in males) and significant locations (via permutation testing), respectively, for the left frontal lobe. Similar patterns exist for all other lobes. Note the larger mass (red) for males in the high-curvedness (top half) regions. (e)-(f) Selected smaller-than-average female and larger-than-average male brains, respectively, adjusted for ICV and painted by C values (red→blue ≡ zero→high) to enable/accentuate visualization of the differences. Thus, based on the meaning of complexity ingrained in Pscale, the female brain appears less “complex”, i.e. more red/yellow. Compare this with Figure 5(j-k).
The two aforementioned cross-sectional clinical studies demonstrate that while the female cortex has a larger number of finer-scale features (which is the meaning of “complexity” in [Luders et al. (2004)]), the male cortex has a disproportionately greater bending in proportion to its larger volume (which is the meaning of “complexity” in [Im et al. (2008); Toro et al. (2008)]). Thus, the results show that folding patterns in males differ from those in females in two fundamental ways: (i) enlargement/scaling of folds and (ii) additional folds or bending. Amazingly, the magnitude of the latter effect is (i) weak enough to keep the bending in female folds, without any adjustment for ICV, more than the bending in males, but (ii) strong enough to reject the hypothesis that folding patterns in the two genders are simply scaled versions of each other. In the context of Figure 1, if we consider the typical cortical surface in females to be S2, then the typical cortical surface in males may be a modified version of S3, say S3*, that is modified with the addition of some finer-scale features. However, the features should be added in such a way that the fraction of the resulting high-curvature features in S3* continues to be less than S2. Furthermore, this paper is perhaps the first to show significant gender differences in gyral/sulcal shape (as indicated by the shape-index variable).
4.2. Folding Differences Between Normal and Abnormal Operculums in Neonates
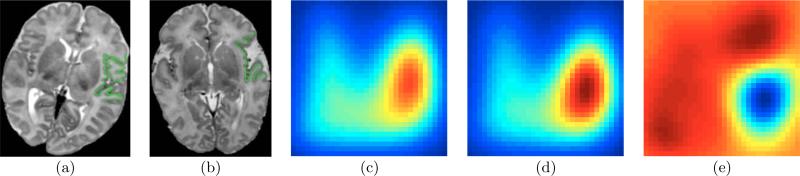
Before studying opercular differences in HLHS and TGA, we first study the differences between normal and abnormal (in CHD) opercular folding. Figure 7 shows the differences in the surface descriptors of two sub-groups of neonates selected by a pediatric neurologist, via visual examination of MR images from the clinical cohort: one sub-group comprises the 2 closest-to-normal operculums (mature, “closed”, folded) and another sub-group comprises the 4 farthest-from-normal operculums (immature, “open”, less folded). Figure 7(e) indicates that opercular maturity leads to (i) a larger fraction of surface patches that have higher curvedness (top half of the plot is red; relating to more complex folding) and (ii) a larger fraction of surface patches that are concave (left half of the plot is red; relating to deeper or more number of sulci and a more “closed” shape). Thus, Figure 7(e) associates meanings/interpretations, in terms of the proposed folding descriptor, to the radiologists’ qualitative interpretation of opercular folding maturity.
Figure 7.
(a)-(b) Example MR images of the normal (mature, “closed”, more folded) and farthest-from-normal (immature, “open”, less folded) operculums, respectively, overlapped with the extracted cortical surfaces. (c)-(d) Average cortical folding descriptors Paverage normal(C, S) and Paverage abnormal(C, S) for 2 closest-to-normal and 4 significantly-abnormal operculums, respectively, chosen by a medical expert based on a subjective clinical scoring protocol [Licht et al. (2009)]. (e) Paverage normal(C, S) − Paverage abnormal(C, S); blue ≡ negative and red ≡ positive values.
The findings from this quantitative study between normal and abnormal operculums are consistent with those from qualitative studies, of immature operculums in neonates [Chen et al. (1996); Childs et al. (2001)], which provide drawings of immature opercular cortical surfaces. These findings are consistent with a subjective clinical study indicating delayed maturation in the operculum in both HLHS and TGA variants of complex CHD [Licht et al. (2009)]. These findings indicate that normal maturational processes in the neonatal human brain affect both cortical complexity (as indicated by the distribution of the curvedness of surfaces patches) as well as cortical shape (as indicated by the distribution of shape indices of surface patches).
4.3. Folding Differences in Opercular Regions Between Neonates with HLHS and TGA
Figure 8 shows the differences in folding complexity and shape index between HLHS and TGA. The group differences in Figure 8(e-f) seem to be dissimilar, and much more subtle, as compared to those between normal and abnormal operculums detected in Figure 7(e). Figures 8(e) and (f) indicate that TGA neonates seem to have a larger fraction of high-curvature patches, while HLHS neonates tend to have a larger fraction of concave low-curvature surface patches. Relating Figures 8(e) and (f) to Figure 7(e) suggests that HLHS and TGA may affect opercular development in different ways, i.e. while HLHS might cause a greater reduction in opercular folding complexity (lower curvedness of patches) as compared to TGA, TGA might cause more “open” operculums via shallower or fewer sulci (smaller fraction of concave patches) as compared to HLHS.
Figure 8.
(a)-(b) Average of cortical folding descriptors P(C, S) in right and left operculums, respectively, for 29 neonates with HLHS. (c)-(d) Average of the descriptors in right and left operculums, respectively, for 13 neonates with TGA. (e)-(f) t-statistic maps; t > 0 ⇒ PHLHS > PTGA. for right and left operculums, respectively. (g)-(h) Significant clusters (via permutation testing) for right and left operculums, respectively.
This study between HLHS and TGA reports differences in multiple aspects (complexity, shape index) of folding patterns. Nevertheless, the differences between HLHS and TGA are subtle and the study can be made more informative via larger sample sizes.
5. Conclusion
This paper presents a novel multivariate high-dimensional (4096-dimensional) folding descriptor that captures multiple aspects of surface geometry. The paper provides new mathematical insights into the different meanings of complexity in the context of ICV differences. The paper exploits these insights to resolve two seemingly-contradictory findings in the state of the art on gender-based cortical folding, i.e. [Luders et al. (2004)] and [Im et al. (2008); Toro et al. (2008)] di er in which gender has higher “complexity”. Recent studies [Sowell et al. (2007)] have found that the female cortex is thicker in some regions even without compensating for lower ICV. After accounting for ICV differences, the entire cortex is significantly thicker in females. The complementary findings concerning cortical complexity, shape, and thickness might help explain the similarities and differences in cognitive skills possessed by both genders.
The results in this paper are perhaps the first to quantitatively demonstrate (i) opercular folding differences between closed and open operculums and (ii) differences between neonates with HLHS and TGA. A cross-sectional folding study between CHD neonates and normal neonates with large sample sizes would be very interesting and probably yield much more significant differences; indeed, this is a part of future work. This paper also demonstrates the potential for proposed cortical folding-pattern descriptor to help quantify the extent of neurodevelopmental delay or abnormality.
The proposed descriptor measures the discrepancy between two surfaces by the differences in the corresponding . However, equivalence of PDFs doesn't necessarily imply equivalence of the surfaces and this is a limitation of the proposed descriptor. Nevertheless, the proposed descriptor (a PDF characterized by thousands of numbers) is significantly more descriptive than typical folding descriptors in the literature that characterize folding by a single number.
The proposed permutation testing approach for multivariate random variables is general and can be applied in a straightforward manner to any clinical study dealing with multivariate data. For instance, the technique has been recently applied in a study of WM tracts incorporating multivariate data in the form of fractional anisotropy and thickness for every point on the tract [Zhang et al. (2009, 2010)]. The proposed cortical surface model can be easily extended to include cortical thickness resulting in a model for the cerebral cortex (cortical sheet) that captures its folding as well as thickness characteristics. The resulting nonparametric cortical sheet model, together with the proposed multivariate permutation-testing approach that continues to be applicable for hypothesis testing with the cortical sheet model, can lead to more probing cortical clinical studies. The proposed surface descriptor can be extended for multiscale analysis by establishing a scale space for the surface geometry. The proposed framework also lends itself to longitudinal studies and correlation studies via modifications of the statistical test at every grid point in the domain of the descriptor; e.g. t test for cross-sectional studies can be replaced by regression tests for correlation or longitudinal studies. The proposed random-field statistical model for cortical surfaces can be extended to be able to incorporate local interactions between surface patches, enabling the modeling and analysis of cortical surfaces as geometric textures.
Acknowledgments
The authors gratefully acknowledge the support of this work by the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Awards Program in Translational Medicine and Therapeutics (NIH funded) at the University of Pennsylvania, NIH grants HD042974, HD046159, NS045839, EB06266, DA14129, DA22807, UL1RR024234, K23 NS052380, NS061111, and K25 AG027785, the Dana Foundation, and the June and Steve Wolfson Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong E, Schleicher A, Omran H, Curtis M, Zilles K. The ontogeny of human gyrification. Cerebral Cortex. 1995;5:56–63. doi: 10.1093/cercor/5.1.56. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Voxel-based morphometry - the methods. NeuroImage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Aubert-Broche B, Collins D, Evans A. Twenty new digital brain phantoms for creation of validation image data bases. IEEE Trans. Med. Imag. 2006;25(11):1410–1416. doi: 10.1109/TMI.2006.883453. [DOI] [PubMed] [Google Scholar]
- Avants B, Gee J. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage. 2004;23(1):139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Awate SP, Gee JC. A fuzzy, nonparametric segmentation framework for DTI and MRI analysis. Proc. Information Processing in Medical Imaging (IPMI) 2007:296–307. doi: 10.1007/978-3-540-73273-0_25. [DOI] [PubMed] [Google Scholar]
- Awate SP, Tasdizen T, Foster NL, Whitaker RT. Adaptive Markov modeling for mutual-information-based unsupervised MRI brain-tissue classification. Medical Image Analysis. 2006;10(5):726–739. doi: 10.1016/j.media.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Awate SP, Win L, Yushkevich P, Schultz RT, Gee JC. 3D cerebral cortical morphometry in autism: Increased folding in children and adolescents in frontal, parietal, and temporal lobes. Proc. Int. Conf. Medical Image Computing and Computer Assisted Intervention. 2008;1:559–567. doi: 10.1007/978-3-540-85988-8_67. [DOI] [PubMed] [Google Scholar]
- Awate SP, Yushkevich PA, Licht DJ, Gee JC. Gender differences in cerebral cortical folding: Multivariate complexity-shape analysis with insights into handling brain-volume differences. Proc. Int. Conf. Medical Image Computing and Computer Assisted Intervention. 2009a;1:200–207. doi: 10.1007/978-3-642-04271-3_25. [DOI] [PubMed] [Google Scholar]
- Awate SP, Yushkevich PA, Song Z, Licht DJ, Gee JC. Multivariate high-dimensional cortical folding analysis, combining complexity and shape, in neonates with congenital heart disease. Proc. Int. Conf. Information Processing in Medical Imaging. 2009b:552–563. doi: 10.1007/978-3-642-02498-6_46. [DOI] [PubMed] [Google Scholar]
- Batchelor P, Castellano-Smith A, Hill D, Hawkes D, Cox T, Dean A. Measures of folding applied to the development of the human fetal brain. IEEE Trans. Med. Imaging. 2002;21(8):953–965. doi: 10.1109/TMI.2002.803108. [DOI] [PubMed] [Google Scholar]
- Chen C, Zimmerman R, Faro S, Parrish B, Wang Z, Bilaniuk L, Chou T. MR of the cerebral operculum: abnormal opercular formation in infants and children. American Journal of Neuroradiology. 1996;17(7):1303–1311. [PMC free article] [PubMed] [Google Scholar]
- Childs A, Ramenghi L, Cornette L, Tanner S, Arthur R, Martinez D, Levene M. Cerebral maturation in premature infants: Quantitative assessment using MR imaging. Amer. J. of Neuroradiology. 2001;22:1577–1582. [PMC free article] [PubMed] [Google Scholar]
- Chow Y, Geman S, Wu L. Consistant cross-validated density estimation. Annals of Statistics. 1983;11(1):25–38. [Google Scholar]
- Davatzikos C, Prince J. An active contour model for mapping the cortex. IEEE Trans. Medical Imaging. 1995;14(1):65–80. doi: 10.1109/42.370403. [DOI] [PubMed] [Google Scholar]
- DoCarmo M. Differential Geometry of Curves and Surfaces. Prentice Hall; 1976. [Google Scholar]
- Fisher RA. The Design of Experiment. Hafner; 1935. [Google Scholar]
- Friston K, Holmes A, Worsley K, Poline J, Frith C, Frackowiak R. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapping. 1995;2:189–210. [Google Scholar]
- Griffin L. The intrinsic geometry of the cerebral cortex. J. Theor. Biol. 1994;166(3):261–73. doi: 10.1006/jtbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- Im K, Lee J, Lytteleton O, Kim S, Evans A, Kim S. Brain size and cortical structure in the adult human brain. Cer. Cor. 2008;18:2181–2191. doi: 10.1093/cercor/bhm244. [DOI] [PubMed] [Google Scholar]
- ITK National Library of Medicine Insight Segmentation and Registration Toolkit (ITK) 2010 URL http://www.itk.org.
- Jones D, Symms M, Cercignani M, Howard R. The effect of filter size on VBM analyses of DT-MRI data. NeuroImage. 2005;26(2):546–554. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Joshi S, Wang J, Miller M, Essen DV, Grenander U. On the differential geometry of the cortical surface. Vision Geometry. 1995;IV:304–311. [Google Scholar]
- Koenderink J, van Doorn A. Surface shape and curvature scales. Image and Vision Computing. 1992;10(8):557–565. [Google Scholar]
- Koenderink JJ. Solid Shape. MIT Press; 1990. [Google Scholar]
- Licht D, Shera D, Clancy R, Wernovsky G, Montenegro L, Nicolson S, Zimmerman R, Spray T, Gaynor W, Vossough A. Brain maturation is delayed in infants with complex congenital heart defects. J. Thorac. Cardiovasc. Surg. 2009;137:529–537. doi: 10.1016/j.jtcvs.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Chen X. Spatial kernel regression estimation: weak consistency. Stat. and Prob. Letters. 2004;68(2):125–136. [Google Scholar]
- Luders E, Narr K, Thompson P, Rex D, Jancke L, Steinmetz H, Toga A. Gender differences in cortical complexity. Nat. Neuro. 2004;7(8):799–800. doi: 10.1038/nn1277. [DOI] [PubMed] [Google Scholar]
- Lyttelton O, Boucher M, Robbins S, Evans A. An unbiased iterative group registration template for cortical surface analysis. NeuroImage. 2007;34:1535–1544. doi: 10.1016/j.neuroimage.2006.10.041. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Prasad R. The fractal dimension of cerebral surfaces using magnetic resonance images. Computers in Physics. 1988;2(6):69–73. [Google Scholar]
- Mangin J, Riviere D, Cachia A, Duchesnay E, Cointepas Y, Papadopoulos-Orfanos D, Scifo P, Ochiai T, Brunelle F, Regis J. A framework to study the cortical folding patterns. NeuroImage. 2004;23(1):S129–S138. doi: 10.1016/j.neuroimage.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Miller S, McQuillen P, Hamrick S, Xu D, Glidden D, Charlton N, Karl T, Azakie A, Ferriero D, Barkovich J, Vigneron D. Abnormal brain development in newborns with congenital heart disease. New Eng. J. Med. 2007;257:1928–1938. doi: 10.1056/NEJMoa067393. [DOI] [PubMed] [Google Scholar]
- Mortamet B, Zeng D, Gerig G, Prastawa M, Bullitt E. Effects of healthy aging measured by intracranial compartment volumes using a designed MR brain database. Med. Imag. Comput. Comp. Assist. Interv. 2005:383–91. doi: 10.1007/11566465_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols T, Holmes A. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human Brain Mapping. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordahl C, Dierker D, Mostafavi I, Schumann C, Rivera S, Amaral D, Van-Essen D. Cortical folding abnormalities in autism revealed by surface-based morphometry. Journal of Neuroscience. 2007;27(43):11725–11735. doi: 10.1523/JNEUROSCI.0777-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Kubick S, Abernathey C. Atlas of the Cerebral Sulci. Thieme Medical. 1990 [Google Scholar]
- Osher S, Paragios N. Geometric Level Set Methods in Imaging, Vision, and Graphics. Springer; 2003. [Google Scholar]
- Papoulis A, Pillai SU. Probability, Random Variables, and Stochastic Processes. 4th Edition McGraw-Hill; 2001. [Google Scholar]
- Parzen E. On the estimation of a probability density function and the mode. Annals of Math. Stats. 1962;33:1065–1076. [Google Scholar]
- Peyre G, Cohen L. Geodesic remeshing using front propagation. Int. J. Computer Vision. 2006;69(1):145–156. [Google Scholar]
- Pienaar R, Fischl B, Caviness V, Makris N, Grant PE. A methodology for analyzing curvature in the developing brain from preterm to adult. Int. J. Imaging Systems Technology. 2008;18(1):42–68. doi: 10.1002/ima.v18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Carranza C, Mukherjee P, Vigneron D, Barkovich J, Studholme C. A framework for in vivo quantification of regional brain folding in premature neonates. Neuroimage. 2008;41(2):462–478. doi: 10.1016/j.neuroimage.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethian J. Level Set Methods and Fast Marching Methods. Cambridge Univ. Press; 1999. [Google Scholar]
- Smith S. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Awate SP, Licht D, Gee JC. Clinical neonatal brain MRI segmentation using adaptive nonparametric data models and intensity-based Markov priors. Proc. Med. Image Computing Comp. Assisted Intervention. 2007;1:883–890. doi: 10.1007/978-3-540-75757-3_107. [DOI] [PubMed] [Google Scholar]
- Sowell E, Peterson B, Kan E, Woods R, Yoshii J, Bansal R, Xu D, Zhu H, Thompson P, Toga A. Sex differences in cortical thickness mapped in 176 healthy individuals between 7 and 87 years of age. Cer. Cor. 2007;17:1550–1560. doi: 10.1093/cercor/bhl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Gerig G. Correction scheme for multiple correlated statistical tests in local shape analysis. SPIE Medical Imaging. 2003:233–240. [Google Scholar]
- Thompson P, Schwartz C, Lin R, Khan A, Toga A. Three-dimensional statistical analysis of sulcal variability in the human brain. Journal of Neuroscience. 1996;16(13):4261–4274. doi: 10.1523/JNEUROSCI.16-13-04261.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Perron M, Pike B, Richer L, Veillette S, Pausova Z, Paus T. Brain size and folding of the human cerebral cortex. Cer. Cor. 2008;18:2352–2357. doi: 10.1093/cercor/bhm261. [DOI] [PubMed] [Google Scholar]
- Van-Essen D. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Van-Essen D, Dierker D. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Van-Essen D, Drury H. Structural and functional analyses of human cerebral cortex using a surface-based atlas. J. Neuroscience. 1997;17(18):7079–7102. doi: 10.1523/JNEUROSCI.17-18-07079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovk U, Pernus F, Likar B. Intensity inhomogeneity correction of multispectral MR images. NeuroImage. 2006;32(1):54–61. doi: 10.1016/j.neuroimage.2006.03.020. [DOI] [PubMed] [Google Scholar]
- VTK The Visualization Toolkit (VTK) 2010 URL http://www.vtk.org.
- Wand M, Jones M. Kernel Smoothing. Chapman and Hall; 1995. [Google Scholar]
- Whitaker RT. A level-set approach to 3D reconstruction from range data. Int. J. Comput. Vision. 1998;29(3):203–231. [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Evans AC. Searching scale space for activation in PET images. Human Brain Mapping. 1996;4(1):74–90. doi: 10.1002/(SICI)1097-0193(1996)4:1<74::AID-HBM5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Yeo B, Yu P, Grant P, Fischl B, Golland P. Shape analysis with overcomplete spherical wavelets. Med. Imag. Comput. Comp. Assist. Interv. 2008:891–899. doi: 10.1007/978-3-540-85988-8_56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Grant PE, Qi Y, Han X, Segonne F, Pienaar R, Busa E, Pacheco J, Makris N, Buckner RL, Golland P, Fischl B. Cortical surface shape analysis based on spherical wavelets. IEEE Trans. Med. Imaging. 2007;26(4):582–597. doi: 10.1109/TMI.2007.892499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P, Piven J, Hazlett H, Smith R, Ho S, Gee J, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved e ciency and reliability. NeuroImage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zhang H, Awate SP, Das SR, Woo JH, Melhem ER, Gee JC, Yushkevich PA. A tract-specific framework for white matter morphometry combining macroscopic and microscopic tract features. Proc. Int. Conf. Medical Image Computing and Computer Assisted Intervention. 2009;1:141–149. doi: 10.1007/978-3-642-04271-3_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Awate SP, Das SR, Woo JH, Melhem ER, Gee JC, Yushkevich PA. A tract-specific framework for white matter morphometry combining macroscopic and microscopic tract features. Medical Image Analysis. 2010 doi: 10.1016/j.media.2010.05.002. Published Online; To Appear in Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Armstrong E, Schleicher A, Kretschmann H. The human pattern of gyrification in the cerebral cortex. Anat. Embryol. 1988;179:173–179. doi: 10.1007/BF00304699. [DOI] [PubMed] [Google Scholar]