Abstract
Catalysis by a class I ribonucleotide reductase (RNR) begins when a cysteine (C) residue in the α2 subunit is oxidized to a thiyl radical (C•) by a cofactor ~ 35 Å away in the β2 subunit. In a class Ia or Ib RNR, a stable tyrosyl radical (Y•) is the C oxidant, whereas a MnIV/FeIII cluster serves this function in the class Ic enzyme from Chlamydia trachomatis (Ct). It is thought that, in either case, a chain of Y residues spanning the two subunits mediates C oxidation by forming transient "pathway" Y•s in a multi-step electron transfer (ET) process that is "gated" by the protein so as to occur only in the ready holoenzyme complex. The drug hydroxyurea (HU) inactivates both Ia/b and Ic β2s by reducing their C oxidants. Reduction of the stable cofactor Y• (Y122•) in Escherichia coli class Ia β2 is faster in the presence of α2 and substrate (CDP), leading to speculation that HU might intercept a transient ET-pathway Y• under these turnover conditions. Here we show that this mechanism is one of two that are operant in HU inactivation of the Ct enzyme. HU reacts with the MnIV/FeIII cofactor to give two distinct products: the previously described homogeneous MnIII/FeIII-β2 complex forms only under turnover conditions (in the presence α2 and the substrate), and a distinct, diamagnetic Mn/Fe cluster forms ~900-fold less rapidly as a second phase in the reaction under turnover conditions and as the sole outcome in the reaction of MnIV/FeIII-β2 only. Formation of MnIII/FeIII-β2 also requires (i) either Y338, its subunit-interfacial ET pathway residue, or Y222, the surface residue that relays the “extra electron” to the MnIV/FeIV intermediate during activation of β2 but is not part of the catalytic ET pathway, and (ii) W51, the cofactor-proximal residue required for efficient ET between either Y222 or Y338 and the cofactor. The combined requirements for the catalytic subunit, the substrate, and, most importantly, a functional surface-to-cofactor electron-relay system imply that HU effects the MnIV/FeIII → MnIII/FeIII reduction by intercepting a Y• that forms when the ready holoenzyme complex is assembled, the ET gate is opened, and the MnIV reversibly oxidizes either Y222 or Y338.
Ribonucleotide reductases (RNRs) catalyze the committed step in the only known pathway for de novo biosynthesis of the deoxyribonucleotide precursors to DNA (1, 2). This reaction proceeds by a free-radical mechanism and is initiated by abstraction of the 3' H-atom of the substrate by a cysteine thiyl radical (C•) (3, 4). The C• is not present in the resting enzyme but is generated in situ by one-electron oxidation of a conserved cysteine residue by an oxidized cofactor. On the basis of the nature of the cofactor employed for generation of the C3'-H-abstracting C•, RNRs have been divided into classes I–III (1, 2). A class I RNR [e.g., those from all mammals, herpes simplex virus I, and the most extensively studied enzyme from Escherichia coli (Ec)] is composed of two homodimeric subunits (1, 5). The catalytic subunit, α2, contains the substrate-binding site, the cysteine residue that is transiently oxidized to the C•, and additional C residues that deliver electrons and a proton to the 2' position of the substrate to reduce it (5). The β2 subunit harbors the C-oxidizing cofactor. In a class Ia or Ib enzyme, the C oxidant is a stable tyrosyl radical (Y•) in close proximity to a carboxylate-bridged dimetal cluster [Fe2III/III in class Ia (6–8) and either Fe2III/III (9, 10) or Mn2III/III (11–13) in class Ib]. The reaction begins with the one-electron oxidation of the C (C439 in the Ec enzyme) in α2 by the Y• (Y122• in the Ec enzyme) in β2 (5). After substrate reduction, the C• reoxidizes the transiently reduced Y back to its radical form. This inter-subunit radical-transfer step has several notable characteristics. First, the Y• in β2 is (in the protoyptical E. coli enzyme) thought to be ~35 Å away from the radical C in α2 (5, 14, 15), a distance too large for C oxidation to proceed at the required rate by a single electron-tunneling step. Evidence indicates that the ET instead proceeds by a multi-step tunneling ("radical-hopping" or "electron-relay") mechanism, in which transient "pathway" radicals form along a chain of conserved, aromatic amino acids (in the Ec enzyme, W48 and Y356 in β2 and Y730 and Y731 in α2) to transfer the electron from the C to the Y• and back again (16–23). Second, the individual ET steps leading to the net ~ 35-Å radical transfer are thought to be coupled to proton transfers (proton-coupled ET or PCET) (5, 24). Third, in the Ec class Ia RNR, the PCET step is gated by the protein so as to occur only in the ready holoenzyme complex, and binding of the substrate to α2 is the key for opening of the gate (20, 22, 23, 25). This conformational gating provides the mechanism by which the potently oxidizing Y122• can be stable for hours, even in the presence of reductants that are sufficiently potent to reduce it (e.g., ascorbate, thiols), but then accept the electron from C439 in tens to hundreds of milliseconds (25) upon assembly of the ready holoenzyme complex and opening of the gate. The gating mechanism for the inter-subunit PCET remains to be defined but is likely to involve dynamic control of one or more of the coupled proton transfer steps in response to protein conformation. In the simplest case, substrate binding to α2 in the α2:β2 complex (with the proper effector also bound) could induce a conformational change favoring a crucial proton transfer (perhaps to the buried cofactor Y• (5, 24); Figure 1A), allowing the oxidizing equivalent to propagate outward onto pathway residues and ultimately onto the C residue in the α2 active site. However, it should be noted that some class I RNRs have been shown to engage in more complex subunit-oligomerization equilibria, involving not only free α2, free β2, and α2β2, but also higher complexes α4β4 and α6β6 (26–33). Because the oligomerization state drastically affects activity and binding of nucleotides affects the oligomerization state (26–33), it is possible that these changes could, in some cases, be involved in the gating transition.
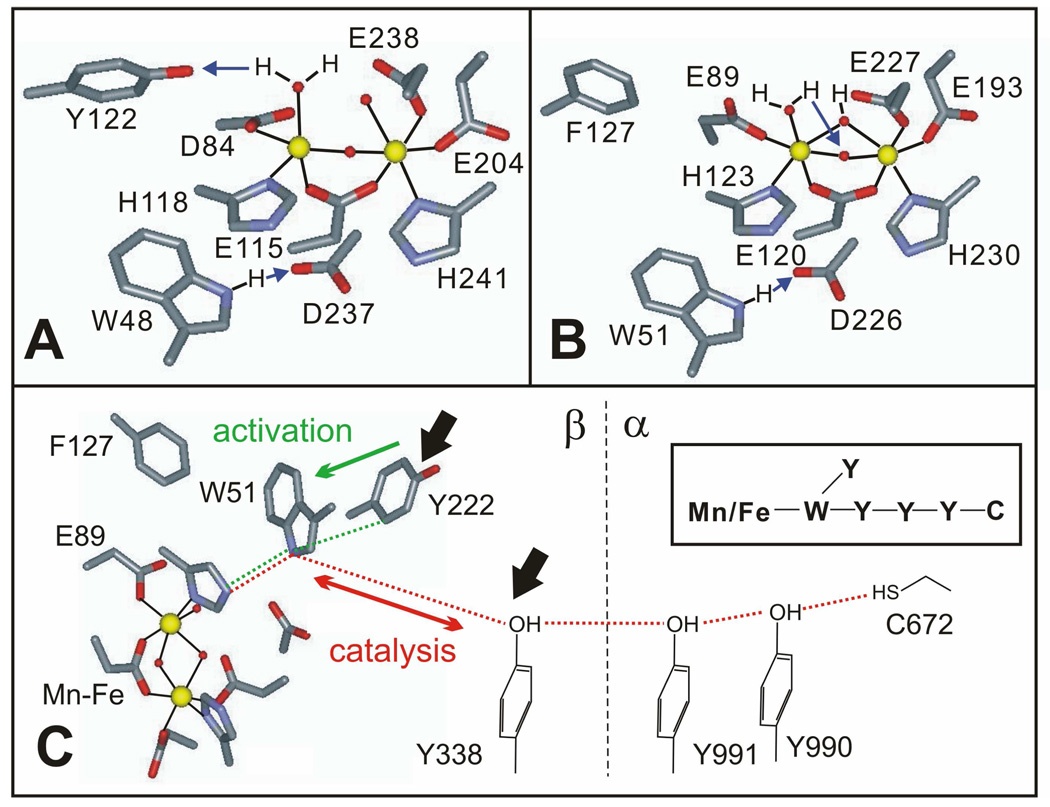
Figure 1.
Structural representations of the Fe2 III/III forms of E. coli β2 (A) and C. trachomatis β2 (B) generated from the crystallographic coordinates available in pdb files 1MXR and 1SYY, respectively. Proposed proton-transfer steps associated with gating of PCET are indicated with blue arrows in A and B. In C, the proposed electron transfer pathways of Ct RNR specific to catalysis (red dotted line) and activation of β2 by formation of its MnIV/FeIII cofactor (green dotted line) are shown. The black arrows indicate amino acid residues important in reduction of the MnIV/FeIII-β2•α2•CDP•ATP complex to the MnIII/FeIII form by hydroxyurea. A symbolic scheme of the residues involved in electron transfer is shown in the inset.
The Y• in a class Ia/b RNR can be reduced by a number of one-electron reductants (6, 34). Because this reaction inactivates RNR to inhibit DNA synthesis, it is of medical importance. One such Y• reductant, hydroxyurea (HU), is used as an anticancer and antiviral agent (35). Its reaction with Ec β2 has been studied by several groups (36–39). HU reduces the Y122• of Ec RNR ~10 times more rapidly in the presence of α2, substrate, and allosteric effector (turnover conditions) than in the absence of these components (36). This observation was interpreted as evidence that Y122• reduction by HU proceeds by two distinct mechanisms: direct access by the drug to the buried Y122• in "resting" β2 (39), and interception of a pathway radical intermediate of the multi-step PCET process under turnover conditions (36, 38). One would expect the first mechanism not to require a functional PCET pathway. This expectation was verified for the class Ia RNR from Mus musculus (40). By contrast, the more interesting second mechanism would be expected to occur only with the PCET pathway at least partially intact, i.e., not in variant β2s with the oxidizing equivalent effectively confined to the cofactor Y• by mutagenesis of one of the cofactor-proximal pathway residues. Surprisingly, this simple and direct test was not subsequently reported for the Ec enzyme or for any other class I RNR. Thus, the notion that HU can intercept a pathway radical intermediate of the multi-step PCET has remained merely interesting speculation.
The class Ic RNR from the obligate intracellular human pathogen, Chlamydia trachomatis (Ct), employs a high-valent, heterodinuclear MnIV/FeIII cofactor to initiate catalysis (41–44). In a structural sense, the MnIV ion replaces one Fe ion from the diiron cluster of a class Ia RNR, and, in a functional sense, it replaces the Y•. The MnIV/FeIII cofactor forms via a MnIV/FeIV intermediate upon reaction of the MnII/FeII form of Ct β2 with O2 (45). The electron needed to convert the FeIV site of the intermediate to the FeIII ion of the stable cofactor is transferred to the cluster site by W51 (the functional counterpart of W48 in Ec β2) and Y222, a residue that has no role in the catalytic PCET process and no functional counterpart in the class Ia/b proteins (Figure 1) (46). Analogy to the well studied Ec enzyme and results of mutagenesis experiments (46) imply that, during catalysis, forward PCET from C672 converts the MnIV/FeIII cluster to the MnIII/FeIII form, which has an S = 1/2 ground state and is therefore EPR-active. The EPR spectrum of MnIII/FeIII–β2 produced by dithionite (DT) reduction of the MnIV/FeIII state in the absence of α2, substrate, and allosteric effector is broad and poorly resolved, suggesting heterogeneity in the structure of the cluster (41). By contrast, reduction by DT in the presence of His6-Δ(1–248)α2 [a catalytically active, affinity tagged, N-terminally truncated form of α2 that is more stable than the wild-type subunit (47) and has been used in our previous mechanistic studies on Ct RNR (41, 46) and throughout this work], substrate, and allosteric effector (turnover conditions) results in a narrower, well-resolved EPR spectrum indicative of a structurally homogeneous state (41). The same well-resolved EPR signal is also observed when the Ct RNR holoenzyme acts on the mechanism-based inactivator, 2'-azido-2'-deoxy-ADP (41), and when it is reduced by HU (48). It seems likely that the structural perturbations to the cofactor revealed by the changes in the EPR spectrum of the MnIII/FeIII redox state reflect the PCET-gating transition.
In this work, we have defined the requirements for generation of the homogeneous MnIII/FeIII state with the resolved EPR spectrum and have probed the mechanism of its production by HU. The data provide direct evidence for two distinct mechanisms for reaction of the MnIV/FeIII cofactor with HU [as previously suggested for the Ec enzyme (36, 38, 39)], with one involving interception of a pathway Y• generated by excursion of the hole from the MnIV site upon opening of the PCET gate, and the other involving direct reaction with the MnIV/FeIII cluster. They indicate that, as in the Ec enzyme (20, 22, 23, 25), binding of the substrate opens the gate. They further suggest that the gate resides close to the cofactor in β2, involving, at most, only W51 among all the PCET pathway residues.
MATERIALS AND METHODS
Construction of Vectors to Over-express the Y991F, Y990F and C672S Variants of His6-Δ(1–248)α2
Residue numbering corresponds to the wild type sequence. The substitutions were introduced by the polymerase chain reaction (PCR) with the pET-28a-CtR1-Δ(1–248)-wt as the template (41). Details are provided in Supporting Information.
Overexpression and Purification of α2 and β2 proteins
Wild type and variant β2 proteins and His6-Δ(1–248)α2 and its site-directed variants were prepared as previously described (41, 46). Residual ND(T)P/dND(T)Ps bound to His6-Δ(1–248)α2 were digested by treatment with calf intestinal phosphatase (CIP) (New England Biolabs) at a concentration of 40 U/ml at ambient temperature for 30 min. The solution was loaded onto a Ni-NTA column again and CIP was washed out. His6-Δ(1–248)α2 was then eluted and concentrated, as previously described (41). The protein was stored in 100 mM HEPES buffer (pH 7.6), 10% (v/v) glycerol. Complete removal of CIP was verified by testing for its activity in the final solution by mass spectrometry.
Preparation of MnIV/FeIII-β2
MnIV/FeIII-β2 was prepared by slow addition of FeII to a solution of MnII and β2 under aerobic conditions, as previously described (49). β2 prepared in this way contains 90% of total Fe in form of the MnIV/FeIII cluster and exhibits a specific activity of 0.4 ± 0.1 s−1 (per β monomer) at 22 ± 2 °C in the presence of a 10-fold molar excess of His6-Δ(1–248)α2.
Stopped-Flow Absorption, Freeze-Quench EPR, and Mössbauer Experiments
The stopped-flow apparatus, the EPR and Mössbauer spectrometers, and the freeze-quench apparatus and procedures have been described (45). Mössbauer spectra were analyzed with the program WMOSS. Absorbance-versus-time traces after equal-volume mixing of a solution of MnIV/FeIII-β2, His6-Δ(1–248)α2, and ATP with a solution of CDP and hydroxyurea were analyzed by nonlinear regression according to equation 1, in which k1 and k2 are apparent first-order rate constants, ΔA1 and ΔA2 are amplitudes for the exponential phases, and A0 is the absorbance at time zero.
| (1) |
RESULTS AND DISCUSSION
Effect of the Presence of Substrate and Allosteric Effector on the EPR Spectrum of the MnIII/FeIII Cluster
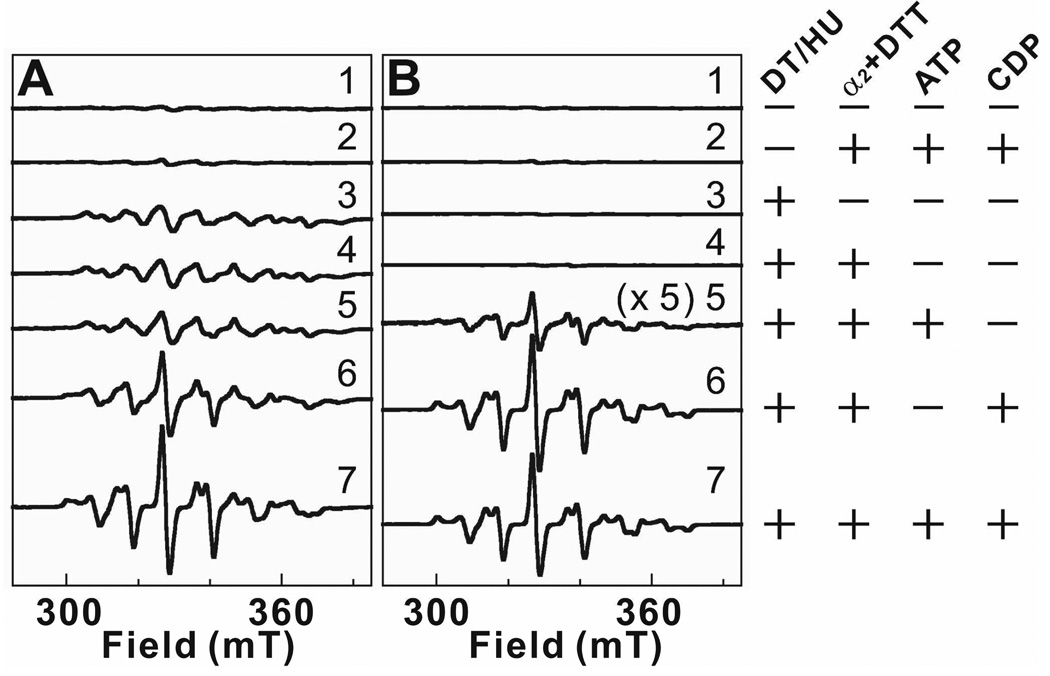
To define requirement(s) for development of the well-resolved EPR signal of the MnIII/FeIII-cluster in β2, components of the presumptive MnIV/FeIII-β2:α2:substrate:effector complex were systematically omitted. These samples were treated with either DT or HU (or no reductant in the control samples) and characterized by EPR spectroscopy (Figure 2). As expected, in the absence of reductant, neither MnIV/FeIII-β2 alone nor the protein in the presence of His6-Δ(1–248)α2, CDP, and ATP exhibits an EPR signal (spectra A1, A2, B1, and B2). Samples in which DT reduction of β2 was carried out in the absence of all other components (spectrum A3), the presence of His6-Δ(1–248)α2 only (spectrum A4), or the presence of His6-Δ(1–248)α2 and ATP (spectrum A5) exhibit nearly identical, broad, heterogeneous EPR spectra. These observations suggest that binding of the substrate, CDP, triggers the transition that permits generation of the resolved, homogeneous EPR signal (spectrum A7). DT reduction in the presence of all components but the allosteric effector, ATP, gives a signal (spectrum A6) that is the superposition of the broad and well-resolved spectra, suggesting that ATP may potentiate the effect of the substrate.
Figure 2.
X-band EPR spectra showing the requirement for different outcomes of reduction at MnIV/FeIII-β2 by dithionite (DT) (A) or hydroxyurea (HU) (B). The presence (+) or absence (−) of the various components is indicated on the right side of the corresponding spectrum. 300 µM β monomer (0.75 equiv MnIV/FeIII) was employed in all samples. [α2] = 900 µM, [DTT] = 10 mM, [CDP] = 4 mM, [ATP] = 1 mM; when present, [DT] = 20 mM and [HU] = 20 mM. All components except DT or HU were quickly premixed, and the reductant was added. Samples were incubated for 5 min at ambient temperature in the absence of O2 before freezing. Spectrum B5 has been multiplied by 5 for comparison. Spectrometer conditions: T = (14.0 ± 0.2) K; ν = 9.45 GHz; P = 200 µW; modulation frequency, 100 kHz; modulation amplitude, 1 mT; scan time, 167 s; time constant, 167 ms.
By contrast to its reduction by DT, treatment of MnIV/FeIII-β2, either alone or in the presence of only His6-Δ(1–248)α2, with HU does not generate an EPR-active species (spectra B3 and B4, respectively). In the presence of both His6-Δ(1–248)α2 and ATP, HU generates only a weak (~10 % of the maximum intensity) resolved signal (spectrum B5), implying that the allosteric effector does not by itself efficiently promote the structural transition permitting generation of the resolved signal.2,3 Again, the presence of substrate, CDP, is the key for the development of the resolved signal (spectrum B7). Indeed, the signal reaches its maximal intensity in the absence of the effector, ATP (spectrum B6), as long as CDP is present. Thus, the results with both DT and HU imply that binding of the substrate triggers the structural change reported by the EPR spectrum of the reduced state.
Kinetics of HU Reduction of MnIV/FeIII-β2 in the Absence and Presence of His6-Δ(1–248)α2, CDP, and ATP
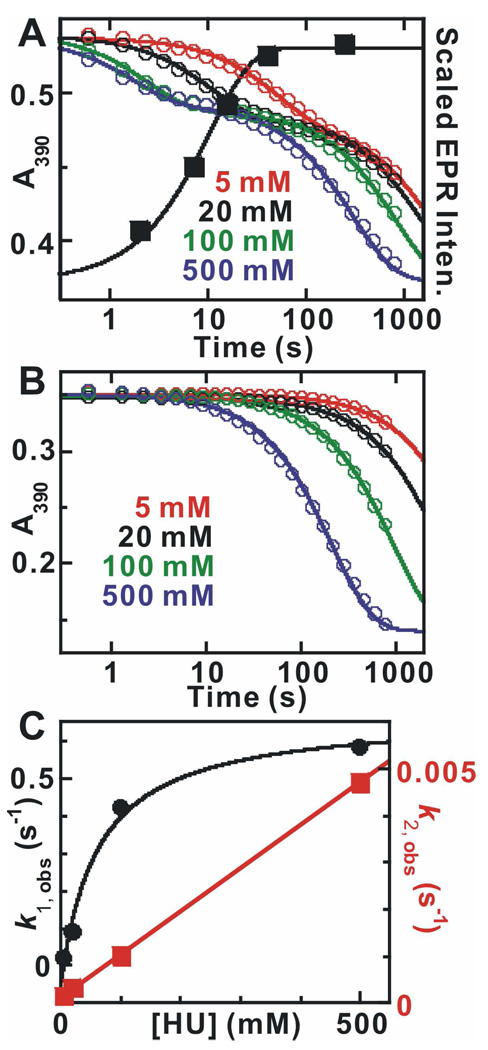
The kinetics of the reactions of MnIV/FeIII-β2 with HU in the absence and presence of His6-Δ(1–248)α2, CDP and ATP were defined in stopped-flow absorption (SF-Abs) and freeze-quench EPR (FQ-EPR) experiments. Both the MnIV/FeIII cluster and the reduction products have broad absorption bands between 300 and 500 nm without prominent peaks, but the features of the products are less intense, resulting in a decrease in absorbance in the 300–500 nm region during the reaction (49). ΔA390-versus-time traces from the reaction of MnIV/FeIII-β2 with HU at (22 ± 2) °C in the presence of His6-Δ(1–248)α2, CDP, and ATP exhibit two well-resolved phases (Figure 3A). The fact that the fast phase is absent in the reaction of only MnIV/FeIII-β2 with HU (Figure 3B) suggests that this phase is associated with formation of the S = 1/2 MnIII/FeIII cluster. This inference is confirmed by the results of freeze-quench EPR experiments at a single HU concentration (20 mM). Development of the well-resolved EPR signal (Figure S1 and solid squares in Figure 3A) is clearly coincident with the fast phase in the SF-Abs trace from the experiment at the same [HU] (black open circles in Figure 3A). The solid line through the experimental EPR intensities (squares) corresponds to a first order rate constant of (0.10 ± 0.03) s−1, in excellent agreement with the apparent first-order rate constant of (0.09 ± 0.02) s−1 obtained by fitting (as described below) the SF-Abs trace from the experiment at the same [HU] (solid line through the open circles). The biphasic SF-Abs traces from the reaction under turnover conditions were analyzed according to equation (1). The apparent first-order rate constant for the fast phase depends hyperbolically on [HU] (black solid circles in Figure 3C), with a limiting rate constant of (0.7 ± 0.1) s−1 and an apparent second-order rate constant of (8 ± 4) M−1s−1. By contrast, the traces from the reaction of MnIV/FeIII-β2 only with HU are monophasic, and a plot of the observed first-order rate constant versus [HU] is linear (red squares in Figure 3C), giving a second-order rate constant (slope) of (9 ± 2) × 10−3 M−1s−1. Thus, the fast phase of the reaction under turnover conditions is ~ 900-fold faster than the reaction of free MnIV/FeIII-β2. The slower phase in the reaction under turnover conditions correlates with the single phase in the reaction of free MnIV/FeIII-β2. The presence of this second phase, even with a two-fold molar excess of His6-Δ(1–248)α2 (which is expected to saturate β2 at these high subunit concentrations), can be explained by invoking asymmetry and a resultant "half-of-sites" reactivity in the Ct RNR holoenzyme complex, as was previously observed for class Ia RNRs (15, 20, 22, 50, 51). Additional evidence for half-of-sites reactivity in Ct RNR is provided below.
Figure 3.
(A) Kinetics of the absorbance changes at 390 nm (left y-axis) during the reaction of MnIV/FeIII–β2 with HU in the presence of α2, CDP, and ATP at ambient temperature (22 ± 2) °C and relative intensities of the EPR signal characteristic of the homogeneous MnIII/FeIII–β2 state (right y-axis). The reaction was carried out by mixing a solution containing 400 µM β monomer (0.75 equiv MnIV/FeIII), 800 µM α monomer, 20 mM DTT, and 2 mM ATP with an equal volume of reaction buffer containing 8 mM CDP and HU at a concentration giving the values of [HU] listed in the figure (after mixing). Solid lines overlaid are fits of the data to equation 1 with two exponential phases. The black solid squares are the EPR signal intensities extracted from the spectra shown in Figure S1. The solid line is the fit according of a single exponential phase to the data, which gives a rate constant of (0.10 ± 0.03) s−1. (B) Kinetics of the reaction of MnIV/FeIII–β2 with HU. The reaction was initiated in the same fashion above but in the absence of α2, CDP, ATP and DTT. (C) Dependence of the apparent first-order rate constants for the reactions on the concentration of HU. k1, obs plotted in black solid circles was extracted from the fast phase in A and the overlaid solid line is the hyperbolic fit to the data, which gives a limiting reduction rate constant (asymptote of hyperbolic fit) of (0.7 ± 0.1) s−1. k2, obs plotted in red solid squares was extracted from the reactions in B. The linear fitting gives the second-order rate constant (slope) of (9.0 ± 2.0) × 10−3 M−1s−1.
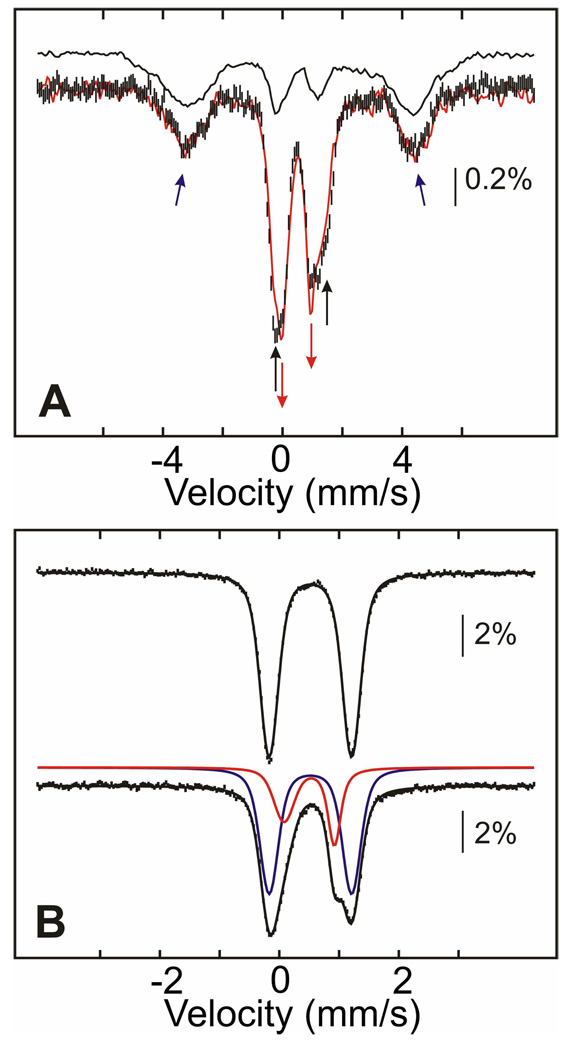
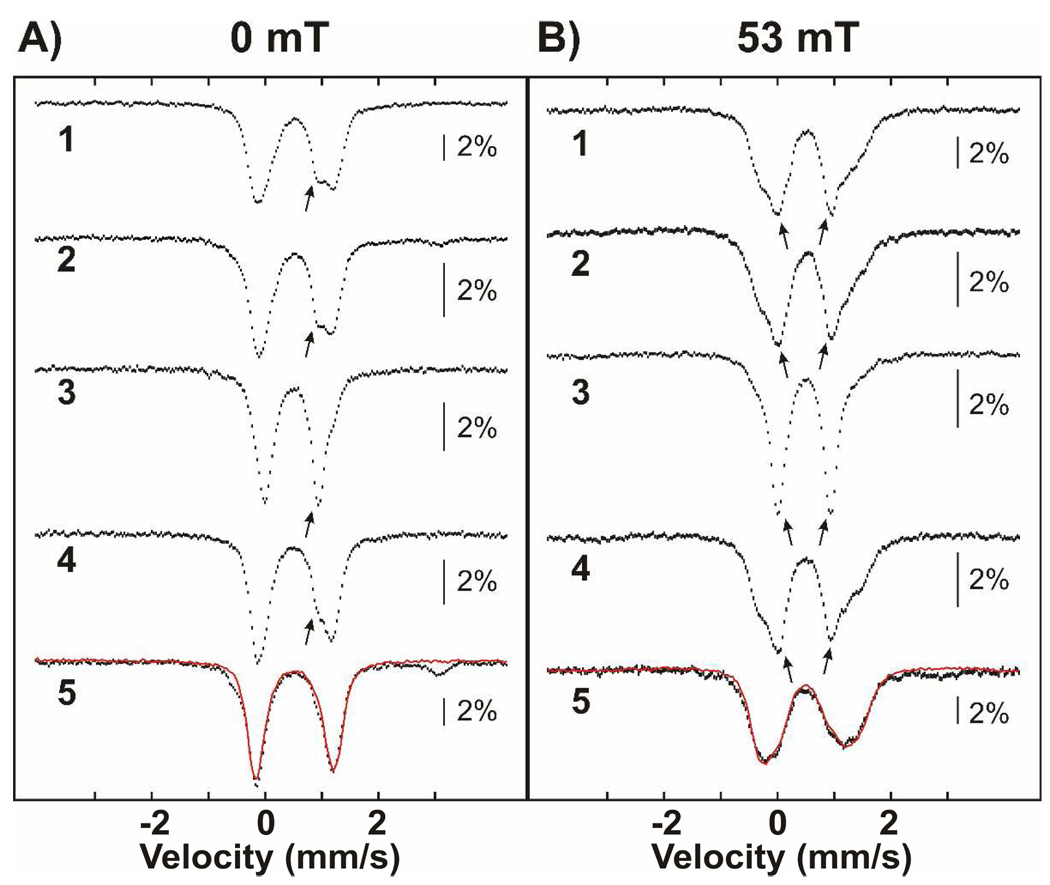
Mössbauer-spectroscopic Evidence for Two Distinct Products from HU Treatment of MnIV/FeIII-β2
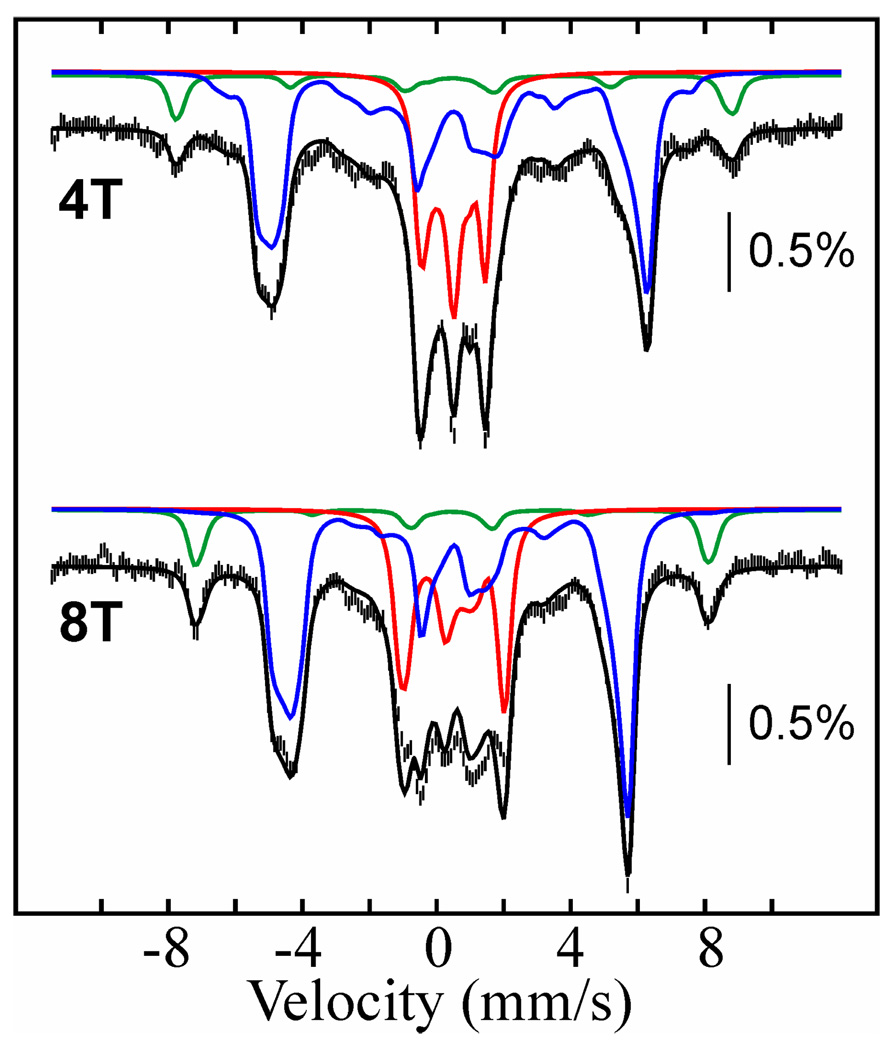
The product of both the reaction of free MnIV/FeIII-β2 with HU and the slow phase of the reaction under turnover conditions is EPR-silent and was therefore characterized by Mössbauer spectroscopy, a method that can provide information about any Fe-containing species, regardless of spin or oxidation state (52). The 4.2-K/53-mT Mössbauer spectra of samples prepared by incubating MnIV/FeIII-β2, His6-Δ(1–248)α2, CDP and ATP with 20 mM HU at ambient temperature for 1 min and 10 min are shown in Figure 4A as black vertical bars and a red line, respectively. These two reaction times correspond to completion of the first, fast phase (1 min) and approximately one quarter of the way through the second phase (10 min). Both spectra display two broad peaks at −5 mm/s and +5 mm/s (blue arrows), which we assign as the outer lines of the magnetically split spectrum of the homogeneous MnIII/FeIII-β2 state formed under turnover conditions. This spectral component accounts for ~45% of the total intensity. Comparably broad and poorly resolved features were previously observed for the heterogeneous MnIII/FeIII-β2 state produced by dithionite reduction of MnIV/FeIII-β2 alone (49), and this spectrum is shown as a solid black line for comparison. Because approximately half of the intensity of the Mössbauer spectrum is attributable to the MnIII/FeIII complex, the product of the fast phase, these results provide additional evidence for half-of-sites reactivity of the presumptive β2:His6-Δ(1–248)α2:CDP:ATP complex. The remaining fraction of the 1-min spectrum is attributable to the features of the MnIV/FeIII-β2 complex (42). Comparison of the spectra of the 1-min and 10-min samples reveals that prolonged incubation with HU yields a small amount (~10%) of a new quadrupole doublet, indicated by red arrows. This quadrupole doublet also develops when MnIV/FeIII-β2 is reacted with HU in the absence of His6-Δ(1–248)α2, CDP and ATP. In the 4.2-K/zero-field Mössbauer spectrum of this β2-only sample (Figure 4B), the contribution from the MnIV/FeIII cluster (blue line) is diminished by 36% relative to that in the starting MnIV/FeIII-β2 complex, and an asymmetric quadrupole doublet with parameters typical of high-spin FeIII (isomer shift δ = 0.49 mm/s and quadrupole splitting parameter |ΔEQ| = 0.88 mm/s, red line) accounts for the lost intensity. In particular, the isomer shift implies that the oxidation state of the iron does not change during the reaction with HU.4 This new species was further characterized by its Mössbauer spectra in applied fields of 4 T and 8 T (Figure 5). Analysis of these spectra reveals that the sample contains three components: MnIV/FeIII-β2 (57% intensity, blue lines), the new FeIII-containing complex (33% intensity, red lines), and a mononuclear high-spin FeIII impurity (10% intensity, green lines). The parameters used for the simulation of the MnIV/FeIII-β2 complex (see legend to Figure 5) are slightly different from (but within 3% of) those previously published (42), perhaps suggesting that this cluster may access slightly different conformations, as we previously suggested on the basis of observed variations in ΔEQ (49). The spectral features of the new FeIII-containing species can be satisfactorily simulated with the parameters (δ and ΔEQ) determined from analysis of the low-field spectra (given above) and the assumption that this complex has a diamagnetic (S = 0) electron-spin ground state.
Figure 4.
Mössbauer spectra illustrating the different outcomes of the reaction of MnIV/FeIII–β2 with HU. (A) 4.2-K/53-mT Mössbauer spectra of samples in which MnIV/FeIII–β2 (800 µM β, 0.75 equiv MnIV/FeIII cluster) was reacted with 20 mM HU at ambient temperature for 1 min (black hashed vertical bars) or 10 min (red solid line) in the presence of 1.2 mM α, 10 mM DTT, 1 mM ATP, and 10 mM CDP. The black solid line is the spectrum of the heterogeneous MnIII/FeIII-β2 state (49). The black and red arrows indicate the conversion of the MnIV/FeIII form to the new FeIII-containing complex. The blue arrows indicate the outer lines of the magnetically split sub-spectrum of the MnIII/FeIII cluster (~45% of the total intensity). The amounts of the MnIII/FeIII cluster in the two samples are identical within the experimental uncertainty. (B) 4.2-K/zero-field Mössbauer spectra of samples prepared by treatment of MnIV/FeIII-β2 (top) with 20 mM HU for 30 min at ambient temperature (bottom). The solid lines on top are simulations as indicated in the text. The blue and red lines represent the contribution of the MnIV/FeIII cluster and the new FeIII-containing, diamagnetic species, respectively.
Figure 5.
4.2-K Mössbauer spectra of the product of reaction of free MnIV/FeIII–β2 and HU recorded in external magnetic fields of 4T (top) and 8T (bottom) applied parallel to the γ-beam. 3.0 mM β (0.75 equiv MnIV/FeIII) was incubated with 60 mM HU at ambient temperature for 25 min before freezing. The contribution of MnIV/FeIII-β2 (57% intensity), the diamagnetic FeIII-containing cluster (33% intensity), and the mononuclear high-spin FeIII impurity (10% intensity) are shown as blue, red, and green lines, respectively. Simulation parameters are: S = 5/2, D = −0.38 cm−1, E/D = 0.31, g = (2.0, 2.0, 2.0), ΔEQ = 0.56 mm/s, δ = 0.46 mm/s, aFe/gNβN = (−22, −22, −22) T, η = −0.19 (green lines); S = 0, ΔEQ = −0.88 mm/s, δ = 0.49 mm/s, η = −0.43 (red lines); S = 1, D = −1.9 cm−1, E/D = 0.33, g = (2.0, 2.0, 2.0), ΔEQ = −1.38 mm/s, δ = 0.52 mm/s, AFe/gNβN = (−39.0, −39.2, −37.6) T, η = −0.23 (blue lines). The black lines overlaid with the experimental spectra are the summations of the three components.
We have considered three possibilities for the identity of the diamagnetic, FeIII-containing species. First and simplest, HU could act as a two-electron reductant, converting the MnIV/FeIII-β2 complex to a MnII/FeIII-β2 form, which would be expected to have a diamagnetic ground state arising from antiferromagnetic coupling between the MnII (SMn = 5/2) and FeIII (SFe = 5/2) ions. However, given that the MnII/FeII and MnIII/FeIII forms of β2 both react efficiently with H2O2 to produce MnIII/FeIII and MnIV/FeIV complexes, respectively (49), we anticipated that a MnII/FeIII form should also react with H2O2, presumably to form the MnIV/FeIII cluster. The diamagnetic complex does not react efficiently with H2O2 (Figure S2). Moreover, treatment of MnIV/FeIII-β2 with a different two-electron reductant, hydrazine (N2H4), does not produce the diamagnetic complex (Figure 6). These considerations weigh against a MnII/FeIII assignment. Second, the diamagnetic, FeIII-containing species could be an adduct between a formally MnIII/FeIII cluster and NO•. The MnIII/FeIII cluster could be generated by one-electron reduction of the MnIV/FeIII form by HU, which would yield an aminoxyl radical (37). The MnIII/FeIII could then react with NO•, which is known to form in solutions containing HU (53), to yield the MnIII/FeIII-NO• adduct. However, attempts to make the complex in high yield by direct exposure of a solution of MnIII/FeIII-β2 to NO• were unsuccessful: analysis of spectra of a sample prepared in this manner (Figure S3) suggests that only ~10% of the total intensity is attributable to the characteristic quadrupole doublet. This observation provides argument against the second possibility.
Figure 6.
Mössbauer spectra of the products of the reaction of MnIV/FeIII–β2 with (1) hydroxyurea, (2) hydroxylamine, (3) N,N-dimethyl-hydroxylamine, (4) acetohydroxamic acid, and (5) hydrazine. Spectra were collected at 4.2 K without applied field (A) and in an external magnetic field of 53 mT orientated parallel to the γ-beam (B). The arrows indicate the position of the quadrupole doublet of the FeIII-containing diamagnetic species. The reaction of MnIV/FeIII–β2 with hydrazine does not result in generation of the FeIII-containing diamagnetic species. To illustrate this, the spectra of the MnIV/FeIII form (with 90% of the total intensity) are overlaid for clarity as solid red lines. Instead, a small amount of a high-spin FeII species (~10%), which is evident from the weak absorption feature at ~3 mm/s, is formed in the reaction with hydrazine.
The third possibility, and the one that we favor, is that the FeIII-containing, diamagnetic complex generated in the reaction of MnIV/FeIII-β2 with HU forms by slow one-electron reduction by HU of the MnIV/FeIII cluster to a MnIII/FeIII form followed by coordination of the HU-derived aminoxyl radical (37) to the cluster, presumably to the FeIII site in order to account for the S = 0 ground state. Such coordination of aminoxyl radicals to transition metals has been reported (54, 55). The diamagnetic ground state could then be rationalized by antiferromagnetic interactions between the FeIII (SFe = 5/2) and MnIII (SMn = 2) sites and between the FeIII site and the aminoxyl radical (Saminoxyl = 1/2). Consistent with this hypothesis, other N-OH-containing compounds [NH2-OH, (CH3)2N-OH, and CH3-(CO)-NH-OH] also support development of the quadrupole doublet characteristic of the diamagnetic FeIII-containing complex when incubated with MnIV/FeIII-β2 (Figure 6). This structural explanation for the diamagnetic state requires direct access of the N-OH-containing reductant to the cofactor, as previously suggested for HU inactivation of Ec β2 in isolation (i.e., not under turnover conditions) (39).
Roles of ET Pathway Residues in the One-Electron Reduction of the MnIV/FeIII Cofactor
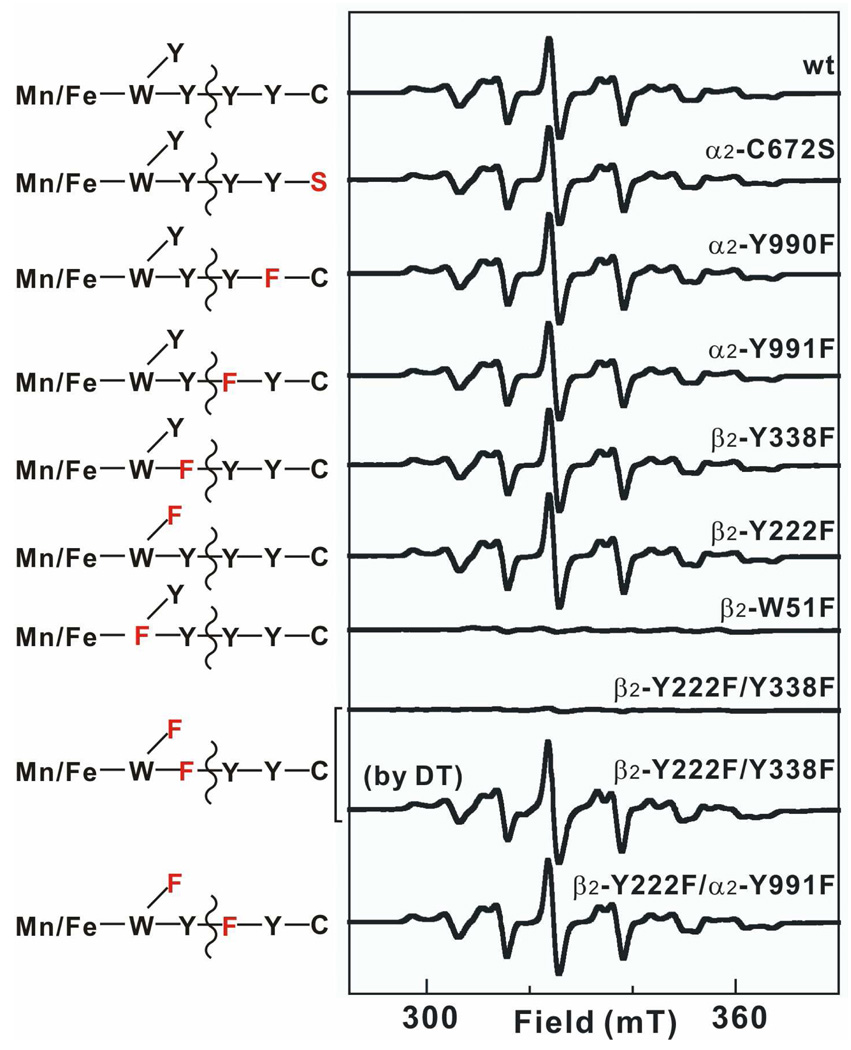
The roles of the catalysis-specific PCET-pathway residues in β2 (W51, Y338) (47, 56) and α2 [Y991, Y990, and C672; residue numbers reflect the sequence of the wild type subunit (47) rather than His6-Δ(1–248)α2] and the activation-specific residue, Y222, in β2 (46) in mediating one-electron reduction of the MnIV/FeIII cofactor by HU were probed by replacing these residues, individually and in combination, by functionally incompetent surrogates (phenylalanine, F, for the W and Y residues and serine, S, for C672) (Figure 1C). With the exception of β2-W51F (discussed below), HU treatment of any of the various single variants in the presence of CDP, ATP, and the wild type partner subunit gives the homogeneous EPR signal (Figure 7). It is tempting to conclude that the tyrosines of the PCET pathway have no role in mediating the one-electron reduction, but the result with the β2-Y222F/Y338F double variant shows otherwise. Very little EPR intensity develops upon HU treatment of this double variant in the presence of His6-Δ(1–248)α2, CDP, and ATP. The fact that the resolved signal is readily generated by treatment of MnIV/FeIII-β2-Y222F/Y338F with the stronger reductant, DT, in the presence of His6-Δ(1–248)α2, CDP, and ATP shows that the double variant is inherently competent to undergo whatever conformational change might be necessary to yield the homogeneous MnIII/FeIII state upon binding to His6-Δ(1–248)α2:CDP:ATP. Thus, either Y222 or Y338 in β2 (but not both) is required for the one-electron reduction outcome effected by HU under turnover conditions. This observation is the best evidence available for the hypothesis [advanced originally in studies of the Ec class Ia RNR (36, 38) but never tested by validation of the expected PCET-pathway dependence] that HU can intercept a Y• under turnover conditions. The inability of β2-W51F to support one-electron reduction of the MnIV/FeIII cluster by HU (although it does support formation of the diamagnetic, formally two-electron-reduced state; Figure S4) is also consistent, as previous studies have shown that proper electron-relay function of both Y222 and Y338 depends on the presence of W51 (46). In the case of this variant, the inability of DT to generate the resolved signal suggests that W51 could have a role in the conformational change required to form the homogeneous MnIII/FeIII state, although caution is warranted in this conclusion because of the unconservative nature of the W51F substitution and the possibility that it might propagate structural changes to other regions of the protein, perhaps preventing formation of the homogeneous state. The fact that none of the pathway residues in α2 is required to support formation of the resolved EPR signal implies that HU can act at the subunit interface (Figure 1C, black arrows), although the possibility that it can also act within α2 cannot be excluded from the available data.
Figure 7.
X-band EPR spectra of samples prepared by reacting wild type or variant MnIV/FeIII–β2 with HU or DT in the presence α2 (wild type or variant), CDP, and ATP. Corresponding schematic of the branched electron-transfer pathways is shown on the left, with redox-active residues in black and redox-inactive ones in red. To make a sample, an O2-free solution containing 300 µM β monomer (0.75 equiv MnIV/FeIII cluster), 900 µM α monomer, 4 mM CDP, 1 mM ATP, and 10 mM DTT was constituted. HU or DT was then added to a final concentration of 20 mM and the solution was incubated for 5 min at ambient temperature before freezing. Spectrometer conditions were: T = (14.0 ± 0.2) K; ν = 9.38 GHz; P = 200 µW; modulation frequency, 100 kHz; modulation amplitude, 1 mT; scan time, 20.97 s; time constant: 5.12 ms.
Evidence for Conformational Gating of PCET in Ct RNR
Among the major outstanding questions concerning the function of class I RNRs are the mechanism of the multi-step PCET process and how it is gated by the protein. The hypothesis of a conformational gate was initially founded on the dichotomy between the remarkable stability (many hours at room temperature) of the potently oxidizing Y122• in Ec β2, even in the presence of compounds such as ascorbate and thiols that are clearly thermodynamically competent to reduce it, and the relatively rapid turnover rate of the enzyme (5–10 s−1), which would seem to require that C439 reduce the Y122• in tens to hundreds of milliseconds (5, 25). Over the last decade, studies by the Stubbe group have provided more direct demonstration of the postulated conformational gating and have shown that substrate binding is the key to opening the gate (20, 22, 23, 25). Kinetic studies suggested that a slow conformational change prevents accumulation of reduced Y122 (the immediate product of PCET) in a single turnover and rate-limits substrate reduction (25). Variants of β2 or α2 containing dihydroxyphenylalanine (Dopa) or 3-aminotyrosine (NH2-Y) in place of one of the PCET pathway Y residues were shown to be capable of accumulating Dopa• or NH2-Y• pathway radicals (20, 22, 23). The reduction potentials of these radicals are less than that of Y•, meaning that reduction of Y122• by the engineered Dopa or NH2-Y pathway residue is thermodynamically favored. Nevertheless, PCET effectively does not proceed or proceeds very sluggishly until substrate is added. Thus, substrate binding opens the PCET gate and allows the favored reaction to proceed at a kinetically competent rate.
Such elegant resolution of steps in the inter-subunit PCET has not yet been achieved for Ct RNR, and so the notion that it is also conformationally gated remains a working hypothesis. Nevertheless, the hypothesis is now supported by a wealth of circumstantial evidence. The Ct enzyme is similar to the Ec protein in that the oxidized cofactor is quite stable (for at least hours) in the free β2 subunit in the presence of thiols but must accept an electron (presumably) from C672 in ~ 1 s to account for the ~ 0.6 s−1 turnover number (46, 47). In addition, CDP is required for production of the MnIII/FeIII complex with the well-resolved EPR signal when either DT or HU is used to reduce the active MnIV/FeIII state, and the latter reductant generates a distinct, diamagnetic product in the absence of substrate. The kinetics of HU reduction under turnover conditions are also consistent with data on Ec RNR suggesting that the opening of the gate is rate-limiting for turnover. The observed "saturation" of the apparent first-order rate constant for the reaction with increasing HU is most simply interpreted according to a mechanism in which (1) CDP (present in the syringe with HU) binds to His6-Δ(1–248)α2 in its complex with β2, (2) a rate-limiting conformational change opens the gate and allows for propagation of the oxidizing equivalent from the MnIV ion of the cofactor into the pathway, and (3) HU then reduces a pathway radical to trap the cofactor in the MnIII/FeIII state. According to this mechanism, saturation with respect to [HU] reflects the approach to 100% efficiency in trapping of the pathway radical by HU, and the limiting value of the rate constant (0.7 ± 0.1 s−1) for HU reduction reflects the rate constant for the gate-opening conformational change. This rate constant is, within error, twice the value of the turnover number under similar reaction conditions (0.4 s−1). If one assumes that only one monomer of the β2 homodimer can be active at a time, as is indicated by the above kinetic and spectroscopic data on the HU reaction, then these two values agree well. On this basis, we tentatively propose that the Ct enzyme may be rate-limited in a single-turnover and the steady-state by a slow conformational change occurring after binding of the substrate, as has been shown more rigorously by Stubbe and co-workers for the Ec enzyme (20, 22, 23, 25).
Location and Nature of the PCET Gate
If the interpretations given above are accepted, then the unique aspects of the Ct enzyme and its reaction with HU provide additional insight concerning the location and nature of the PCET gate. In the reaction of the Ec enzyme with HU, a ~ 10-fold difference in the rate of Y122• reduction under resting and turnover conditions (36) is the sole experimental manifestation of the postulated Y•-interception mechanism. Thus, one anticipates a 10-fold diminution (at most) in the rate of the reaction in a pathway-disrupted variant, were similar experiments to be performed on the Ec enzyme. By contrast, the Ct reaction has two spectroscopically distinguishable products and a ~ 900-fold difference in the rates of the reaction pathways leading to them. These features permit clearer definition of the requirements for the Y•-interception mechanism and opening of the PCET gate. More importantly, the existence of two intersecting electron-relay systems connecting the surface of β2 to the buried cofactor (the W51-Y338 pathway employed in catalytic PCET and the W51-Y222 pathway involved only in activation), a feature that is apparently unique to the Ct system among all class I RNRs that have been studied to date, provides a unique probe for the location of the gate. Although conformational transduction of the binding of the substrate to the opening of the PCET gate almost certainly involves an extensive network of interactions spanning both subunits, the actual gating mechanism could involve (i) primarily the electron accepting MnIV/FeIII cofactor (e.g., by promoting an increase in its effective reduction potential), (ii) primarily the electron donating C672 (e.g., by promoting an increase in its effective oxidation potential), (III) primarily the mediating pathway (e.g., by modification of the disposition of component residues to engage the pathway), or (iv) some combination of these elements. The observation that substituting any of the pathway Y residues does not prevent generation of the MnIII/FeIII product by HU implies that none of these residues plays a critical role in PCET gating. More specifically, the simplest interpretation of the observation that Y222 supports formation of MnIII/FeIII-β2 in the absence of the subunit-interfacial PCET pathway residue, Y338, in a substrate-gated fashion (Figure S5) is that opening of the gate permits propagation of the oxidizing equivalent from the MnIV ion onto the off-pathway Y222, generating a Y222• that is quenched by HU. This conclusion is somewhat surprising, as Y222 has no role in catalytic PCET [as evidenced by the absence of an effect of the Y222F substitution on the catalytic activity (46)] and would not necessarily be expected to communicate with the cofactor in a gated fashion. It is thus also quite informative, implying that gate opening induces a "pull" from the MnIV that is sufficiently strong to draw an electron from the off-pathway residue, if the intended target (Y338) is not present. Thus, the gating mechanism probably involves an increase in the effective reduction potential of the cofactor, and the gate is probably located close to the cofactor.
What is the nature of the gate? Stubbe, Nocera, and co-workers have proposed for Ec RNR that a proton from the Fe1-coordinated water is transferred to the Y122 phenolic oxygen to increase the effective reduction potential of the Y• and the proton from the indole N-H of W48 is transferred to D237 to decrease the potential of the W48 radical (blue arrows in Figure 1A), permitting migration of the radical from its resting position on Y122 onto W48 (5, 24). We have proposed a derivative mechanism by which dynamic control of proton transfers might also gate PCET in the Mn/Fe-dependent Ct RNR [Figure 1B (44)]. This mechanism is founded on X-ray absorption data, which revealed the presence of a (μ-oxo)(μ-hydroxo)-bridged MnIV/FeIII cluster in Ct β2 (57). We proposed that protonation of the μ-oxo-group yielding a bis-(μ-hydroxo)-bridged cluster could increase the reduction potential of the MnIV site to open the gate. By contrast, a recent computational study proposed that forward PCET is coupled to protonation of the hydroxo bridge, resulting in an (μ-oxo)(μ-aqua)-MnIII/FeIII cluster (58). Experiments to distinguish among these two models and other alternatives using electron nuclear double resonance (ENDOR) spectroscopy on the well-resolved EPR signal of the MnIII/FeIII state are ongoing. The pathway and mechanism by which the substrate-binding event is transduced the estimated 35 Å across the subunit interface to control the relevant proton transfers and open the gate remain to be elucidated.
Supplementary Material
Abbreviations
- ATP
adenosine-5’-triphosphate
- C•
cysteinyl radical
- CDP
cytidine-5'-diphosphate
- Ct
Chlamydia trachomatis
- dCDP
2'-deoxycytidine-5’-diphosphate
- DT
dithionite
- DTT
dithiothreitol
- Ec
Escherichia coli
- EPR
electron paramagnetic resonance
- ET
electron transfer
- HU
hydroxyurea
- PCET
proton-coupled electron transfer
- RNR
ribonucleotide reductase
- Y•
tyrosyl radical
Footnotes
This work was supported by the National Institutes of Health (GM-55365 to JMB and CK), the National Science Foundation (REU-DBI 05-21443 was used to acquire the EPR spectrometer used in this study), and the Dreyfus Foundation (Teacher Scholar Award to CK).
It is likely that the weak signal generated in the presence of ATP but absence of CDP reflects the effect of an ADP contaminant in the ATP solution. (ADP is a substrate for the enzyme.) Analysis of the ATP solution by mass spectrometry confirmed that ~16% of ATP had been hydrolyzed to ADP.
In the study of Roshick et al., truncation of residues 1–248 of α2, which removes a region that is marked by its sequence as a dATP binding site, did not appear to affect the behavior of the subunit toward the activator, ATP (47). However, because a potential nucleotide binding site that is present in the wild type subunit is absent in our His6-Δ(1–248)α2, caution is warranted in extending to the wild type α2 subunit the conclusion that ATP binding plays a relatively minor role in His6-Δ(1–248)α2 in promoting the structural transition that permits formation of the MnIII/FeIII cluster with the well-resolved EPR spectrum.
Alternatively, the novel features can be simulated with two symmetric quadrupole doublets of equal intensity. The parameters (δ1 = 0.55 mm/s, ΔEQ1 = 0.76 mm/s, δ2 = 0.46 mm/s, and ΔEQ2 = 0.94 mm/s) also support the assignment as high-spin FeIII for both sub-sites.
SUPPORTING INFORMATION AVAILABLE. Procedure for construction of the vectors to express the His6-affinity-tagged, N-terminally truncated α2 subunit [His6-Δ(1–248)α2] and its site-directed variants; stopped-flow absorption data from the reaction between HU and different redox forms of β2; Mössbauer spectra of samples prepared by exposing MnIII/FeIII-β2 to nitric oxide; Mössbauer spectra of samples prepared by reacting MnIV/FeIII-β2-W51F with HU; and EPR spectra showing that production of MnIII/FeIII-β2-Y338F by HU requires the presence of substrate. This material is available free of charge at the journal website http://pubs.acs.org.
Contributor Information
Carsten Krebs, Email: ckrebs@psu.edu.
J. Martin Bollinger, Jr, Email: jmb21@psu.edu.
REFERENCES
- 1.Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Stubbe J. Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol. 2000;10:731–736. doi: 10.1016/s0959-440x(00)00153-6. [DOI] [PubMed] [Google Scholar]
- 3.Stubbe J, Ackles D. On the mechanism of ribonucleoside diphosphate reductase from Escherichia coli. J. Biol. Chem. 1980;255:8027–8030. [PubMed] [Google Scholar]
- 4.Mao SS, Yu GX, Chalfoun D, Stubbe J. Characterization of C439SR1, a mutant of Escherichia coli ribonucleotide diphosphate reductase: evidence that C439 is a residue essential for nucleotide reduction and C439SR1 is a protein possessing novel thioredoxin-like activity. Biochemistry. 1992;31:9752–9759. doi: 10.1021/bi00155a031. [DOI] [PubMed] [Google Scholar]
- 5.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer? Chem. Rev. 2003;103:2167–2202. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 6.Atkin CL, Thelander L, Reichard P, Lang G. Iron and free radical in ribonucleotide reductase. Exchange of iron and Mössbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J. Biol. Chem. 1973;248:7464–7472. [PubMed] [Google Scholar]
- 7.Nordlund P, Sjöberg B-M, Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 8.Stubbe J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr. Opin. Chem. Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 9.Jordan A, Pontis E, Atta M, Krook M, Gibert I, Barbe J, Reichard P. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12892–12896. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huque Y, Fieschi F, Torrents E, Gibert I, Eliasson R, Reichard P, Sahlin M, Sjöberg B-M. The active form of the R2F protein of class Ib ribonucleotide reductase from Corynebacterium ammoniagenes is a diferric protein. J. Biol. Chem. 2000;275:25365–25371. doi: 10.1074/jbc.M002751200. [DOI] [PubMed] [Google Scholar]
- 11.Willing A, Follmann H, Auling G. Ribonucleotide reductase of Brevibacterium ammoniagenes is a manganese enzyme. Eur. J. Biochem. 1988;170:603–611. doi: 10.1111/j.1432-1033.1988.tb13740.x. [DOI] [PubMed] [Google Scholar]
- 12.Abbouni B, Oehlmann W, Stolle P, Pierik AJ, Auling G. Electron paramagnetic resonance (EPR) spectroscopy of the stable-free radical in the native metallo-cofactor of the manganese-ribonucleotide reductase (Mn-RNR) of Corynebacterium glutamicum. Free Radical Res. 2009;43:943–950. doi: 10.1080/10715760903140568. [DOI] [PubMed] [Google Scholar]
- 13.Cotruvo JA, Stubbe J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry. 2010;49:1297–1309. doi: 10.1021/bi902106n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 15.Bennati M, Robblee JH, Mugnaini V, Stubbe J, Freed JH, Borbat P. EPR distance measurements support a model for long-range radical initiation in E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2005;127:15014–15015. doi: 10.1021/ja054991y. [DOI] [PubMed] [Google Scholar]
- 16.Ekberg M, Sahlin M, Eriksson M, Sjöberg B-M. Two conserved tyrosine residues in protein R1 participate in an intermolecular electron transfer in ribonucleotide reductase. J. Biol. Chem. 1996;271:20655–20659. doi: 10.1074/jbc.271.34.20655. [DOI] [PubMed] [Google Scholar]
- 17.Ekberg M, Pötsch S, Sandin E, Thunnissen M, Nordlund P, Sahlin M, Sjöberg B-M. Preserved catalytic activity in an engineered ribonucleotide reductase R2 protein with a nonphysiological radical transfer pathway. The importance of hydrogen bond connections between the participating residues. J. Biol. Chem. 1998;273:21003–21008. doi: 10.1074/jbc.273.33.21003. [DOI] [PubMed] [Google Scholar]
- 18.Rova U, Goodtzova K, Ingemarson R, Behravan G, Gräslund A, Thelander L. Evidence by site-directed mutagenesis supports long-range electron transfer in mouse ribonucleotide reductase. Biochemistry. 1995;34:4267–4275. doi: 10.1021/bi00013a016. [DOI] [PubMed] [Google Scholar]
- 19.Rova U, Adrait A, Pötsch S, Gräslund A, Thelander L. Evidence by mutagenesis that Tyr370 of the mouse ribonucleotide reductase R2 protein is the connecting link in the intersubunit radical transfer pathway. J. Biol. Chem. 1999;274:23746–23751. doi: 10.1074/jbc.274.34.23746. [DOI] [PubMed] [Google Scholar]
- 20.Seyedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenylalanine in the β2 subunit of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 21.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. pH rate profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli ribonucleotide reductase: Evidence that Y356 is a redox-active amino acid along the radical propagation pathway. J. Am. Chem. Soc. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 22.Seyedsayamdost MR, Stubbe J. Forward and reverse electron transfer with the Y356DOPA-β2 heterodimer of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2007;129:2226–2227. doi: 10.1021/ja0685607. [DOI] [PubMed] [Google Scholar]
- 23.Seyedsayamdost MR, Xie J, Chan CTY, Schultz PG, Stubbe J. Site-specific insertion of 3-aminotyrosine into subunit α2 of E. coli ribonucleotide reductase: direct evidence for involvement of Y730 and Y731 in radical propagation. J. Am. Chem. Soc. 2007;129:15060–15071. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 24.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Philos. Trans. Royal. Soc., B. 2006;361:1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge J, Yu G, Ator MA, Stubbe J. Pre-steady-state and steady-state kinetic analysis of E.coli class I ribonucleotide reductase. Biochemistry. 2003;42:10017–10083. doi: 10.1021/bi034374r. [DOI] [PubMed] [Google Scholar]
- 26.Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 27.Scott CP, Kashlan OB, Lear JD, Cooperman BS. A quantitative model for allosteric control of purine reduction by murine ribonucleotide reductase. Biochemistry. 2001;40:1651–1661. doi: 10.1021/bi002335z. [DOI] [PubMed] [Google Scholar]
- 28.Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42:1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- 29.Radivoyevitch T, Kashlan OB, Cooperman BS. Rational polynomial representation of ribonucleotide reductase activity. BMC Biochem. 2005;6:8. doi: 10.1186/1471-2091-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Lohman GJS, Stubbe J. Enhanced subunit interactions with gemcitabine-5'-diphosphate inhibit ribonucleotide reductases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassan AQ, Wang Y, Plate L, Stubbe J. Methodology to probe subunit interactions in ribonucleotide reductases. Biochemistry. 2008;47:13046–13055. doi: 10.1021/bi8012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasrayan A, Birgander PL, Pappalardo L, Regnstrom K, Westman M, Slaby A, Gordon E, Sjoberg B-M. Enhancement by effectors and substrate nucleotides of R1–R2 interactions in Escherichia coli class Ia ribonucleotide reductase. J. Biol. Chem. 2004;279:31050–31057. doi: 10.1074/jbc.M400693200. [DOI] [PubMed] [Google Scholar]
- 33.Rofougaran R, Crona M, Vodnala M, Sjöberg B-M, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J. Biol. Chem. 2008;283:35310–35318. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- 34.Stubbe J. Adv. Enzymol. Relat. Areas Mol. Biol. Wiley; 1990. Ribonucleotide reductases; pp. 349–419. [DOI] [PubMed] [Google Scholar]
- 35.Donehower RC. An overview of the clinical experience with hydroxyurea. Semin. Oncol. 1992;19:11–19. [PubMed] [Google Scholar]
- 36.Karlsson M, Sahlin M, Sjöberg B-M. Escherichia coli ribonucleotide reductase. Radical susceptibility to hydroxyurea is dependent on the regulatory state of the enzyme. J. Biol. Chem. 1992;267:12622–12626. [PubMed] [Google Scholar]
- 37.Lassmann G, Thelander L, Gräslund A. EPR stopped-flow studies of the reaction of the tyrosyl radical of protein R2 from ribonucleotide reductase with hydroxyurea. Biochem. Biophys. Res. Comm. 1992;188:879–887. doi: 10.1016/0006-291x(92)91138-g. [DOI] [PubMed] [Google Scholar]
- 38.Gerez C, Fontecave M. Reduction of the small subunit of Escherichia coli ribonucleotide reductase by hydrazines and hydroxylamines. Biochemistry. 1992;31:780–786. doi: 10.1021/bi00118a020. [DOI] [PubMed] [Google Scholar]
- 39.Sneeden JL, Loeb LA. Mutations in the R2 subunit of ribonucleotide reductase that confer resistance to hydroxyurea. J. Biol. Chem. 2004;279:40723–40728. doi: 10.1074/jbc.M402699200. [DOI] [PubMed] [Google Scholar]
- 40.Davydov A, Öhrström M, Liu A, Thelander L, Gräslund A. Chemical reduction of the diferric/radical center in protein R2 from mouse ribonucleotide reductase is independent of the proposed radical transfer pathway. Inorg. Chim. Acta. 2002;331:65–72. [Google Scholar]
- 41.Jiang W, Yun D, Saleh L, Barr EW, Xing G, Hoffart LM, Maslak M-A, Krebs C, Bollinger JM., Jr A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316:1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- 42.Jiang W, Bollinger JM, Jr, Krebs C. The active form of Chlamydia trachomatis ribonucleotide reductase R2 protein contains a heterodinuclear Mn(IV)/Fe(III) cluster with S = 1 ground state. J. Am. Chem. Soc. 2007;129:7504–7505. doi: 10.1021/ja072528a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang W, Yun D, Saleh L, Bollinger JM, Jr, Krebs C. Formation and function of the manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Biochemistry. 2008;47:13736–13744. doi: 10.1021/bi8017625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bollinger JM, Jr, Jiang W, Green MT, Krebs C. The manganese(IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase: structure, assembly, radical initiation, and evolution. Curr. Opin. Struct. Biol. 2008;18:650–657. doi: 10.1016/j.sbi.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Jiang W, Hoffart LM, Krebs C, Bollinger JM., Jr A manganese(IV)/iron(IV) intermediate in assembly of the manganese(IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase. Biochemistry. 2007;46:8709–8716. doi: 10.1021/bi700906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang W, Saleh L, Barr EW, Xie J, Gardner MM, Krebs C, Bollinger JM., Jr Branched activation- and catalysis-specific pathways for electron relay to the manganese/iron cofactor in ribonucleotide reductase from Chlamydia trachomatis. Biochemistry. 2008:8477–8484. doi: 10.1021/bi800881m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roshick C, Iliffe-Lee ER, McClarty G. Cloning and characterization of ribonucleotide reductase from Chlamydia trachomatis. J. Biol. Chem. 2000;275:38111–38119. doi: 10.1074/jbc.M006367200. [DOI] [PubMed] [Google Scholar]
- Voevodskaya N, Lendzian F, Ehrenberg A, Gräslund A. High catalytic activity achieved with a mixed manganese-iron site in protein R2 of Chlamydia ribonucleotide reductase. FEBS Lett. 2007;581:3351–3355. doi: 10.1016/j.febslet.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 49.Jiang W, Xie J, Nørgaard H, Bollinger JM, Jr, Krebs C. Rapid and quantitative activation of Chlamydia trachomatis ribonucleotide reductase by hydrogen peroxide. Biochemistry. 2008;47:4477–4483. doi: 10.1021/bi702085z. [DOI] [PubMed] [Google Scholar]
- 50.Uppsten M, Färnegårdh M, Domkin V, Uhlin U. The first holocomplex structure of ribonucleotide reductase gives new insight into its mechanism of action. J. Mol. Biol. 2006;359:365–377. doi: 10.1016/j.jmb.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 51.Seyedsayamdost MR, Chan CTY, Mugnaini V, Stubbe J, Bennati M. PELDOR spectroscopy with DOPA-β2 and NH2Y-β2s: Distance measurements between residues involved in the radical propagation pathway of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2007;129:15748–15749. doi: 10.1021/ja076459b. [DOI] [PubMed] [Google Scholar]
- 52.Krebs C, Price JC, Baldwin J, Saleh L, Green MT, Bollinger JM., Jr Rapid freeze-quench 57Fe Mössbauer spectroscopy: Monitoring changes of an iron-containing active site during a biochemical reaction. Inorg. Chem. 2005;44:742–757. doi: 10.1021/ic048523l. [DOI] [PubMed] [Google Scholar]
- 53.King SB. The nitric oxide producing reactions of hydroxyurea. Curr. Med. Chem. 2003;10:437–452. doi: 10.2174/0929867033368213. [DOI] [PubMed] [Google Scholar]
- 54.More KM, Eaton GR, Eaton S. Metal-nitroxyl interactions. 52. EPR spectra of nitroxyl radicals coordinated to manganese(III) tetraphenylporphyrin via the nitroxyl oxygen. Inorg. Chem. 1987;26:2618–2620. [Google Scholar]
- 55.Dikalov SI, Vitek MP, Maples KR, Mason RP. Amyloid β peptides do not form peptide-derived free radicals spontaneously, but can enhance metal-catalyzed oxidation of hydroxylamines to nitroxides. J. Biol. Chem. 1999;274:9392–9399. doi: 10.1074/jbc.274.14.9392. [DOI] [PubMed] [Google Scholar]
- 56.Högbom M, Stenmark P, Voevodskaya N, McClarty G, Gräslund A, Nordlund P. The radical site in Chlamydial ribonucleotide reductase defines a new R2 subclass. Science. 2004;305:245–248. doi: 10.1126/science.1098419. [DOI] [PubMed] [Google Scholar]
- 57.Younker JM, Krest CM, Jiang W, Krebs C, Bollinger JM, Jr, Green MT. Structural analysis of the Mn(IV)/Fe(III) cofactor of Chlamydia trachomatis ribonucleotide reductase by extended X-ray absorption fine structure spectroscopy and density functional theory calculations. J. Am. Chem. Soc. 2008;130:15022–15027. doi: 10.1021/ja804365e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roos K, Siegbahn PEM. Density functional theory study of the manganese-containing ribonucleotide reductase from Chlamydia trachomatis: Why manganese is needed in the active complex. Biochemistry. 2009;48:1878–1887. doi: 10.1021/bi801695d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.