Abstract
The marine-derived macrolides latrunculins A (1) and B, from the Red Sea sponge Negombata magnifica, have been found to reversibly bind actin monomers, forming a 1:1 complex with G-actin and disrupting its polymerization. The microfilament protein actin is responsible for several essential functions within the cell such as cytokinesis and cell migration. One of the main binding pharmacophores of 1 to G-actin was identified as the C-17 lactol hydroxyl moiety that binds arginine 210 NH. Latrunculin A-17-O-carbamates 2–6 were prepared by reaction with the corresponding isocyanates. Latrunculin A (1) and carbamates 4–6 displayed potent anti-invasive activity against the human highly metastatic human prostate cancer PC-3M cells in a Matrigel™ assay at a concentration range of 50 nM-1 µM. Latrunculin A (1, 500 nM) decreased the disaggregation and cell migration of PC-3M-CT+ spheroids by three-fold. Carbamates 4 and 5 were two and half and five-fold more active than 1, respectively, in this assay with less actin binding affinity. Latrunculin A (1, IC50 6.7 µM) and its 17-O-[N-(benzyl)carbamate (6, IC50 29 µM) suppress hypoxia-induced HIF-1 activation in T47D breast tumor cells.
Latrunculin A (1) and B are macrolides reported by Kashman and coworkers from the Red Sea sponge Negombata magnifica Kelly-Borges and Vacelet (Podospongiidae).1 Latrunculins are reported to decrease intraocular pressure and increase outflow facility without corneal effects in monkeys.2,3 Latrunculin B and analogs showed antiangiogenic, antimetastatic, and antimicrobial activities.4 The most important biological effects of latrunculins are their abilities to disrupt microfilament organization and inhibit microfilament-mediated processes without affecting the organization of the microtubular system.5 The latrunculins bind reversibly to the cytoskeleton actin monomers, forming 1:1 complexes with G-actin and disrupting polymerization.5 Actin-active agents are attracting more attention in the field of cancer chemotherapy because microfilament and microtubule proteins form versatile dynamic polymers that can define cell polarity, organize cytoplasmic organelles, control cell shape and promote stable cell-cell and cell-matrix adhesions, and generate protrusive forces required for migration.6–8 These functions usually fail and become abnormal in cancer cells. 6–8
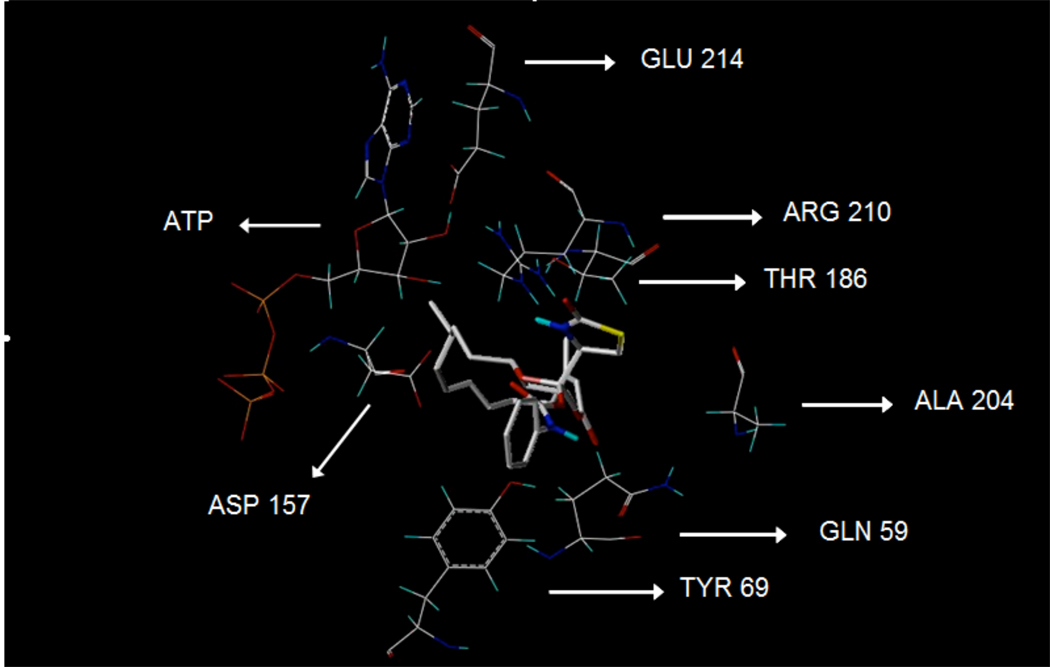
Based on X-ray crystallography, the binding site of 1 has been located between subdomains II and IV, in the vicinity of the ATP binding cleft of the protein target in the actin monomer.9,10 The binding pharmacophores of 1 to G-actin were identified as follows: C-1 carbonyl oxygen through water to glutamate 214 carboxy, C-17 lactol hydroxyl to arginine 210 NH (major binding), C-17 pyran oxygen to tyrosine 69 hydroxy, thiazolidinone NH to aspartate 157 carboxy, and thiazolidinone C-20 carbonyl oxygen to threonine 186 hydroxy.9,10 Only the thiazolidinone NH group acts as a hydrogen bonding donor while the rest of the binding functions act as hydrogen bonding acceptors.9,10
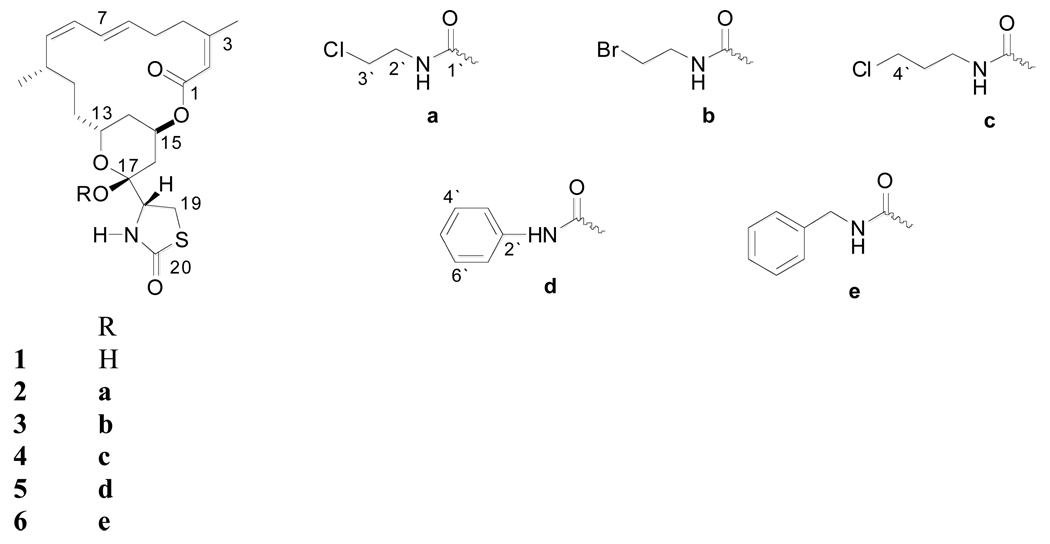
Semi-synthetic carbamoylation products of the C-17 lactol group in 1 were produced to study the pharmacological effects of the addition of hydrogen bond donors and acceptors at this key position. This study reports the anti-invasive and HIF-1 inhibitory activities of latrunculin A (1) and its semi-synthetic analogs (2–6).
Results and Discussion
Reflux of latrunculin A (1) in toluene with chloroethyl, bromoethyl, chloropropyl, benzyl, and phenyl isocyanates in the presence of catalytic amounts of triethylamine afforded the C-17-O-carbamates, 2–6, respectively.
The HRMS analysis of compound 2 revealed the molecular formula, C25H35ClN2O6S, with [M]+ and [M+2]+, 3:1, isotopic clusters characteristic for a monochlorinated compound. The downfield shift of the C-17 carbon signal in 2 (+1.1 ppm) compared with that of the starting material 1 suggested possible carbamoylation at this position.11 The 1H and 13C NMR data further supported this fact and were closely comparable to those of 1 except in the signals of an additional C-17-O-[N-(2-chloroethyl)carbamoyl] moiety.11 The carbonyl carbon at δC 152.5 was assigned to C-1´. This was based on its 3J-HMBC correlation with the methylene multiplet H2-2´ (δH 3.61), which, in turn, showed 1H-1H COSY coupling with the H2-3´ multiplet (δH 3.63).
Interpretation of the HRMS data of 3 suggested the molecular formula, C25H35BrN2O6S, with characteristic isotopic clusters for a monobrominated compound. The 1H and 13C NMR data were similar to those of 2 with the replacement of chlorine at C-3´ by bromine. The methylene multiplet H2-3´ (δH 3.69), which correlated with the methylene carbon at δC 42.1, was assigned based on its 1H-1H COSY coupling with the H2-2´ multiplet (δH 3.47, δC 48.0).
Analysis of the HRMS and 1H and 13C NMR data indicated 4 to be nearly identical in structure to 2, except for the presence of an additional methylene carbon in the carbamate side chain. The broad methylene multiplet H2-2´ (δH 3.47, δC 37.7) showed 1H-1H COSY coupling with the H2-3´ multiplet (δH 1.42, δC 31.8). The latter protons showed a 1H-1H COSY coupling with the H2-4´ triplet (δH 3.59, δC 42.3).
Interpretation of the HRMS and 1H and 13C NMR data of 5 indicated the presence of a 17-O-[N-(phenyl)carbamoyl] side chain. The broad doublet H2-3´/7´ (δH 7.47, δC 121.5) showed COSY coupling with the H2-4´/6´ double doublet (δH 7.33, δC 129.1). The latter protons showed coupling in the COSY spectrum with the H-5´ multiplet (δH 7.15, δC 124.5).
Analysis of the HRMS and 1H and 13C NMR data indicated that the structure of 6 is closely similar to 5, except that the phenyl group in 5 was replaced by a benzyl group in the carbamate side chain. The methylene doublet H2-2´ (δH 4.39) showed a 3J HMBC correlation with the carbonyl carbon at δC 153.5 (C-1´), confirming the 17-O-[N-(benzyl)carbamoyl] side chain.
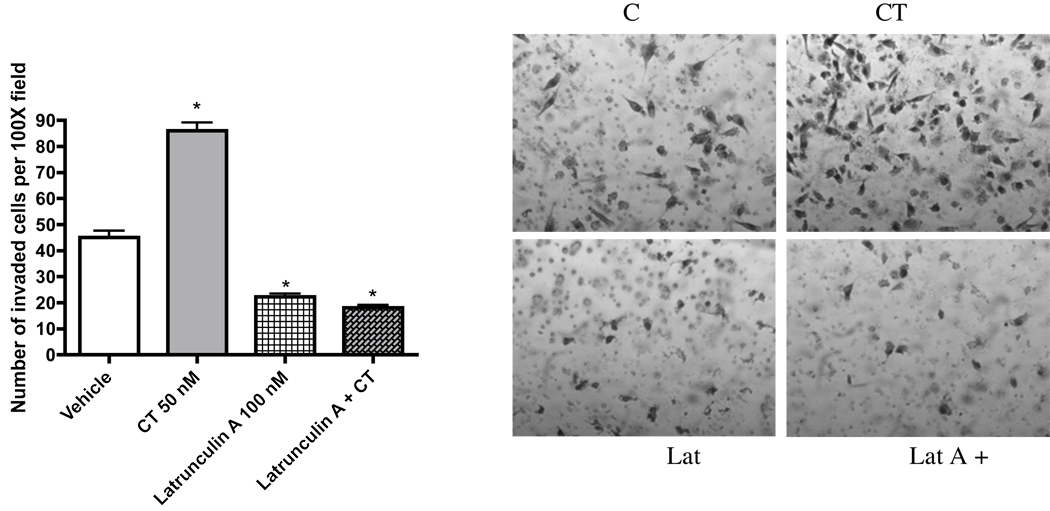
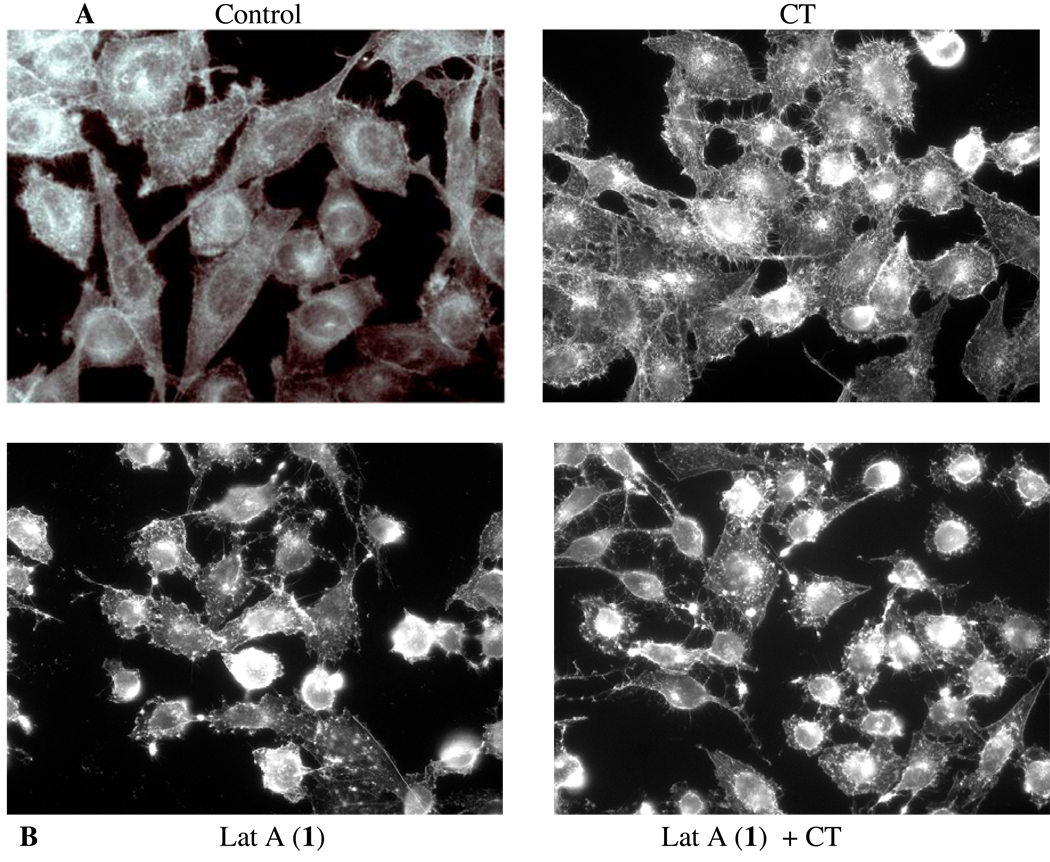
Metastasis is the predominant cause of cancer mortality. The potential antimetastatic effects of compounds 1–6 against a human highly metastatic human prostate cancer PC-3M-CT+ cell line overexpressing calcitonin (CT) were evaluated using two in vitro models: linear invasion of cells through the Matrigel™ barrier; and spheroid disaggregation.13a–d Latrunculin A (1) (100 nM) remarkably inhibited baseline and CT-stimulated invasion in the linear Matrigel™ assay (Figure 1). However, 1 was cytotoxic at doses higher than 500 nM (data not shown). There is evidence to suggest that cell migration and invasion involve multiple processes regulated by various signaling molecules.14a–d The actin cytoskeleton and its regulatory proteins are crucial for cell migration in most cells.15a,b During cell migration, the actin cytoskeleton is dynamically remodeled, and this reorganization produces the force necessary for cell migration.15a,b Since latrunculin A (1) attenuated invasiveness of PC-3M cells, its effect was examined on the actin cytoskeleton of PC-3M cells. PC-3M cells display an invasive phenotype and they usually attach to the surface, exhibiting stretched actin fibers with long exon-type extensions (Figure 2A). Upon stimulation with 50 nM CT, the cells displayed rapid reorganization of the actin cytoskeleton, leading to visible changes in cell morphology within 10 minutes. The CT-stimulated cells demonstrated long actin-rich extensions resembling filopodia or microspikes or web-like actin-containing fibers linking these filopodium-like structures. However, the addition of latrunculin A (1) resulted in rapid changes in cell morphology and actin distribution as characterized by the remarkable shrinkage of actin stress fibers, possibly caused by the depolymerization disruption of actin (Figure 2B). Compound 1 also abolished CT-induced changes in the actin cytoskeleton. These results are consistent with the actions of latrunculin on PC-3M cell invasion (Figure 1), and suggest that anti-invasive actions of 1 are mediated through the disruption of actin cytoskeleton remodeling.
Figure 1.
Anti-invasive activity of latrunculin A against PC-3M-CT+ cells in a Matrigel™ assay.
Figure 2.
Figure 2A. PC-3M cells displaying stretched actin fibers with long exon-type extensions (Control). CT (50 nM)-stimulated cells exhibit long actin-rich extensions resembling filopodia or web-like actin-containing fibers linking these filopodium-like structures (CT).
Figure 2B. Addition of latrunculin A (50 nM) resulted in rapid changes in cell morphology and actin distribution as characterized by the remarkable shrinkage of actin stress fibers (Lat A and Lat A+CT).
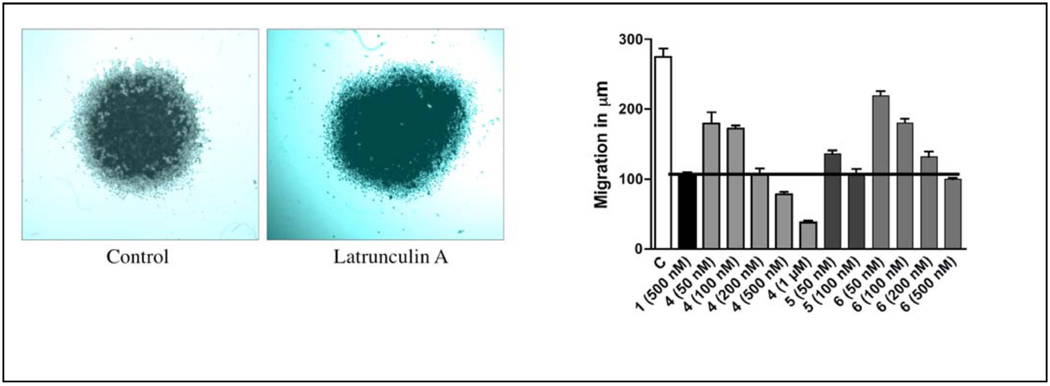
Spheroid disaggregation provides the measure of cell disaggregation as well as cell migration, and this model simulates the process of tissue disaggregation and invasion of cells in vivo.13b,15c This assay is based on disaggregation of cancer cell spheroids, and radial migration of released cells on extracellular matrix (ECM).13a–c,14d Current evidence has shown that tumor cells from primary tumors in vivo are generally released in clumps, which attach to a favorable ECM and are later released gradually to migrate in all directions.13a–c,14d Therefore, the spheroid disaggregation model is closer to in situ tumor metastasis than the the linear Matrigel™ invasion assays. The results shown in Figure 3A demonstrate that latrunculin A (1, 500 nM) decreased disaggregation and cell migration of PC-3M-CT+ spheroids three-fold. Figure 3B shows the antimetastatic actions of latrunculin A (1) with analogs 4–6 at multiple doses. Analogs 4–6 also attenuated PC-3M-CT+ spheroid disaggregation/cell migration, and this response was dose-dependent in all three analogs tested. Based on the results shown in Figure 3B, analog 5 was the most potent. Only a dose of 100 nM of analog 5 was required to generate inhibition of PC-3M-CT+ spheroid disaggregation/cell migration to the extent of that produced by 500 nM of 1. Compound 4 required 200 nM to produce the equivalent effect. Analog 6 was equipotent to latrunculin A. Halogenated ethylcarbamates 2 and 3 were inactive, suggesting the significance of conjugation and extension of the carbamate side chain. The phenylcarbamate 5 was two-fold more potent than benzylcarbamate 6, suggesting the importance of optimum distance between the aromatic ring and the carbamate functionality for activity.
Figure 3.
Figure 3A. Spheroid disaggregation of PC-3M-CT+ cells using 500 nM latrunculin A (1).
Figure 3B. Spheroid disaggregation of PC-3M-CT+ cells comparison of different doses of carbamates 4–6 with the activity of latrunculin A.
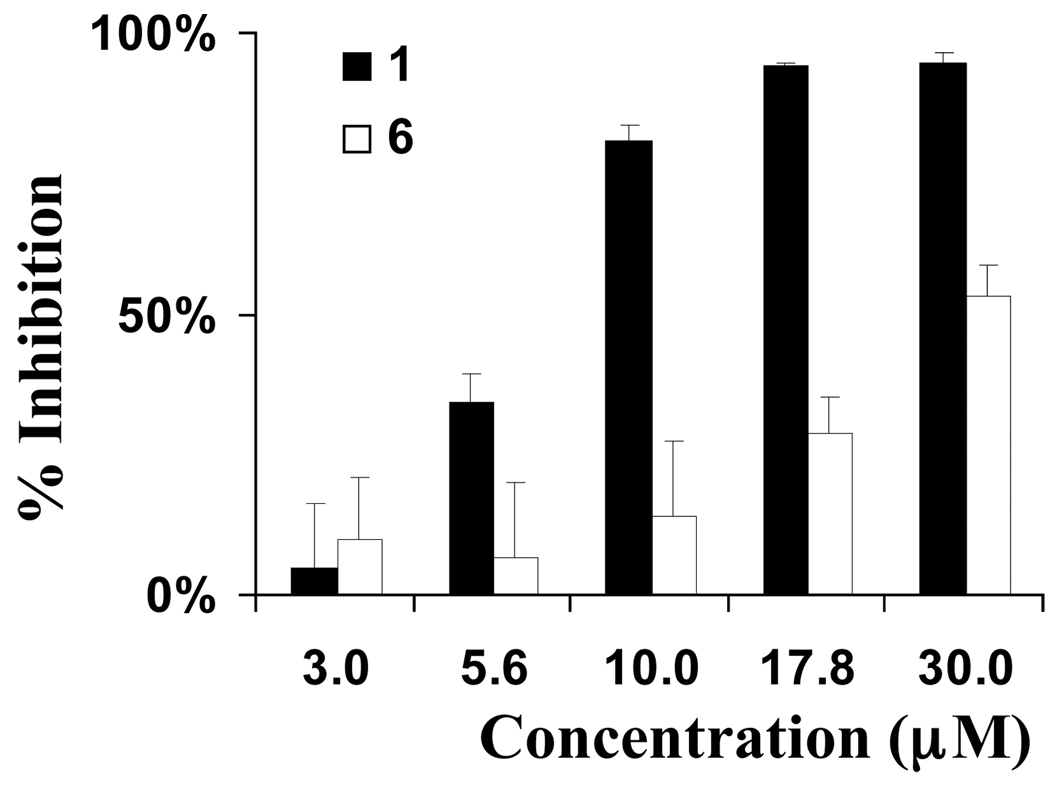
Relatively uncontrolled proliferation of cells within a tumor outstrip the capacity of existing vasculature to supply oxygen. This results in the formation of hypoxic regions within tumors. The occurrence of tumor hypoxia is associated with a poor prognosis in cancer patients.16a No drugs have been approved clinically to specifically target hypoxic tumor cells. The transcription factor hypoxia-inducible factor-1 (HIF-1) regulates hypoxia-induced gene expression in tumor cells that are subjected to hypoxic conditions.16b Disruption of HIF-1-mediated hypoxic adaptation/survival reduces tumor growth in animal models.16c–i HIF-1 inhibitors represent potential anticancer drug leads that may selectively target hypoxic tumor masses.16j–k The effects of 1–6 on hypoxia (1% O2)-induced HIF-1 activation were examined using a T47D human breast carcinoma cell-based luciferase reporter assay,17 and compared to those observed on 1,10-phenanthroline-induced HIF-1 activation. Compound 1 inhibited hypoxia-induced activation (IC50 6.7 µM, Figure 4) and 1,10-phenanthroline induced HIF-1 activation (IC50 25 µM, data not shown). Compound 6 weakly inhibited hypoxia-induced HIF-1 activation (IC50 29 µM). None of the other compounds (2–5) exerted greater than 50% inhibition of hypoxia-induced HIF-1 activation at the highest concentration tested (30 µM). The effects of 1–6 on the expression of luciferase from a control construct were examined in a T47D cell-based reporter assay and hypoxic cell viability/proliferation in a sulforhodamine B-based cell viability assay. None of the latrunculin analogs significantly inhibited the activity of a constitutively expressed pGL3 control reporter or significantly suppressed T47D cell viability under experimental conditions (less than 20% inhibition at the concentration of 30 µM). Therefore, latrunculin-based actin depolymerization disruptors appear to have the ability to selectively inhibit hypoxia-induced HIF-1 activity in tumor cells at concentrations below those evident for the inhibition of other critical cellular processes. The role of actin in HIF-1 signaling has never been documented. These results represent the first observation of the ability of actin inhibitors to inhibit HIF-1 activation at concentrations that also inhibit actin polymerization and these findings may indicate that actin polymerization plays a vital role in HIF-1 signaling.
Figure 4.
Effects of latrunculin A and 6 on hypoxia-induced HIF-1 activation under hypoxic conditions in T47D breast carcinoma cells.
To further correlate the anti-invasive activity with the actin binding affinity, docking of 1–6 toward rabbit muscle δ-actin was implemented using the SYBYL 7.3.4 and SurFlex-Dock Programs. Docking results were expressed in three functions: total score, crash, and polar. The total score was expressed in -log(Kd) units to represent binding affinities. Crash is the degree of inappropriate penetration by the ligand into the protein and of interpenetration between ligand atoms that are separated by rotatable bonds. Crash scores close to 0 are favorable. Negative numbers indicate penetration. The Polar score is the contribution of the polar non-hydrogen bonding interactions to the total score, may be useful for excluding docking results that involve no hydrogen bonds. The docking procedure was validated using the same conditions to dock latrunculin A (1) into the binding pocket of α-actin. The docking simulation resulted in a very close model to the crystallographic structure.9,10 The docking data are summarized in Table 1.
Table 1.
1H NMR Data of Compounds 2–6.a
| position | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| δH | δH | δH | δH | δH | |
| 1 | 1.88, m | 1.76, m | 1.61, m | 1.70, m | 1.70, m |
| 2 | 5.75, dd (9.5, 15.4) | 5.52, dd (9.1, 15.0) | 5.57, dd (8.4, 15.4) | 5.52, dd (9.1, 15.4) | 5.52, dd (9.1, 15.4) |
| 3 | 5.64, d (15.4) | 5.35, d (15.0) | 5.44, d (15.4) | 5.41, d (15.4) | 5.42, d (15.4) |
| 5 | 2.07, dd (3.6, 16.8) | 2.12, dd (5.4, 14.6) | 2.72, dd (5.4, 14.6) | 2.20, dd (5.8, 14.3) | 2.22, dd (5.8, 14.3) |
| 1.97, dd (5.4, 16.8) | 1.94, dd (2.9, 14.6) | 2.42, dd (2.9, 14.6) | 1.80, dd (2.5, 14.3) | 1.80, dd (2.5, 14.3) | |
| 6 | 5.56, m | 5.66, m | - | 4.60, m | 4.60, m |
| 7 | 5.55, d (15.0) | 5.35, d (15.0) | 6.03, s | 5.57, br d (9.5) | 5.56, br d (9.8) |
| 9 | 2.30, 2H, m | 2.25, m; 1.82, m; | 3.65, m; 2.02, m | 2.00, 2H, m | 1.85,2H, m |
| 10 | 1.58, 2H, m | 1.45, 2H, m | 2.22, m; 2.02, m | 2.33, m 1.95, m | 2.24, m 1.82, m |
| 11 | 3.43, d (10.2) | 4.44, dd (3.6, 11) | 4.96, dd (6.2, 6.2) | 4.46, dd (6.2, 10.2) | 4.31, dd (6.2, 9.8) |
| 13 | 1.71, m 1.39, m | 1.35, 2H, m | 2.10, m 1.90, m | 2.29, m 1.90, m | 2.15, m 1.75, m |
| 14 | 1.85, m 1.35, m | 1.73,2H, m | 1.49, 2H, m | 1.65, m 1.45, m | 1.65, m 1.35, m |
| 15 | 1.40, m | 1.65, m | 1.51, m | 1.59, m | 1.60, m |
| 16 | 0.79, 3H, d (6.6) | 0.85, 3H, d (6.6) | 0.79, 3H, d (6.6) | 0.83, 3H, d (6.6) | 0.85, 3H, d (6.6) |
| 17 | 0.90, 3H, d (6.6) | 0.88, 3H, d (6.6) | 0.82, 3H, d (6.6) | 0.86, 3H, d (6.6) | 0.88, 3H, d (6.6) |
| 18 | 1.31, 3H, s | 1.31, 3H, s | 1.27, 3H, s | 1.28, 3H, s | 1.26, 3H, s |
| 19 | 1.65, 3H, s | 1.72, 3H, s | 1.84, 3H, d (1.4) | 1.63, 3H, s | 1.62, 3H, s |
| 20 | 1.51, 3H, s | 1.37, 3H, s | 1.47, 3H, s | 4.99, s; 5.18, s | 5.00, s; 5.15, s |
| Ac-6 | 2.02, 3H, s | 2.00, 3H, s | |||
| NH | 7.20, s | ||||
In CDCl3, 400 MHz. Coupling constants (J) are in Hz.
The parent compound, latrunculin A (1), as well as the phenylcarbamate (5), showed the highest scores and molecular interaction, consistent with the observed anti-invasive activity (Table 1, Figure 5). The carbonyl oxygen of the carbamoyl moiety in 5 makes a strong H-bonding interaction with the guanidine amino group of arginine (ARG) 210 of actin (Figure 5). A similar interaction has been observed for carbamates 3 and 6. Therefore, ARG 210 seems to play a central role in the binding of latrunculin A (1) and analogs within the actin monomers.9,10 Although structure 6 showed anti-invasive activity, the scoring was the lowest among latrunculin A derivatives. Such deviation could be attributed to the activity of structure 6 on protein target(s) other than actin.
Figure 5.
Detailed view of docked structure 5 with the corresponding rabbit muscle α-actin interacting amino acids.
Experimental Section
General Experimental Procedures
Measurements of optical rotation were carried out on a Rudolph Research Analytical Autopol III polarimeter. IR spectra were recorded on a Varian 800 FT-IR spectrophotometer. The 1H and 13C NMR spectra were recorded in CDCl3, using TMS as an internal standard, on a JEOL Eclipse NMR spectrometer operating at 400 MHz for 1H and 100 MHz for 13C. The HREIMS experiments were conducted at the University of Michigan on a Micromass LCT spectrometer. TLC analysis was carried out on precoated silica gel 60 F254 500 µm TLC plates, using the developing systems n-hexane/EtOAc (1:1) or CHCl3-MeOH (9:1). For column chromatography, silica gel 60 (particle size 63–200 µm) or Bakerbond octadecyl (C18), 40 µm were used. For Sephadex LH-20 column chromatography, n-hexane-CHCl3 (1:3), CHCl3, and CHCl3-MeOH (9:1) systems were used. For column chromatography, silica gel 70–230 mesh was used.
Biological Material
The sponge Negombata magnifica Kelly-Borges and Vacelet (order Poecilosclerida, sub-order Mycalina, family Podospongiidae) was collected as red long finger-like strips by SCUBA from the sand-covered bottom at −10 to −15 m at Hurghada, on the Egyptian Red Sea coast, in June 2003.18,19 The sponge was identified by Mr. Tamer Helmy, Suez Canal University. A voucher specimen (03RS3) was deposited in the Department of Basic Pharmaceutical Sciences, College of Pharmacy, University of Louisiana at Monroe, Louisiana.
Extraction and Isolation
The frozen sponge (6 kg) was coarsely minced and extracted with CHCl3 (5 × 1000 mL) at room temperature. The CHCl3 extract was then concentrated under vacuum and subjected to liquid chromatography, as described elsewhere, to afford 1, 1.2 g (0.0002%).1,4,11
Preparation of Carbamates 2–6
To solutions of 50 mg of 1 in toluene (2 mL), 59 µL of 2-chloroethyl isocyanate, 60 µL of 2-bromoethyl isocyanate, 60 µL of 3-chloropropyl isocyanate, 40 µL of phenyl isocyanate, or 36 µL of benzyl isocyanate were added, respectively, and separately mixed with 10 µL of Et3N. Each solution was separately stirred at room temperature for 1 h (12 h for benzyl isocyanate only). Water (10 mL) was then added and the product of each reaction mixture was extracted with EtOAc. Each EtOAc extract was dried over anhydrous Na2SO4 and concentrated under reduced pressure. Crude products were then purified by column chromatography on silica gel 60 using EtOAc-n-hexane 1:9 to give compounds 2 (20.0 mg, 40% Rf 0.60, silica gel, CHCl3-MeOH 9.5:0.5) and 4 (13.2 mg, 26.4%, Rf 0.64),or EtOAc-n-hexane 2:8 to give compounds 3 (13.3 mg, 26.6%, Rf 0.57) and 6 (19.4 mg, 38.8%, Rf 0.55) or using CHCl3-MeOH (9.5:0.5) to afford compound 5 (13.6 mg, 27.2%, Rf 0.51).
17-O-[N-(2-Chloroethyl)carbamoyl]-latrunculin A (2)
colorless oil, (c 0.51, CHCl3); IR νmax (neat) 3564, 3331, 2953-2856, 1761, 1708, 1680, 1542, 1355, 1164 cm−1; 1H NMR (CDCl3) δH 3.61 (2H, m, H2-2´), 3.63 (2H, m, H2-3´), 8.48 (1H, brs, exchangeable, NH); 13C NMR δC 152.5 (qC, C-1´), 42.2 (CH2, C-2´), 43.2 (CH2, C-3´); HRESIMS m/z 549.1808 (calcd for C25H35ClN2O6SNa, 549.1802 [M+Na]+).
17-O-[N-(2-Bromoethyl)carbamoyl]-latrunculin A (3)
colorless oil, (c 0.55, CHCl3); IR νmax (neat) 3563, 3327, 2995-2857, 1735, 1713, 1560, 1356, 1301, 1128 cm−1; 1H NMR (CDCl3) δH 3.47 (2H, m, H2-2´), 3.69 (2H, m, H2-3´), 8.49 (1H, t, J = 5.2 Hz, exchangeable, NH); 13C NMR δ 152.2 (qC, C-1´), 48.0 (CH2, C-2´), 42.1 (CH2, C-3´); HRESIMS m/z 593.1306 (calcd for C25H35BrN2O6SNa, 593.1297 [M+H]+).
17-O-[N-(3-Chloropropyl)carbamoyl]-latrunculin A (4)
colorless oil, (c 0.47, CHCl3); IR νmax (neat) 3564, 3335, 2953-2856, 1740, 1704, 1545, 1356, 1288, 1163 cm−1; 1H NMR (CDCl3) δH 3.47 (2H, m, H2-2´), 1.42 (2H, m, H2-3´), 3.59 (2H, t, J = 6.2 Hz, H2-4´), 8.23 (1H, t, J = 5.8 Hz, exchangeable, NH); 13C NMR δC 152.4 (qC, C-1´), 37.7 (CH2, C-2´), 31.8 (CH2, C-3´), 42.3 (CH2, C-4´); HRESIMS m/z 563.1953 (calcd for C26H37ClN2O6SNa, 563.1959 [M+H]+).
17-O-[N-(Phenyl)carbamoyl]-latrunculin A (5)
colorless oil, (c 0.14, CHCl3); IR νmax (neat) 3570, 3286, 2927-2853, 1710, 1601, 1680, 1554, 1446, 1296, 1160 cm−1; 1H NMR (CDCl3) δH 7.47 (2H, brd, J = 8.1 Hz, H-3´/H-7´), 7.33 (2H, dd, J = 8.1, 8.0 Hz, H-4´/H2-6´), 7.15 (1H, m, H-5´); 13C NMR δC 149.6 (qC, C-1´), 137.1 (qC, C-2´), 121.5 (CH, C-3´/C-7´), 129.1 (CH, C-4´/C-6´), 124.5 (CH, C-5´); HRESIMS m/z 563.2200 (calcd for C26H37N2O6SNa, 563.2192 [M+Na]+).
17-O-[N-(Benzyl)carbamoyl]-latrunculin A (6)
colorless oil, (c 0.55, CHCl3); IR νmax (neat) 3564, 3334, 2953-2857, 1730, 1703, 1680, 1542, 1455, 1357, 1282, 1166 cm−1; 1H NMR (CDCl3) δ 4.39 (2H, d, J = 5.4 Hz, H2-2´), 7.27 (2H, brd, J = 8.0 Hz, H-4´/H-8´), 7.30 (2H, dd, J = 8.0, 7.8 Hz, H-5´/H2-7´), 7.32 (1H, dd, J = 8.0, 7.8 Hz, H-6´); 13C NMR δC (151.2, qC, C-1´), 41.5 (CH2, C-2´), 141.5 (qC, C-3´), 127.0 (CH, C-4´/C-8´), 129.1 (CH, C-5´/C-7´), 126.5 (CH, C-6´); HRESIMS m/z 577.2360 (calcd for C30H38N2O6SNa, 577.2348 [M+Na]+).
Invasion Assay
These experiments were conducted in 24-well, two compartment, Matrigel™ invasion chambers (Becton Dickinson, Bedford, MA).13a–13d The PC-3M-CT+ cells were grown exponentially under serum-starved conditions for 24 h [basal RPMI medium containing no serum or growth factors but containing 0.1% BSA, 10 mM HEPES, 4 mM L-glutamine, 100 IU mL−1 penicillin G, and 100 mg mL−1streptomycin]. The cells were then harvested, and seeded at a density of 25 × 103 cells per well in the upper insert of the Matrigel™ invasion chamber. The lower chamber received the chemo-attractant medium, which consisted of 90% basal RPMI medium and 10% conditioned medium from the cultures of PC-3M-CT+ cells expressing constitutively active Gas protein. The incubation was carried out for 24 h, after which the Matrigel™ (along with non-invading cells) was scraped off with cotton swabs, and outer side of the insert was fixed and stained using Diff Quick staining (Dade Behring Diagnostics, Aguada, Peurto Rico). The number of cells migrated on the outer bottom side of the insert counted under the microscope in six or more randomly selected fields (magnification: 100×). Final results are expressed as mean +/− SEM per 100× field. Each experiment was performed in triplicate, and the experiment was repeated twice.13a–13d
Growth Correction
Since some cell lines can exhibit high proliferation rates, it is likely that the cells migrating during the early part of the 24 h incubation period could proliferate during the remaining period of incubation, leading to a slight overestimation of the final results. To correct this probability, the growth rate of PC-3M cells was determined under identical culture conditions. Twenty-five thousand cells were plated at hourly intervals in six-well dishes and cultured with/without CT (50 nM) for 1–24 h. Mean percent increase in the cell number was determined at the end of the incubation period by counting the net increase in the number of cells. The relative CT-induced increase of the pooled results of all time points was found to be 1.19 (vehicle control = 1). This correction was applied to the results of invasion assays.13b,15d,e
Spheroid Disaggregation Assay
Spheroids were prepared from single cell suspension of prostate cell lines as described before.13b,13d In brief, 5 × 104/mL cells in RPMI 1640 serum-free medium were placed on 96-well low-attachment tissue culture plates. The plates were rocked on a gyrorotatory shaker in a CO2 incubator at 37 °C for 2 days, at the end of which the spheroids measuring 150–300 µm in diameter (~4 × 104 cells/spheroid) were formed. A single spheroid was then placed in the center of each well of extracellular matrix (ECM)-coated 24-well microplate in 200 mL of serum-free medium. From previous studies, it was determined that 1 h is an appropriate time for spheroids to begin adhering to an ECM. Thus t = 0 was set as 1 h from initial plating, so that if the plate was not disturbed, the spheroids would not move from their location at the time of plating. Spheroids were photographed digitally at t = 0, cultured at 37 °C for 48 h, and then re-photographed. The spheroids were then fixed, stained with Diff-Quik™ (Dade Behring, Newark, DE) and examined under light microscopy. The diameter of the area covered with cells migrated from the spheroids was measured in a microscope calibrated with a stage and ocular micrometer. The radial distance of migration was calculated after subtraction of the mean initial spheroidal diameter at t = 0. Values shown represent the average percent increase in surface area of spheroids.
Cell Proliferation/Viability Assay
Human breast carcinoma T47D cells were grown in DMEM/F12 medium with L-glutamine (Mediatech) supplemented with 10% (v/v) fetal calf serum (FCS, Hyclone), 50 units mL−1 penicillin G (Na salt), and 50 µg mL−1 streptomycin sulfate (referred to as "Pen/Strep") (Invitrogen) in a humidified atmosphere (5% CO2/95% air) at 37 °C. Exponentially grown cells were plated at a density of 30,000 cells per well into 96-well tissue culture plates (Corning) in a volume of 100 µL of DMEM/F12 medium with 10% FCS and Pen/Strep. The cells were incubated at 37 °C overnight.17,20 Test compounds were diluted in DMEM/F12 medium with Pen/Strep and added in a volume of 100 µL per well. Following a 30-min incubation, the compound treatment continued for another 16 h (or 48 h) at 37 °C under hypoxic (5% CO2/1% O2/94% N2) or normoxic (5% CO2/95% air) conditions. Cell proliferation/viability (performed in triplicate) was determined as previously described.17,20 The absorbance at 515 nm was measured on a BioTek Synergy HT microplate reader with correction wavelength at 690 nm. The data were normalized to the untreated control. The following formula was used to calculate % inhibition of cell proliferation/viability: % inhibition = 1 - OD515(treated)/OD515(control).
Cell-Based Reporter Assay for HIF-1 Activity
The transfection, compound treatment, exposure to hypoxic conditions (5% CO2/1% O2/94% N2), normoxic conditions (5% CO2/95% air), and a hypoxia mimetic (10 µM 1,10-phenanthroline), and luciferase activity determination were performed as previously described.20 Emetine was used as a positive control (IC50 0.11 µM).
Molecular Modeling
Docking and scoring modeling studies were performed using SYBYL 7.3.4 (Tripos Discovery Informatics, St. Louis, MO) installed on a Dell desktop workstation equipped with a 1.86 GHz Intel® Xeon® processor and the Red Hat Enterprise Linux (version 4) operating system. The 3D coordinates of rabbit muscle α-actin complex with latrunculin A (1) was retrieved from the Protein Data Bank (PDB code: 1esv). The selected structure is of the best 3D resolution (2.0 Å). The protein structure was utilized in subsequent docking experiments without energy minimization. Explicit water molecules were removed from the structure. Chemical structures of 1–6 were drawn in SYBYL 7.3.4 and assigned Gasteiger partial charges and energy minimized using Energy Force Field. Docking simulation of structures 1–6 were carried out using SurFlex-Dock (version 2.1). SurFlex-Dock identifies the active site of the protein and constructs a docking target (protomol) to which molecules match. Protmol was generated by the ligand based method, setting the Threshold and Bloat parameters as default value (0.5 and 0.0, respectively).
Figure 6.
Table 2.
13C NMR Data of Compounds 2–6.a
| position | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| δC | δC | δC | δC | δC | |
| 1 | 123.5, CH | 123.5, CH | 126.5, CH | 128.3, CH | 128.3, CH |
| 2 | 141.1, qC | 139.7, qC | 160.3, qC | 136.7, qC | 136.9, qC |
| 3 | 35.4, CH2 | 34.6, CH2 | 31.5, CH2 | 37.0, CH2 | 36.1, CH2 |
| 4 | 23.2, CH2 | 25.9, CH2 | 25.7, CH2 | 29.4, CH2 | 29.4, CH2 |
| 5 | 70.3, CH | 91.4, CH | 123.5, CH | 56.8, CH | 64.1, CH |
| 6 | 62.8, qC | 64.0, qC | 134.7, qC | 148.5, qC | 148.0, qC |
| 7 | 30.3, CH2 | 33.8, CH2 | 36.2, CH2 | 35.6, CH2 | 34.7, CH2 |
| 8 | 27.5, CH2 | 27.3, CH2 | 29.0, CH2 | 28.8, CH2 | 28.6, CH2 |
| 9 | 32.6, CH | 32.8, CH | 32.2, CH | 33.3, CH | 33.3, CH |
| 10 | 20.7, CH3 | 19.4, CH3 | 20.0, CH3 | 19.3, CH3 | 19.3, CH3 |
| 11 | 21.1, CH3 | 21.0, CH3 | 20.2, CH3 | 21.0, CH3 | 20.9, CH3 |
| 12 | 29.7, CH3 | 32.0, CH3 | 30.0, CH3 | 32.5, CH3 | 32.4, CH3 |
| 13 | 15.9, CH3 | 15.9, CH3 | 25.0, CH3 | 15.6, CH3 | 15.6, CH3 |
| 14 | 27.2, CH3 | 25.4, CH3 | 15.0, CH3 | 113.3, CH3 | 113.5, CH3 |
| 15 | 123.5, CH | 123.5, CH | 126.5, CH | 128.3, CH | 128.3, CH |
| 16 | 141.1, qC | 139.7, qC | 160.3, qC | 136.7, qC | 136.9, qC |
| 17 | 35.4, CH2 | 34.6, CH2 | 31.5, CH2 | 37.0, CH2 | 36.1, CH2 |
| 18 | 23.2, CH2 | 25.9, CH2 | 25.7, CH2 | 29.4, CH2 | 29.4, CH2 |
| 19 | 70.3, CH | 91.4, CH | 123.5, CH | 56.8, CH | 64.1, CH |
| 20 | 62.8, qC | 64.0, qC | 134.7, qC | 148.5, qC | 148.0, qC |
| 21 | 111.9, qC | 187.5, qC | |||
| Ac-6 | 170.3, qC | 169.3, qC | |||
| 21.4, CH3 | 21.3, CH3 |
In CDCl3, 100 MHz. Carbon multiplicities were determined by DEPT135° or APT experiments. qC = quaternary, CH = methine, CH2 = methylene, CH3 = methyl carbons.
Table 3.
Virtual Binding Affinity of Compounds 1–6 for Rabbit Muscle α-Actin Complex.a
| compound | score | crash | polar |
|---|---|---|---|
| 1 | 6.24 | −0.96 | 1.14 |
| 2 | 4.37 | −2.52 | 0.00 |
| 3 | 4.60 | −3.38 | 0.02 |
| 4 | 5.93 | −2.80 | 0.01 |
| 5 | 6.12 | −1.54 | 0.01 |
| 6 | 3.89 | −2.97 | 2.46 |
Using SYBYL 7.3.4, SurFlex-Dock 2.1.
Acknowledgment
This investigation was made possible through the support of NIH grant number P20RR16456 from the BRIN Program of the National Center for Research Resources, grant R01CA96534 (G.V.S.), and grant CA098787 (D.G.N.-Y.D.Z.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
Dedicated to Dr. G. Robert Pettit of Arizona State University for his pioneering work on bioactive natural products.
References and Notes
- 1.Kashman Y, Groweiss A, Shmueli U. Tetrahedron Lett. 1980;21:3629–3632. [Google Scholar]
- 2.Okka M, Tian B, Kaufman PL. Arch. Ophthalmol. 2004;122:1482–1488. doi: 10.1001/archopht.122.10.1482. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman PL, Geiger B. 772,412. U. S. Patent. 2002
- 4.El Sayed KA, Youssef DTA, Marchetti D. J. Nat. Prod. 2006;69:219–223. doi: 10.1021/np050372r. [DOI] [PubMed] [Google Scholar]
- 5.Spector I, Shochet NR, Blasberger D, Kashman Y. Cell Motil. Cytoskel. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 6.Newman DJ, Cragg GM. J. Nat. Prod. 2004;67:1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 7.Nakaseko Y, Yanagida M. Nature. 2001;412:291–292. doi: 10.1038/35085684. [DOI] [PubMed] [Google Scholar]
- 8.Gachet Y, Tournier S, Millar JB, Hyams JS. Nature. 2001;412:352–355. doi: 10.1038/35085604. [DOI] [PubMed] [Google Scholar]
- 9.Yarmola EG, Somasundaram T, Boring TA, Spector I, Bubb MR. J. Biol. Chem. 2000;275:28120–28127. doi: 10.1074/jbc.M004253200. [DOI] [PubMed] [Google Scholar]
- 10.Morton WM, Ayscough KR, McLaughlin PJ. Nat. Cell. Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 11.Groweiss A, Shmueli U, Kashman Y. J. Org. Chem. 1983;48:3512–3516. [Google Scholar]
- 12.Blasberger D, Carmely S, Cojocaru M, Spector I, Shochet NR, Kashman Y. Liebigs Ann. Chem. 1989;12:1171–1188. [Google Scholar]
- 13.(a) Shah GV, Noble MJ, Austenfeld M, Weigel J, Deftos LJ, Mebust WK, Winston K. Prostate. 1992;21:87–97. doi: 10.1002/pros.2990210202. [DOI] [PubMed] [Google Scholar]; (b) Thomas S, Chigurupati S, Anbalagan M, Shah G. Mol. Endocrinol. 2006;20:1894–1911. doi: 10.1210/me.2005-0284. [DOI] [PubMed] [Google Scholar]; (c) Chien J, Ren Y, Wang YQ, Bordelon W, Thompson E, Davis R, Rayford W, Shah G. Mol. Cell Endocrinol. 2001;181:69–79. doi: 10.1016/s0303-7207(01)00530-5. [DOI] [PubMed] [Google Scholar]; (d) Thomas S, Chiriva-Internati M, Shah GV. Clin. Exp. Metastasis. 2007;24:363–377. doi: 10.1007/s10585-007-9073-y. [DOI] [PubMed] [Google Scholar]
- 14.(a) Cooper CR, Chay CH, Pienta KJ. Neoplasia. 2002;4:191–194. doi: 10.1038/sj.neo.7900224. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ghosh PM, Ghosh-Choudhury N, Moyer ML, Mott GE, Thomas CA, Foster BA, Greenberg NM, Kreisberg JI. Oncogene. 1999;18:4120–4130. doi: 10.1038/sj.onc.1202792. [DOI] [PubMed] [Google Scholar]; (c) Chen Y, Wang Y, Yu H, Wang F, Xu W. Exp. Biol. Med. 2005;230:731–741. doi: 10.1177/153537020523001006. [DOI] [PubMed] [Google Scholar]; (d) Alroy I, Yarden Y. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]; (d) Hoevel T, Macek R, Swisshelm K, Kubbies M. Int. J. Cancer. 2004;108:374–383. doi: 10.1002/ijc.11571. [DOI] [PubMed] [Google Scholar]
- 15.(a) Byers HR, Etoh T, Doherty JR, Sober AJ, Mihm MC. Am. J. Pathol. 1991;139:423–435. [PMC free article] [PubMed] [Google Scholar]; (b) Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cascone I, Giraudo E, Caccavari F, Napione L, Bertotti E, Collard JG, Serini G, Bussolino F. J. Biol. Chem. 2003;278:50702–50713. doi: 10.1074/jbc.M307234200. [DOI] [PubMed] [Google Scholar]; (d) Yoshioka K, Nakamori S, Itoh K. Cancer Res. 1999;59:2004–2010. [PubMed] [Google Scholar]; (e) Gondi CS, Lakka SS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Tung CH, Weissleder R, Rao JS. Cancer Res. 2004;64:4069–4077. doi: 10.1158/0008-5472.CAN-04-1243. [DOI] [PubMed] [Google Scholar]
- 16.(a) Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KS, Chapman JD, Eckelman WC, Fyles AW, Giaccia AJ, Hill RP, Koch CJ, Krishna MC, Krohn KA, Lewis JS, Mason RP, Melillo G, Padhani AR, Powis G, Rajendran JG, Reba R, Robinson SP, Semenza GL, Swartz HM, Vaupel P, Yang D, Croft B, Hoffman J, Liu G, Stone H, Sullivan D. Int. J. Radiat. Biol. 2006;82:699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]; (b) Semenza GL. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]; (c) Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Ryan HE, Lo J, Johnson RS. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]; (f) Kung AL, Wang S, Klco JM, Kaelin WG, Livingston DM. Nat. Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]; (g) Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J, Memmert K, Naegeli HU, Petersen F, Eck MJ, Bair KW, Wood AW, Livingston DM. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]; (h) Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E, Katschinski DM, Wenger RH. Oncogene. 2003;22:3213–3220. doi: 10.1038/sj.onc.1206385. [DOI] [PubMed] [Google Scholar]; (i) Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]; (j) Semenza GL. Expert Opin. Ther. Targets. 2006;10:267–280. doi: 10.1517/14728222.10.2.267. [DOI] [PubMed] [Google Scholar]; (k) Melillo G. Cancer Metastasis Rev. 2007;16:341–352. doi: 10.1007/s10555-007-9059-x. [DOI] [PubMed] [Google Scholar]
- 17.Dai J, Liu Y, Zhou Y-D, Nagle DG. J. Nat. Prod. 2007;70:130–133. doi: 10.1021/np0604883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly-Borges M, Vacelet J. Mem. Queensl. Mus. 1995;38:477–503. [Google Scholar]
- 19.Antunes EM, Copp BR, Davies-Coleman MT, Samaai T. Nat. Prod. Rep. 2005;22:62–72. doi: 10.1039/b407299p. [DOI] [PubMed] [Google Scholar]
- 20.Hodges TW, Hossain FC, Kim Y-P, Zhou Y-D, Nagle DG. J. Nat. Prod. 2004;67:767–771. doi: 10.1021/np030514m. [DOI] [PubMed] [Google Scholar]