Abstract
Membrane glycoproteins play vital roles in many fundamental physiological and pathophysiological processes in the central nervous system and represent important targets for pharmaceuticals and biomarker discovery. However, their isolation and characterization has been greatly limited. Lectin affinity chromatography (LAC) has evolved as a powerful method to enrich glycoproteins in biofluid and cell/tissue lysate. However, its use in the hydrophobic fraction of the samples has rarely been explored. In this study, we have conducted a systematic investigation on the lectin binding efficiency in the presence of four commonly used detergents. We have found that under certain concentrations, detergents can minimize the nonspecific bindings and facilitate the elution of hydrophobic glycoproteins. With the Detergent Assisted Lectin Affinity Chromatography (DALAC), a total of 1491 proteins were identified with low numbers of false positives from two lectins. 699 proteins were identified with at least two unique peptides, of which 219 are membrane glycoproteins. Compared to the traditional methods, the DALAC approach significantly increased the recovery of plasma membrane and glycoproteins. NP-40 is recommended as a well rounded detergent for DALAC, but the conditions for enriching certain target proteins need to be empirically determined. This study represents the first global identification of the murine brain glycoproteome.
Keywords: Brain, Detergent, Glycoproteins, Lectin Affinity Chromatography, Membrane Proteins, Mass Spectrometry
INTRODUCTION
It is well recognized that the cell membrane and its constituents play crucial roles in many fundamental physiological and pathophysiological processes within the dynamic protein networks. It is therefore not surprising that membrane proteins and their ligands represent important targets for more than two thirds of contemporary drugs and promising sources for potential disease biomarkers1. The proteins on mammalian cell surfaces are generally decorated with a dense layer of carbohydrates. After the oligosaccharides are assembled and modified in the endoplasmic reticulum (ER) and Golgi apparatus, the glycoconjugates are then transported to the plasma membrane, secretory granules, or lysosomes2. The membrane glycoproteins are responsible for many important biological functions, including differentiation, embryogenesis, inflammatory response, and cancer progression3.
Recent studies have suggested that glycans are pivotal in the regulation of development and functions in the central nervous system4. For example, fucose α(1–2) galactose carbohydrates have been implicated in modulating neuronal outgrowth and morphology5. Glycosylation modulates cell signaling processes and has been suggested to be involved in the memory consolidation pathways6. Recent research has shed light on the impact of glycosylation on the biosynthesis and structure of prion protein (PrP), whose change of conformation is responsible for the onset and development of prion diseases7. The congenital disorders of glycosylation, also known as CDG syndrome, are a type of rare inborn multisystemic diseases that often cause major nervous system impairment, resulting in mental retardation, underdeveloped cerebellum along with other severe developmental abnormalities. It has been found that the major type of CDG syndrome is caused by a defect in the gene encoding phosphomannomutase, an enzyme responsible for the biosynthesis of the oligosaccharide precursor necessary for N-glycan biosynthesis, highlighting the significance of glycosylation on the health of the central nervous system8.
Access to these structurally defined glycoconjugates is a prerequisite for revealing their functions. Despite the importance of membrane glycoproteins in the central nervous system, their isolation and characterization has been greatly limited for several reasons. Most integral membrane proteins are amphipathic and are located in the lipid bilayer. Due to the inefficient solublization and protease digestion, the membrane proteome is generally underrepresented in conventional shotgun proteomics strategy. Glycosylated membrane proteins are of low abundance and are usually heterogeneous because of the presence of multiple glycosylation sites, wide variety of glycan structures and less than 100% stoichiometry of modification. Glycoproteomics studies have been greatly facilitated with the use of lectin affinity chromatography (LAC), in which glycoproteins are enriched by binding to immobilized lectins on a solid phase via lectin-glycan interactions. However, LAC has previously been conducted mainly in blood plasma, cerebrospinal fluid (CSF) and tissue or culture extracts to enrich the soluble glycoproteins9–13. Its use in the hydrophobic fraction of samples has rarely been explored, mainly because of the dilemma whether to use detergents in the LAC of membrane glycoproteins. On one hand, it is a common practice to solublize the membrane proteins by suitable detergents after the membrane protein enriched pellets are obtained by subcellular fractionation. On the other hand, since lectins in many cases are composed of subunits that are held together by noncovalent interactions, detergents and chaotropic agents may lower the binding efficiency of lectins by causing conformational changes and subunits denaturation. By conducting a series of physicochemical measurements, Ahmad et al. has shown that the lectin concanavalin A (ConA) undergoes structural transition from β-sheet to α-helix in the presence of detergents sodium dodecyl sulphate (SDS), an anionic detergent, and cetyl trimethyl ammonium bromide (CTAB), a cationic detergent; but retains its conformation with low concentrations of 3-[(3-cholamidopropyl) dimethyl-ammonio]-1-propanesulphonate (CHAPS), a zwitterionic detergent, and Brij-35, a nonionic detergent14. Lotan et al. have studied the effect of detergents on the binding capacity of immobilized lectins with standard glycoproteins, and have shown that the adverse effect on binding are detergent- and lectin-dependent15. Both studies have concluded that nonionic and zwitterionic detergents are more suitable for LAC than ionic detergents. In this study, we perform systematic investigation on the lectin binding and elution efficiency in the presence of a few commonly used detergents. We then explore and evaluate their utility for detergent assisted LAC to enrich and characterize mouse brain membrane glycoproteins.
RESULTS
Binding Study
In order to determine the optimal conditions for LAC in the presence of detergents, we first measured the binding efficiencies of two lectins, namely, ConA and wheat germ agglutinin (WGA) affected by four nonionic and zwitterionic detergents commonly used in membrane protein solublization. ConA and WGA are widely used in glycoproteomics studies and are well known for their broad selectivity towards glycan structures, and can serve as a starting point for the application of LAC in membrane protein samples. Among the four detergents used in our study, Triton X-100, NP-40 (also known as Sigma Igepal CA-630) and Brij-35 are nonionic, whereas CHAPS is zwitterionic. Triton X-100 is structurally similar to but slightly more hydrophilic than NP-40. Brij-35 has applications in preventing nonspecific binding to gel filtration and affinity chromatography supports. Nonionic detergents are known to be mild and nondenaturing because they disrupt protein-lipid and lipid-lipid interactions rather than protein-protein interactions, whereas zwitterionic detergents such as CHAPS is a bit more harsh and can sometimes disrupt protein-protein interactions16. Chicken ovalbumin and bovine fetuin were used as standard glycoproteins for the binding study of ConA and WGA respectively. Instead of the radioactivity measurements utilized by Lotan et al15, we applied the newly developed Pierce 660nm protein assay to measure the quantity of standard proteins in bound and unbound fractions. This assay is more linear than coomassie-based Bradford assays and is compatible with higher concentrations of most detergents and reducing agents, which overcomes the potential interference caused by the use of detergents and reducing inhibitory sugars in the elution buffers.
In both ConA and WGA studies, the flow through, wash and elution fractions were collected separately. The binding efficiency (BE) is calculated by BE = E/(E+U), where E is the quantity of standard protein in the elution, and U is the total unbound quantity with the combined mass of flow through and wash fractions. The elution efficiency (EE) is calculated by EE = E/GP, where GP is the mass of glycoprotein loaded, in this case, 200μg. The calculated BE and EE values from detergent experiments are expressed as relative efficiencies [(efficiencies in detergent)/(efficiencies in absence of detergent)x100%] and presented in Supporting Information (SI) Figure S1. For each detergent, four concentrations of 0.1, 0.25, 0.5 and 1% w/v were tested in triplicates. Although the spin column format of the LAC used in this study may have higher variability than the PEEK column format driven by LC pumps10, it offers greater flexibility, easier and faster operation, and great reproducibility, as suggested by a study that employed similar LAC protocol17. In our study, the relative standard deviation (%RSD) of the BE ranges from 0.44% to 12.78%, with an average %RSD of 4.70% and 5.27% for ConA and WGA studies respectively; the %RSD of the EE ranges from 2.21% to 23.50%, with an average value of 7.50% and 8.55% for ConA and WGA studies respectively. It is not surprising that the BE has less variability because it is determined by the relative recovery of the elution to the total recovery of all the fractions, whereas EE is solely dependent on the absolute recovery of elution. The fact that our protocol has an average %RSD of only about 5% provides a solid foundation based on which we can accurately compare the performance of individual detergent at each concentration.
As shown in SI Figure S1, the binding efficiencies for ConA have been decreased to roughly 80% of their original capacity in all conditions, whereas WGA has retained its complete capacity under low concentrations of Triton X-100 and Brij-35. A general trend observed is that increasing detergent concentrations significantly lower the binding efficiencies for both lectins, which is in agreement with the conclusion drawn in the previous study15. It is interesting to note that while the recoveries of elution from WGA are significantly reduced due to the compromised binding efficiency, the ConA counterparts have about the same or higher EE with the use of detergents, which is likely attributed to the independent hydrophobic binding domain on ConA molecules18. Despite the adverse effect on binding, the use of detergents throughout the whole protocol not only largely disrupted the undesirable hydrophobic interactions, but also enhanced the elution of hydrophobic glycoproteins, resulting in improved EE. The choices of concentration of each detergent (listed in the Supporting Information Experimental Method) used for brain membrane protein extraction and DALAC binding are made based on considerations of both BE and EE values. It is noted that these choices are empirical to some extent because the different solublizing power of a particular detergent at different concentrations should also be taken into consideration, which is beyond the scope of the current study.
Brain Membrane Glycoproteome
In spite of the increasing need for the isolation of membrane glycoproteins from the central nervous system for drug targets and biomarker discovery research, a detailed investigation of compatible detergents to enrich membrane glycoproteins using LAC on the brain samples is currently lacking. Coupled with two dimensional gel electrophoresis and peptide mass fingerprinting by matrix-assisted laser desorption/ionization – time-of-flight (MALDI-TOF) mass spectrometry, Owen et al. recently investigated the differences in the ConA-associated proteins of hippocampus and inferior parietal lobule in subjects with Alzheimer’s disease (AD)19. However, this work focused on the whole homogenate of the sample and several differentially expressed individual protein spots on the gel. A more general study of the total glycoproteome, and specifically the membranous fraction, would be desirable. To the best of our knowledge, our present study is the first to examine the global membrane glycoproteome of brain using both ConA and WGA.
By using a modified version of the tissue subcellular fractionation protocol developed by Cox et al.20, we obtained the membrane protein-enriched pellets by ultracentrifugation and high pH wash, followed by extracting the hydrophobic proteins using lectin binding buffer containing the corresponding detergents. Only 0.13% of the original mass of the wet tissue could be extracted into the lectin binding buffer without detergent, whereas 0.35%-0.72% was recovered by using detergents (SI Table S1), illustrating the necessity of detergents in membrane protein solublization. Despite the slightly different concentrations utilized in each extraction, the recovery from NP-40 extraction was significantly higher than those from the other detergents. The same proportions of the extracted samples were loaded on the lectin columns to assess the relative overall recoveries. Again, the final recoveries of membrane glycoproteins from NP-40 extraction were remarkably higher than the other detergent counterparts (SI Table S1). It is noted that, in contrast to our previous binding study on soluble glycoproteins, the EE’s of control were evidently lower than those of detergent extractions from the brains. This is largely because the majority of the glycoproteins in cells are located on the membrane, which in the first place were more efficiently extracted into the solution phase by detergents. This is also supported by the observation that the EE’s of the LAC using the supernatant fraction, which contains mostly the cytoplasmic proteins, were much lower than using the pellets. The EE’s of WGA DALAC were constantly higher than those of ConA, suggesting that the brain membrane glycoproteins are more enriched with N-acetylglucosamine (GlcNAc) or sialic acid type of glycans than high mannose type. To assess if detergents facilitate the elution of hydrophobic glycoproteins, we compared the elution fractions of WGA LAC with or without detergents. The binding and washing buffers both contained the same amount of detergents. Both gel electrophoresis and BCA assay data (SI Figures S2 & S3) implicate that hydrophobic interactions also occur on the WGA beads and detergents enhance the recovery of hydrophobic glycoproteins.
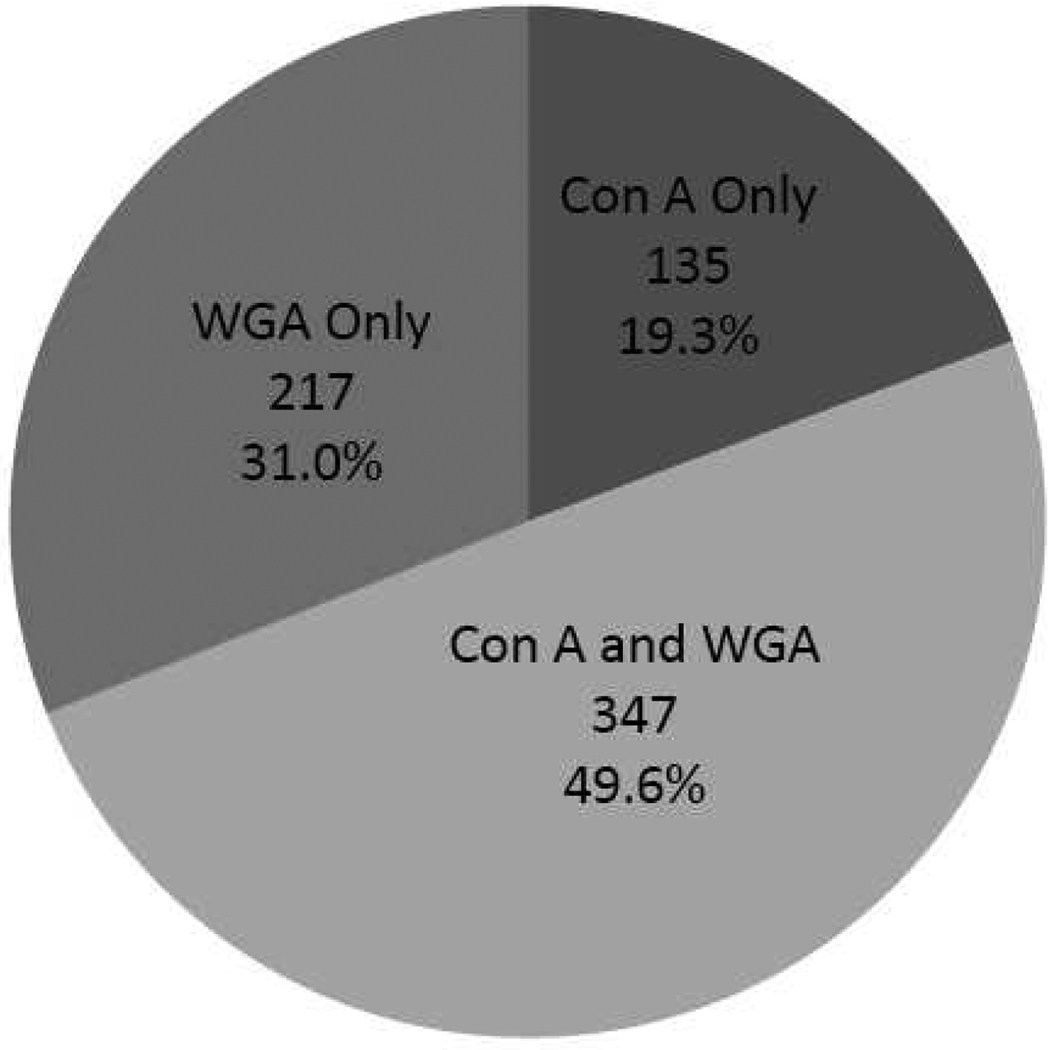
The duplicated samples were separately subjected to LC-MS/MS analyses on LTQ linear ion trap, and the protein identifications from the SEQUEST database search were combined. As a result, 1491 unique proteins were identified with reasonably low false discovery rate (Peptide FDR = 0.64%, Protein FDR = 2.0%), of which 958 were from ConA and 1166 were from WGA DALAC. 482 proteins from ConA and 564 proteins from WGA DALAC were identified with at least two unique peptides with a zero FDR. We choose to limit our discussion to the 699 unique identifications with at least two unique peptides, about a half of which was identified with both lectins (Figure 1). WGA DALAC has enabled identifications of more proteins, again indicating that more glycoproteins are sialyated or contain GlcNAc subunits.
Figure 1.
Distribution of proteins identified by ConA and WGA DALAC. Overall 699 proteins were identified with at least two unique peptides and zero FDR.
The acetone precipitation and ProteaseMAX-assisted trypsin digestion played important roles in LC-MS/MS detection of protein mixtures. Acetone precipitation removes the chromatography- and MS-unfriendly detergents, whereas ProteaseMAX, a novel MS compatible detergent, ensures the high recovery of hydrophobic proteins into the solution phase and allows complete digestion in three hours. It is expected that by using multidimensional separation and/or multiple MS compatible surfactants during digestion21, even greater number of protein identifications can be achieved.
The enrichment of membranous glycoproteins has long been a challenge. As a matter of fact, two previous studies have made effort to analyze cell membrane glycoproteins using 1% NP-40 to help membrane protein extraction22, 23. However, no subcellular fractionation was performed to enrich the membrane proteins, and no detergent was used to facilitate the elution of hydrophobic glycoproteins, which led to the low recovery of membrane glycoproteins. Only approximately 30% of the proteins isolated was predicted to be integral membrane proteins without specifying the number of glycoproteins in the first study22; in the second study, only 22–28% from ConA and 14–16% from WGA LAC were glycoproteins, and the number of membrane proteins was not given23. In our study, ultracentrifugation and high pH buffer wash provided highly enriched membrane fraction, and the use of detergents throughout the protocol not only prevented non-specific binding, but also facilitated the elution of hydrophobic glycoproteins. Even when no perfusion was conducted at the time of sampling to remove blood contamination, very few blood glycoproteins were observed. As a result, over 60% of all identified proteins were membrane proteins, as high as 80% were from plasma membrane, whereas only a minor fraction was from organelles such as mitochondrion, lysosome and ER (Table 1). About half of all identified proteins were annotated as glycoproteins, among which 54–79% from ConA and 72–89% from WGA DALAC were membrane glycoproteins. Overall, a total of 219 unique membrane glycoproteins were identified. It is noted that the actual number of glycoproteins may be higher, as some glycosylation sites have not yet been annotated. NP-40 has consistently outperformed the other three detergents, with 74%/76% membrane protein recovery, 57%/50% glycoprotein recovery, and 116/129 membranous glycoprotein identifications, from ConA/WGA DALAC, respectively. This is likely attributed to better extraction efficiency resulted from high hydrophobicity of NP-40 and its mildness that retains the lectin activity.
Table 1.
Categorization of proteins identified in ConA and WGA DALAC, traditional LAC, and membrane fraction without lectin enrichment. The cellular localization and glycosylation information are obtained from ExPASy proteomics server.
| Conditions |
Con A |
WGA |
Membrane | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DALAC | LAC | DALAC | LAC | ||||||||
| Properties | Tri-X-100 | NP-40 | Brij-35 | CHAPS | Control | Tri-X-100 | NP-40 | Brij-35 | CHAPS | Control | |
| Plasma Membrane | 138 | 165 | 134 | 103 | 64 | 125 | 194 | 172 | 157 | 112 | 72 |
| Organelle Membrane | 38 | 52 | 56 | 42 | 7 | 33 | 47 | 46 | 87 | 13 | 40 |
| Total Membrane | 176 | 217 | 190 | 145 | 71 | 158 | 241 | 218 | 244 | 125 | 112 |
| Organelle | 67 | 40 | 76 | 53 | 49 | 44 | 24 | 66 | 45 | 30 | 50 |
| Cytoplasm | 24 | 20 | 35 | 20 | 24 | 42 | 37 | 57 | 40 | 26 | 74 |
| Extracellular | 31 | 18 | 36 | 39 | 31 | 25 | 15 | 33 | 34 | 19 | 14 |
| Glycoprotein | 176 | 168 | 173 | 145 | 106 | 118 | 160 | 152 | 139 | 109 | 30 |
| Membrane Glycoprotein | 110 | 132 | 102 | 78 | 48 | 86 | 143 | 115 | 100 | 78 | 21 |
| Total ID | 298 | 295 | 337 | 257 | 175 | 269 | 317 | 374 | 363 | 200 | 250 |
DISCUSSION
Compared to the previous studies, our method is significantly advantageous in enriching both membrane proteins and glycoproteins. Although a similar percentage of membrane protein and glycoprotein identifications could be achieved using traditional LAC approach without any detergent, the membrane protein extraction efficiency was only 18–38% of those using detergents, and the final eluted mass recovery was even worse, only 3–17% compared to DALAC approaches. This low recovery greatly limited the practical use of traditional LAC in isolating hydrophobic glycoproteins, especially when very limited samples are available, or enriching low abundance targets is desirable.
Because the elution buffers contain the same concentrations of detergents as in the binding and washing buffers, the elution process can be considered as solely driven by the inhibitory sugars. Therefore, the existence of protein complexes between glyco-and non-glycoproteins is responsible for the identification of non-glycoproteins in the final results. To further assess the performance of our approach, a membrane pellet was extracted with NP-40 and directly subjected to acetone precipitation and trypsin digestion without performing the DALAC. Not surprisingly, only 12% proteins identified were glycosylated and 45% were membrane proteins, of which only 64% belonged to plasma membrane.
The use of different detergents yields complementary information, resulting in higher coverage of the whole membrane glycoproteome. In ConA DALAC, only 30% were commonly identified with all four detergents, whereas 32% were uniquely identified in one detergent; in WGA DALAC, only 25% were commonly identified (SI Figure S4). This feature not only increases the coverage of the whole membrane glycoproteome, but also provides guidance on which detergent to choose if the enrichment of certain targeted proteins is the goal. Interestingly, a single peptide from prion protein (PrP) was repeatedly detected in all detergent extracts except for NP-40 (SI Figure S5), suggesting that NP-40 is not an ideal detergent for PrP extraction.
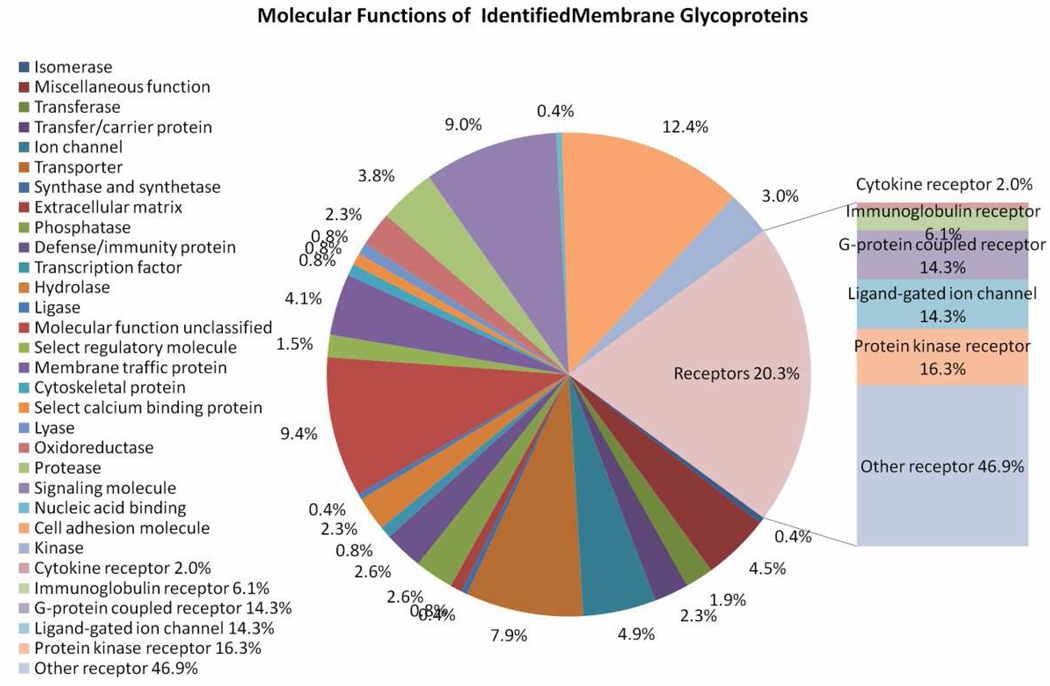
The 219 unique membrane glycoproteins covered a wide range of molecular functions (Figure 2), biological processes and pathways, as suggested by the PANTHER database24. The top molecular functional categories include receptor (26%), cell adhesion molecule (16%), signaling molecule (11%), and transporter (10%). A number of the receptors are protein kinase receptors, ligand-gated ion channels and G-protein coupled receptors. Relatively high proportions of the membrane glycoproteins are involved in signal transduction (39%), developmental processes (25%) and neuronal activities (24%). Some molecules are involved in disease-related pathways, such as amyloid beta (A4) precursor protein (APP) and a disintegrin and metallopeptidase domain 10 (ADAM10) in the Alzheimer’s disease-amyloid secretase pathway. This result indicates that the DALAC technology could provide a more comprehensive view of the membrane glycoproteome of the central nervous system19.
Figure 2.
The PANTHER molecular functions of the 219 unique membrane glycoproteins identified. The composition of the receptors is displayed on the right.
In addition to our DALAC method, hydrazide chemistry serves as a great alternative to target the cell surface glycoproteins. It is generally observed that lectin and hydrazide approaches provide distinct and complementary coverage of the glycoproteome25. In spite of the undesirable periodate oxidation of the secreted and intracellular components, and the non-specific binding of abundant proteins to the hydrazide resin that cannot be completely removed by high salt wash25, careful manipulation of the labeling conditions can maintain maximum viability of the cells, minimizing endocytosis and exocytosis, and thus greatly increase the labeling specificity on the surface26. However, the success of hydrazide approach depends on obtaining viable cells in solution. Therefore, application of this approach is generally limited to cultured and primary cells and in some cases tissues, only when live single-cell suspensions are available26. It can be foreseen that by combining our DALAC approach with site-specific approaches such as PNGase F27 and pronase protocols28, more glycosylation sites can be determined, including the novel ones.
Although it would be interesting to evaluate the specificity of DALAC towards glycosylation types by examining the glycan components, it is extremely challenging to do so because (1) there is currently no complete database of glycan structures due to the heterogeneity and posttranslational nature of this modification; (2) the relatively low recovery rate of membrane glycoproteins from mouse brain tissues prevented performing PNGase F digestion to generate reasonable amount of glycans for subsequent MS characterization; and (3) even if we had enough samples for glycan analysis, the assignment of glycan structures to any particular glycosylation site would be ambiguous due to the heterogeneous nature of membrane glycoproteins.
In conclusion, our investigation suggests that under certain concentrations, mild non-ionic and zwitterionic detergents can be used in LAC to facilitate the solublization and elution of hydrophobic glycoproteins. The optimum conditions need to be determined empirically with regards to the nature of detergents, lectins, and samples. The DALAC approach enables enrichment and characterization of membranous glycoproteins from complex samples with high recovery and specificity. NP-40 is recommended for the general isolation purpose, while other detergents may meet the need to specifically enrich certain target proteins and generate complementary information. This work lays the foundation for further studies such as site-specific characterization and glycomics analysis of the membrane glycoproteins, upon which important insights can be gained towards the discovery of biomarkers and drug targets.
Supplementary Material
ACKNOWLEDGMENT
We thank the University of Wisconsin Human Proteomics Program for access to the LTQ instrument, and Dr. Jeffrey Johnson for access to the spectrophotometer. This research was supported in part by National Institutes of Health through grant AI0272588 and 3R21AI072588-02S1 (NIAID Summer Undergraduate Research Administrative Supplement). L.L. acknowledges an Alfred P. Sloan Research Fellowship and a Vilas Associate Fellowship.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Yildirim MA, Goh K-I, Cusick ME, Barabasi A-L, Vidal M. Nat Biotech. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 2.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 3.Lotan R, Nicolson GL. Biochimica Et Biophysica Acta. 1979;559:329–376. doi: 10.1016/0304-4157(79)90010-8. [DOI] [PubMed] [Google Scholar]
- 4.Murrey HE, Hsieh-Wilson LC. Chem Rev. 2008;108:1708–1731. doi: 10.1021/cr078215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalovidouris SA, Gama CI, Lee LW, Hsieh-Wilson LC. J Am Chem Soc. 2005;127:1340–1341. doi: 10.1021/ja044631v. [DOI] [PubMed] [Google Scholar]
- 6.Murphy KJ, Regan CM. Neurobiology of Learning and Memory. 1998;70:73–81. doi: 10.1006/nlme.1998.3839. [DOI] [PubMed] [Google Scholar]
- 7.Rudd PM, Merry AH, Wormald MR, Dwek RA. Curr Opin Struct Biol. 2002;12:578–586. doi: 10.1016/s0959-440x(02)00377-9. [DOI] [PubMed] [Google Scholar]
- 8.Van Schaftingen E, Jaeken J. FEBS Letters. 1995;377:318–320. doi: 10.1016/0014-5793(95)01357-1. [DOI] [PubMed] [Google Scholar]
- 9.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Clin Chem. 2006;52:1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 10.Madera M, Mechref Y, Klouckova I, Novotny MV. J Chromatogr B. 2007;845:121–137. doi: 10.1016/j.jchromb.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 11.Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin J, Li JG, Zhu D, Pan C, Zhang J. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 12.Atwood JA, Minning T, Ludolf F, Nuccio A, Weatherly DB, Alvarez-Manilla G, Tarleton R, Orlando R. J Proteome Res. 2006;5:3376–3384. doi: 10.1021/pr060364b. [DOI] [PubMed] [Google Scholar]
- 13.Wei X, Li L. Brief Funct Genomic Proteomic. 2009;8:104–113. doi: 10.1093/bfgp/eln053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad E, Naeem A, Javed S, Yadav S, Khan RH. J Biochem. 2007;142:307–315. doi: 10.1093/jb/mvm133. [DOI] [PubMed] [Google Scholar]
- 15.Lotan R, Beattie G, Hubbell W, Nicolson GL. Biochemistry. 1977;16:1787–1794. doi: 10.1021/bi00628a004. [DOI] [PubMed] [Google Scholar]
- 16.Speers AE, Wu CC. Chem Rev. 2007;107:3687–3714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- 17.Madera M, Mann B, Mechref Y, Novotny MV. J Sep Sci. 2008;31:2722–2732. doi: 10.1002/jssc.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelman GM, Wang JL. J Biol Chem. 1978;253:3016–3022. [PubMed] [Google Scholar]
- 19.Owen JB, Di Domenico F, Sultana R, Perluigi M, Cini C, Pierce WM, Butterfield DA. J Proteome Res. 2008;8:471–482. doi: 10.1021/pr800667a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox B, Emili A. Nat Protocols. 2006;1:1872–1878. doi: 10.1038/nprot.2006.273. [DOI] [PubMed] [Google Scholar]
- 21.Chen EI, McClatchy D, Park SK, Yates JR. Anal Chem. 2008;80:8694–8701. doi: 10.1021/ac800606w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh D, Krokhin O, Antonovici M, Ens W, Standing KG, Beavis RC, Wilkins JA. J Proteome Res. 2004;3:841–850. doi: 10.1021/pr049937f. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Ao X, Vuong H, Konanur M, Miller FR, Goodison S, Lubman DM. J Proteome Res. 2008;7:4313–4325. doi: 10.1021/pr8002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald CA, Yang JY, Marathe V, Yen T-Y, Macher BA. Mol Cell Proteomics. 2009;8:287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Nat Biotech. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaji H, Yamauchi Y, Takahashi N, Isobe T. Nat Protocols. 2007;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- 28.An HJ, Peavy TR, Hedrick JL, Lebrilla CB. Anal Chem. 2003;75:5628–5637. doi: 10.1021/ac034414x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.