Abstract
Ion Mobility Spectrometry–Time-of-Flight Mass Spectrometry (IMS-TOFMS) has been increasingly used in analysis of complex biological samples. A major challenge is to transform IMS-TOFMS to a high-sensitivity high-throughput platform for e.g. proteomics applications. In this work, we have developed and integrated three advanced technologies, including efficient ion accumulation in the ion funnel trap prior to IMS separation, multiplexing (MP) of ion packet introduction into the IMS drift tube and signal detection with an analog-to-digital converter (ADC), into the IMS-TOFMS system for the high-throughput analysis of highly complex proteolytic digests of e.g. blood plasma. To better address variable sample complexity, we have developed and rigorously evaluated a novel dynamic MP approach that ensures correlation of the analyzer performance with an ion source function, and provides the improved dynamic range and sensitivity throughout the experiment. The MP IMS-TOF MS instrument has been shown to reliably detect peptides at a concentration of 1 nM in the presence of highly complex matrix, as well as to provide a three orders of magnitude dynamic range and a mass measurement accuracy of better than 5 ppm. When matched against human blood plasma database, the detected IMS-TOF features were found to yield ~ 700 unique peptide identifications at a false discovery rate (FDR) of ~ 7.5 %. Accounting for IMS information gave rise to a projected FDR of ~ 4 %. Signal reproducibility was found to be greater than 80 %, while the variations in the number of unique peptide identifications were < 15 %. A single sample analysis was completed in 15 min that constitutes almost an order of magnitude improvement compared to a more conventional LC-MS approach.
Introduction
The coupling of electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) to a system incorporating Ion Mobility Spectrometry (IMS) Time-of-Flight Mass Spectrometry (TOF MS) has extended the IMS capabilities to proteomics and other system biology applications.1–4 In experiments aimed at discovery of candidate biomarkers in human blood plasma for early cancer detection, many proteins of interest are expected at abundance levels far below that of higher abundance proteins, representing a significant analytical challenge for multidimensional separations coupled to MS.5 To facilitate detection of this interesting part of the proteome, different forms of enrichment/fractionation prior to sample analysis are employed. These include the removal of high-abundance proteins with immunoaffinity chromatography,6–7 fractionation using stacked capture columns filled with different sorbents8–9 and organic solvent extraction.10 The challenge is to perform in-depth high-throughput proteomic studies involving multiple fractions of hundreds of individual human blood plasma samples. Shortening the timescale of condensed phase separation, such as reverse-phase capillary liquid chromatography (RPLC), generally entails reduction in separation power that is mitigated by a decrease in the stationary phase particle size accompanied by an increase in the chromatographic column back pressure.11,12 The latter incurs practical limitations for routine analysis. An elegant and more efficient approach for increasing the overall peak capacity of a system incorporating fast RPLC would be to add orthogonal and complementary separation stages. As such, IMS can potentially increase the overall peak capacity of the RPCL-based platform by an order of magnitude without affecting the analysis speed and better address both the depth of coverage and throughput needs. The higher peak capacities achieved by LC-IMS-TOF MS could provide a basis for detection and reliable identification of candidate biomarkers, and additional criteria for the enhanced screening and differentiation of disease states.
Though IMS enables separation of isobaric peptides13 and increases the linear dynamic range of a system incorporating TOF MS, its incorporation into an LC-MS platform poses a number of technical challenges. IMS is an inherently pulsed analytical technique that uses a weak electric field to rapidly separate ions traversing a drift tube filled with a homogenous neutral gas. IMS is initiated with injection of a discrete ion packet through an ion gate into the drift tube. Spatial separation of gas-phase ions is achieved due to a combination of a drag force exerted on an ion by the medium and the electric field strength. Under the condition of negligible space charge, the temporal full width at half magnitude (FWHM) of the IMS signal is governed by: 14
| (1) |
| (2) |
where Rd is thermal-diffusion limited maximum resolution, tgate is the IMS gate pulse width, tdrift is the ion’s drift time, L is the drift tube length, E is the electric field strength, q is the ion’s charge, kb is Boltzmann’s constant, and T is the absolute temperature. Equations 1–2 shows that higher IMS resolving power (and peak capacity) is obtained with a shorter ion gate pulse width. Practical considerations that account for ion drift velocity across the ion gate result in a typical IMS gate pulse width of 0.1–0.2 ms. Therefore, 50–100 ms IMS separation would exhibit a duty cycle of only 0.1–0.4 % of the ion production that severely restricts the sensitivity of an LC-IMS-TOF platform. Importantly, no ion losses caused by ion dispersion in the drift tube and in the TOF MS interface region are considered in the above analysis for the signal averaging IMS approach. Though drastically minimized due to high-field focusing elements at the drift tube exit15 and the ion funnel interface,16 the transmission losses between the IMS ion gate and TOF MS further deteriorate the instrument detection limit.
To increase ion utilization efficiency while maintaining the optimum IMS resolving power, ion accumulation prior to IMS analysis is performed. Both Paul4 and linear ion trap17 arrangements have been explored in the pressure regions of 5×10−3 − 10−4 Torr, and up to a 200-fold increase in the signal-to-noise ratio (SNR) has been reported when comparing to signal levels obtained in the conventional (no ion trap) configuration. Though low amol limit of detection was estimated, no experimental data with low concentration samples (~1–10 nM) were reported. The use of either a Paul or linear ion trap at a pressure of few mTorr implies the need to drive ions against a strong drag force into the higher pressure region (2–5 Torr) by a high-amplitude electric field pulse that results in ion dispersion and is likely to limit instrument sensitivity in analysis of low abundance analytes.
We have recently reported on the development of a high-charge-capacity ion funnel trap (IFT) capable of operating in a pressure region of several Torr.18,19 The IFT performance has been rigorously evaluated with both TOF MS18 and IMS-TOF MS,19 and the IFT space charge limit was measured to be ~ 107 charges. Given an incoming ion current of 1 nA typically observed in analysis of complex biological mixtures with our ion funnel interface and an IFT trapping efficiency of 50 %, such a trap is filled to its capacity in ~3 ms, limiting the duty cycle of 60 ms IMS separation to ~ 5 %.
Further improvements in IMS-TOF duty cycle are attainable due to a combination of ion accumulation with a multiplexing scheme, where multiplexing implies the capability of introducing multiple ion packets into the drift tube on the time scale of a single IMS separation. One of the most promising multiplexing approaches that give rise to sensitivity enhancement for all species in the course of signal detection experiment is based on Hadamard transform (HT).20 In general, if signals are simultaneously detected in N measurements characterized by the uncorrelated noise, the theoretical increase in SNR over a single measurement is expected to be . By modulating a continuous beam from the ESI source with a Bradbury-Nielsen gate in an atmospheric-pressure IMS instrument (without MS), up to 6-fold increase in sensitivity has recently been reported in HT-IMS measurements.21–22 We have recently developed a novel multiplexing approach (MP) that enables encoding and reconstruction of IMS-TOF MS signals in combination with ion accumulation prior to IMS.23 The use of a weighing scheme for signal reconstruction resulted in nearly perfect ion utilization efficiency and yielded an increase in the SNR by a factor of 10 as compared to the signal averaging experiment. The weight coefficients accounted for the durations of variable accumulation periods prior to ion packet releases into the IMS drift tube. The MP IMS-TOF MS approach has been demonstrated with a mixture of peptides of known concentrations that simplified a choice of the weighing scheme in the inverse transform. In proteomic experiments, a priori knowledge of analyte abundances is often unavailable. To further improve robustness of signal reconstruction in the MP IMS-TOF MS measurements, fixed accumulation periods throughout the encoding sequence have recently been explored.24 Though exhibiting a reduced duty cycle of 50 %, this encoding scheme was found to be practical and applicable to signals of arbitrary complexity.
Using ion trap instrumentation, large variations in signal intensities from a complex biological sample have been most efficiently mitigated by using an automated gain control (AGC) algorithm.25 Since our IMS-TOF system incorporates an IFT, a similar approach would be beneficial for eliminating space-charge-caused ion discrimination26 and detecting lower abundance species with high mass measurement accuracy. In the context of a multiplexing experiment, variable accumulation times imply the use of different encoding sequences that are dynamically selected based on the source function. Here we report on development and implementation of the novel dynamic multiplexing approach with an IMS-TOF MS instrument. The instrument performance has been rigorously evaluated using highly complex proteolytic digests.
Experimental
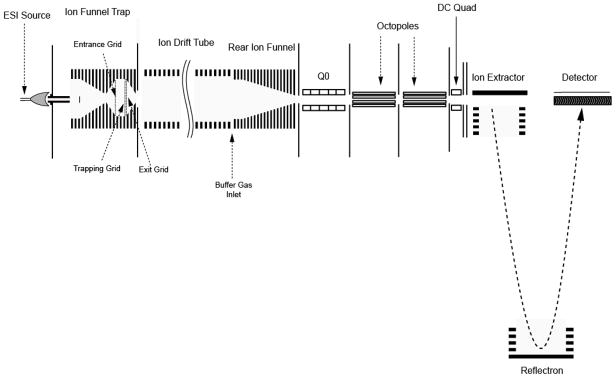
A schematic diagram of our IFT-IMS-TOF MS instrument is shown in Figure 1. The IMS configuration has been described elsewhere19 and only the details relevant to the current configuration are presented here. Multiple samples (or sample fractions) were loaded on a 96-well plate of an automated sample delivery system (TriVersa NanoMate™, Advion Biosciences, Ithaca, NY) and directly infused into the IMS–TOFMS instrument using an ESI-chip based interface. Positively charged electrospray droplets were desolvated in a 508 μm i.d., 10-cm-long stainless steel inlet capillary interface, heated to ~ 160°C.
Figure 1.
Schematic diagram of the IMS-TOFMS instrument. Ion accumulations in the ion funnel trap (IFT) and signal encoding were accomplished by modulating potentials applied to the front, trapping and exit grids of the IFT. The entrance grid represents the IFT entrance gate, while the trapping and exit grids encompass the IFT exit gate. Q0 is the segmented quadrupole designed to interface the IMS instrument to an Agilent 6210 TOFMS.
Ion Funnel Trap
Ions exiting the heated inlet capillary were captured by an electrodynamic ion funnel operated at a pressure of ~ 4 Torr and then injected into the IFT for further accumulation using a pre-determined time sequence. The electrodynamic ion funnel and the IFT consist of 98 brass ring-electrodes, 0.5 mm thick, of variable inner diameters and separated by 0.5 mm-thick Teflon spacers. The trapping volume of the IFT is confined between the entrance, trapping and exit grids, respectively. The grids were made of 95%-transmission electroformed nickel mesh (InterNet Inc., Minneapolis, MN). A 180° phase-shifted RF waveform was applied to the adjacent electrodes of the funnel and IFT. A controllable DC gradient was generated with a resistor chain and superimposed onto the RF field to assist ion transmission through the funnel and IFT. The resistor and capacitor chains were mounted each on separate printed circuit boards, which were attached to the funnel through custom-made zero insertion force connectors (Tactic Electronics, Plano, TX). The RF field applied to the device produced an effective potential that confined ions radially, while axial confinement in the IFT region was established by applying the appropriate potentials to the grids. Both the ion funnel and IFT operated at a peak-to-peak amplitude of 125 Vpp and a frequency of 520 kHz. The DC gradients in the ion funnel and IFT regions were 20 V/cm and 4 V/cm, respectively. To reduce gas load and minimize introduction of large droplets and neutrals into the IFT, a brass disk of 6.4 mm in diameter (a jet disrupter) was positioned ~20 mm downstream of the funnel entrance. An independent DC voltage (~ −25 V offset from the funnel entrance DC voltage) was applied to the jet disrupter. Ions exiting the IFT were separated in a built-in-house 84-cm-long IMS drift tube, captured by an electrodynamic ion funnel terminating the IMS, and injected into an Agilent 6210 TOFMS instrument (Agilent Technologies, Santa Clara, CA) via a custom-made 6-cm-long segmented quadrupole interface. The latter was designed to assist ion transmission through an intermediate pressure region (~ 200 mTorr) into the TOFMS. The IMS drift tube was filled with high purity nitrogen at a pressure of 4 Torr (equal to that of the IFT). A small negative pressure gradient of ~10 mTorr was established between the drift tube to eliminate neutral introduction into the system. To enable gas phase separations, an axial DC gradient of 20 V/cm was generated in both the IMS drift tube and the terminal ion funnel. The segmented quadrupole is comprised of 6 sections, each 20-mm in length, and has an inscribed radius of 2.8 mm. The quadrupole was driven by a custom-made high-Q head at a peak-to-peak amplitude of 270 Vpp and a frequency of 725 kHz, and an axial DC gradient of 5 V/cm was superimposed onto the RF field. TOFMS was operated at a repetition rate of 8 kHz and had a typical mass resolving power of 7000.
Signal encoding and detection
The timing for ion introduction and ejection from the IFT was determined by a pseudo-random binary sequence (PRS) applied to the entrance, trapping and exit grids. The analog waveforms were generated with a 4-channel NI-6711 PCI board (National Instruments, Austin, TX) and applied to the grids using fiber-optic couplers. PRS sequences were derived as described elsewhere.20 A typical 6-bit PRS comprises a combination of digital ‘1’ and ‘0’ elements as follows:
000001000011000101001111010001110010010110111011001101010111111
An element of the above sequence will be referred to as modulation bin. In the IMS drift tube, ion packets disperse both axially and radially due to thermal diffusion and space charge repulsion. To minimize the detrimental effect of ion packet broadening upon signal reconstruction and increase ion utilization efficiency, the conventional PRS shown above has been extended by incorporating additional zeros, so that each ‘0’ and ‘1’ modulation bins were represented as 0000000000 and 1000000000, respectively.23 The triggering of each element in the extended PRS was synchronous with the rising edge of a TOF extraction pulse, while the duration of an element was equal to TOF spectrum acquisition. The elements of the extended PRS will be referred to as sub-modulation bins. Since each modulation bin in the extended 6-bit PRS encompasses 10 sub-modulation bins, the extended encoding sequence represents a 10-fold oversampling.
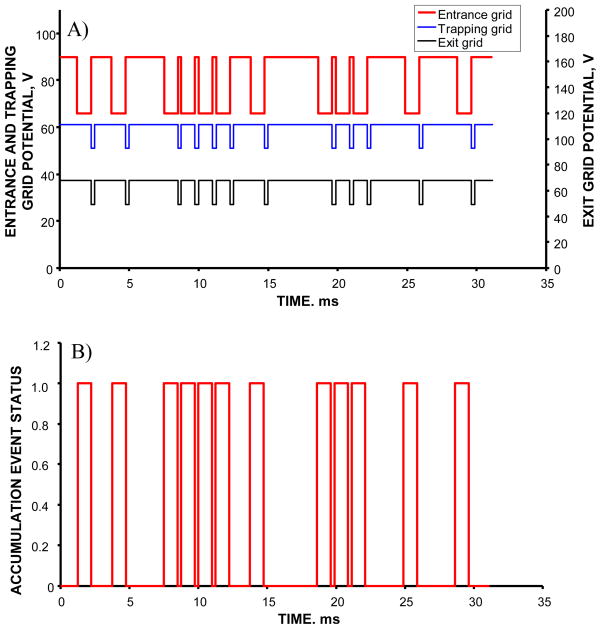
Figure 2 shows a small portion of one of the sequences used for signal encoding. Manipulation of potentials at the grids enabled three distinct events during the experiment: blocking of the continuous ion beam at the entrance grid, accumulation of ions in the IFT and gating of the accumulated ion packets into the IMS drift tube. These three events will be referred to as blocking, accumulation and gating. Figure 2a shows the timing diagram of the IFT events. Blocking was achieved by applying potentials of 90 V, 61 V and 68 V to the entrance, trapping and exit grids, respectively. Accumulation was enabled by reducing the entrance grid potential to 66 V, while keeping the trapping and exit grid voltages at the same levels as during blocking. Ejection was performed by a synchronous increase in the entrance grid potential to 90 V and a decrease in the trapping and exit grid potentials to 51 V and 49 V, respectively. As previously described,24 synchronization of the waveforms applied to the front and trapping/exit grids ensured constant accumulation periods throughout a multiplexed IMS-TOF MS experiment that simplified signal reconstruction procedure and made it robust in analysis of complex samples. Each accumulation period was equal to the shortest interval between two adjacent ion gating events within PRS, as shown in Figure 2b. Four different PRS were employed in the dynamic multiplexing experiment, corresponding to 6-, 5-, 4-, and 1-bit encoding. The last sequence represents the conventional signal averaging mode, with the accumulation time in the IFT equal to the duration of IMS-TOFMS separation. In order for the multiplexed IMS-TOFMS acquisition to match the timescale of that performed in the signal averaging mode, the oversampling factors for the 6-, 5- and 4-bit encoding sequences were chosen to be 10, 20 and 40, respectively. As described above, the minimum oversampling factor of 10 was determined by the ion packet broadening in the IMS drift tube. Given the TOF spectrum duration of 125 μs, the accumulation periods throughout the 6-, 5- and 4-bit encoding sequences were 1.25 ms, 2.5 ms and 6 ms, respectively. Since the number of sub-modulation bins in an N-bit extended PRS equals (2N−1)× oversampling _ factor, the durations of 6-, 5-, 4- and 1-bit IMS-TOFMS acquisitions were 78.8, 77.5, 75 and 75 ms, respectively.
Figure 2.
A) Timing diagrams of potentials applied to the entrance, trapping and exit grids. The entrance grid is referred to as the IFT entrance gate, while a combination of the trapping and exit grids constitutes the IFT exit gate. The diagrams show a small portion of the signal encoding with a 6-bit PRS. For each grid, higher potential corresponds to ion beam blocking, while lower potential accomplishes ion transmission. Note that lower potentials at the exit gate correspond to ion releases to the IMS drift tube, while lower potentials at the entrance gate enable ion accumulation events in the IFT. B) Accumulation events as a function of the elapsed time. The events are correlated with potential distributions in A. ‘1’ corresponds to ion accumulation in the IFT, while ‘0’ occurs during ion beam blocking at the entrance gate and ion gating to the IMS drift tube.
Signal detection was performed with a 1 GS/s 8-bit AP240 analog-to-digital converter (ADC) board with a sustained sequential recording (SSR) option (Acqiris, Geneva, Switzerland). A custom C# code was developed to incorporate the ADC board into the IMS-TOFMS data acquisition system. In earlier work,19,23 we employed a 10 GS/s time-to-digital converter (TDC) board (Model 9353, Ortec, Oak Ridge, TN) for multiplexed IMS-TOFMS signal detection. There were at least two drawbacks associated with TDC signal acquisition. First, the dynamic range per single TOF acquisition was limited to unity, thus reducing the benefits of ion accumulation in the IFT. The use of ADC-based acquisition system expands the dynamic range of IMS-TOFMS instrument by more than two-orders of magnitude. Second, TDC acquisition is prone to systematic mass error shifts due to variable signal intensities. Though improved due to the use of a Constant Fraction Discriminator (CFD), the TDC-based acquisition system remains inferior to that of the ADC with respect to mass measurement accuracy. Detailed comparison of the two IMS-TOFMS data acquisition systems will be reported in a separate publication.
Sample Preparation
Proteolytic digestions of bovine serum albumin (BSA) and phosphorylase B (PHB) (Pierce Biotechnology, Rockford, IL) were conducted using sequencing grade trypsin (Promega, Madison, WI), following previously described procedures.27 Control human blood plasma samples (Sigma-Aldridge, St. Louis, MO) were depleted of 12 most abundant proteins28 using a pre-packed Seppro mixed IgY12 LC5 flow-through immunoaffinity column (Genway, San Diego, CA). The depleted samples were denatured in 8 M urea, reduced with 10 mM dithiothreitol at 37 °C during 1 hour, alkylated with 50 mM iodoacetamide at room temperature for 45 min and digested with sequencing grade modified trypsin (1:50 enzyme:substrate). The digested sample (800 μg) was spiked with 14 peptides at concentrations ranging from 1 nM to 10 μM and fractionated with a 250×2 mm reverse-phase analytical column packed with 5 μm C18 particles (Jupiter, Phenomenex, Torrance, CA). A total of 25 fractions were collected, dried down and then reconstituted in water:acetonitrile:formic acid (70:30:1 v%) electrospray buffer.
Data analysis
Identification of peptides in the multiplexed IMS-TOFMS experiment was based on the previously reported accurate mass and time (AMT) tag approach29,30 and only a brief summary of the procedure is described below. The approach relies on the tagging of each species with an accurate mass and elution time and then matching the tagged species to a database that was previously generated using peptide identifications from thousands of LC-MS/MS runs. In addition to other parameters, the database contains information on peptide masses, elution times, and their sequence. The encoded raw signals were reconstructed at a resolution of 1 ns using the earlier reported inverse transform algorithm.23 The reconstructed IMS-TOFMS features were “deisotoped” to determine species charge states and the corresponding neutral masses. This process was automated with the developed in-house Decon2LS software.31 The neutral masses and drift times were then clustered according to neutral mass deviation (within a pre-determined mass accuracy) and IMS profile continuity into the so-called “Unique Mass Classes (UMCs)”.32 The clustered data were aligned in elution time and m/z domains with the corresponding entries from the AMT tag database to obtain the best fit to the database. Features that fit within alignment criteria were then searched for peptide identification within a user-defined mass accuracy and normalized elution time. Since a peptide may be matched to more than one UMC, the identified peptides were filtered for redundancy to obtain the “unique” peptides list.
Results and Discussion
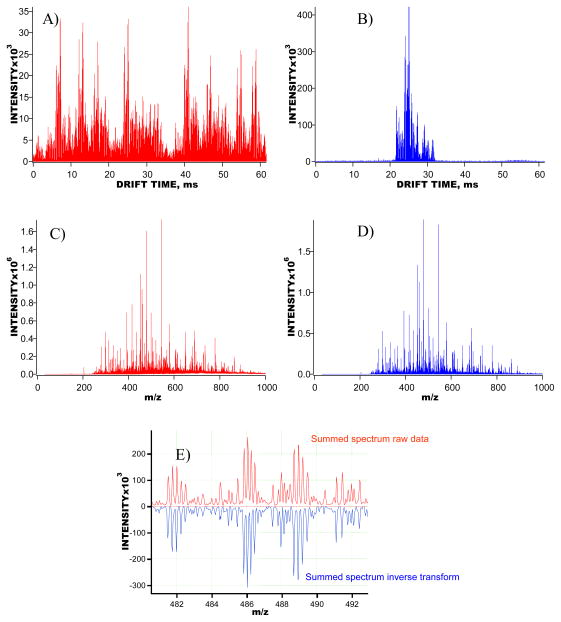
Accurate mass information is essential for peptide identifications based on the AMT tag approach. In MP IMS-TOF experiments, signal encoding is performed in the IMS domain, while the m/z domain remains unchanged as compared to that of the signal averaging experiments. Therefore, it is important that the mass spectrum obtained as a result of summation of all the TOF mass spectra of the reconstructed IMS-TOF signal (the summed reconstructed mass spectrum) matches that of the encoded IMS-TOF signal (the summed encoded mass spectrum). High fidelity of signal reconstruction, i.e. the ability to decode multiplexed IMS-TOF signals at a digitizer resolution without distortion of the original signal vector and a loss of information, ensures improved signal-to-noise ratio (SNR) for the multiplexed signals at a mass accuracy characteristic of the signal averaging approach. Figure 3a–b shows the 5-bit encoded and reconstructed IMS-TOF MS data vectors obtained with a 200 nM solution of phosphorylase B (PHB) tryptic digest. Figure 3c-d illustrates comparison of the summed mass spectra derived from the signals in Figure 3a–b. Detailed examination of the summed encoded and reconstructed mass spectra, as shown in Figure 3e, revealed a correlation near unity between the isotopic envelopes of the same species. The correlation observed in the summed mass spectra was further confirmed by matching the neutral masses derived from the encoded and reconstructed summed mass spectra with those of theoretically predicted tryptic peptides from a PHB in-silico digest. As a result, the same tryptic peptides were identified at mass accuracy tolerances of 10 ppm and 20 ppm. Both the examination of the summed mass spectra and analysis of peptide identifications evidenced the high-fidelity of our signal reconstruction algorithm developed for the de-multiplexing of complex IMS-TOF signals recorded with ADC data acquisition system.
Figure 3.
Multiplexed IMS-TOFMS signals obtained for a 200 nM phosphorylase B solution. A) IMS-TOFMS signal encoded with a 5-bit PRS and averaged over 200 acquisitions. All 600 TOF spectra taken during IMS separation are represented as one concatenated data vector. B) Reconstructed IMS-TOFMS signal using data in A. Similar to A, all TOF spectra acquired during IMS separation are represented as one data vector. C) Summed mass spectrum obtained using encoded data in A. D) Summed mass spectrum generated using reconstructed signal in B. E) A small region of the panels in C–D, showing detailed comparison of the encoded and reconstructed summed mass spectra.
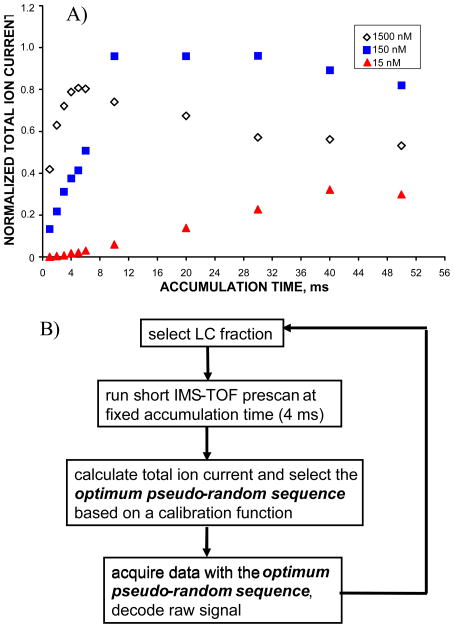
Figure 4a shows the total ion chromatogram (TIC) obtained for BSA solutions at three different sample concentrations as a function of the ion accumulation time in the IFT. At higher sample concentrations, the TIC curve reaches a plateau at accumulation times of only a few ms, while at low concentration the dependence of the TIC on the accumulation time approximates with a linear function. To enhance the instrument dynamic range in analysis of higher concentration samples and to eliminate undesirable m/z discrimination, ion clouds need to be released from in the IFT prior to reaching the charge capacity of the trap. TIC levels that correspond to the IFT space charge limit were examined as a function of the accumulation time in the IFT and correlated to the appropriate encoding sequences to obtain a calibration function. The use of calibration function ensured that high abundance signals were acquired with higher-bit encoding sequences that yield shorter accumulation time, while the low abundance signals were recorded with a low-bit PRS corresponding to longer accumulation times. Figure 4b illustrates the flow control diagram of an automated dynamic multiplexing measurement. Per single sample (or sample fraction) analysis, multiplexed IMS-TOFMS acquisition was conducted in the following sequence. First, a short IMS-TOF pre-scan was performed in the signal averaging mode for 3 s. Ions were accumulated for a fixed period of 4 ms prior to each gating event and spectra were averaged over 50 acquisitions. Second, a total ion signal (TIS), that is the sum of intensities for all the TOF bins throughout the IMS separation, was calculated and the optimum PRS was automatically selected based on the calibration function. Third, multiplexed IMS-TOF acquisition was performed for 15 s (that corresponds to 200 averages), using the optimum encoding sequence determined in step 2. Steps 1 through 3 were run in a cycle, so that IMS-TOFMS signals from multiple fractions were acquired in fully automated fashion without operator interference.
Figure 4.
A) The normalized total ion currents (TIC) from bovine serum albumin (BSA) solutions at concentrations of 15 nM, 150 nM and 1500 nM as functions of the ion accumulation time in the IFT. TIC dependence at high concentration exhibits saturation at ~ 4 ms accumulation time in the IFT, indicating the need for multiple ion gating events throughout 60 ms IMS separation. TIC curve at low concentration approximates with a linear function, implying that the IFT charge capacity has not been reached on the timescale of IMS separation. B) Flow control diagram of the automated dynamic multiplexing experiment with IMS-TOFMS. The optimum encoding sequence is selected based on the IMS-TOFMS pre-scan signal.
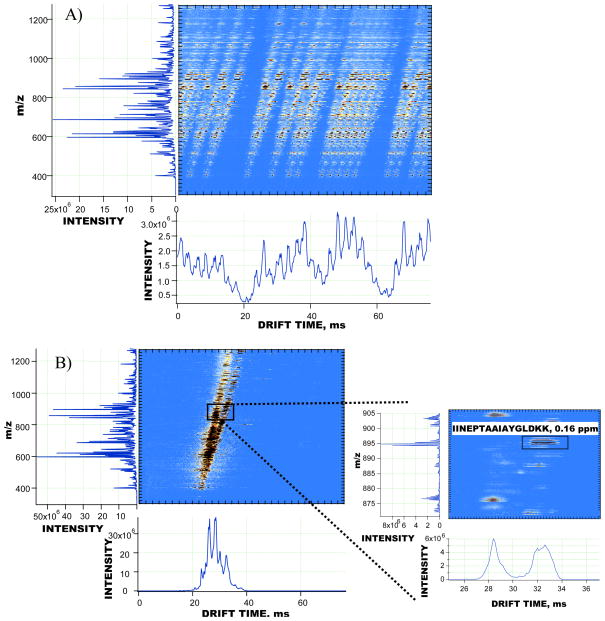
The dynamic multiplexing approach has been applied to the study of a 0.5 mg/mL sample of depleted human blood plasma. The sample was fractionated off-line into 25 RP fractions. Guided by the total signal chromatogram obtained with a ultra-violet light detector, fractions from 6 to 25 were studied in 10 separate experiments. One experiment encompasses a fully automated analysis of all the selected depleted human blood plasma fractions. In a fully automated fashion, IMS-TOF signals from each fraction were initially acquired over a short pre-scan and then encoded with the optimum PRS, as described above. Following data acquisition, the encoded signals from all RP fractions were reconstructed by applying the inverse transform algorithm in the batch processing mode. Table 1 shows the results of matching of the human plasma peptides against the IMS-TOFMS features detected from different fractions at a mass accuracy of 5 ppm and a normalized retention time tolerance of 4 %. Identifications per fraction are shown along with the encoding sequences used in the automated experiment. As seen in Table 1, the majority of identifications were acquired from fractions 10 to 16. The performance of the multiplexed IMS-TOFMS instrument is illustrated in analysis of data obtained in one of the experiments (#4). Figure 5a–b shows an example of signal encoding and reconstruction for the human blood plasma fraction 14. The Inset in Figure 5b demonstrates the IMS profile and isotopic envelope of one of the identified peptides (peptide sequence: IINEPTAAAIAYGLDKK). Detailed analysis of the reconstructed IMS-TOF MS signals in Figure 5b indicated that the dynamic range of the acquired mass spectra exceeded three orders of magnitude, resulting in detection of more than 2,000 IMS-TOF features per IMS separation. Table 2 in the Supporting Information includes only those IMS-TOF features in Figure 5b that were matched to the human blood plasma peptides at a mass accuracy of 5 ppm and a normalized LC retention time tolerance of 4 %. Table 2 entries are sorted in a descending order based on the IMS-TOF feature abundance. For a few IMS-TOF features in Table 2, the mass measurement accuracy and retention time tolerances used in data processing were insufficiently stringent to unambiguously assign the detected IMS-TOF feature to a single human plasma peptide; e.g. the feature detected at m/z 739.3803 was matched to VAVPPLTEEDVLFNVNSDTR and YNPVVIDFEMQPIHEVLR peptides at mass errors of −0.28 ppm and 0.85 ppm, respectively. Therefore, all the human plasma peptides that deviate from the detected feature within the pre-determined mass measurement error and retention time tolerance were considered valid matches and included in Table 2. Higher confidence in peptide identifications is expected due to the use of targeted tandem MS information for the matched peptides. When combined with mass measurement accuracy and retention time tolerance, peptide sequence information to be obtained from fragmentation spectra is an additional parameter that will further reduce false positive identifications and minimize ambiguity in matching the database peptides to a detected IMS-TOF feature.33
Table 1.
Results of analysis of a 0.5 mg/mL depleted human blood plasma sample with the MP IMS-TOFMS instrument. From left to right, the columns represent the sample fraction number, the encoding sequence used in the dynamic multiplexing experiment and the number of identified unique peptides from human plasma database that matched the detected IMS-TOFMS features at a mass measurement accuracy of 5 ppm and a normalized retention time tolerance of 4 %.
| FRACTION | PRS, bits | MATCHES |
|---|---|---|
| 6 | 1 | 0 |
| 7 | 1 | 0 |
| 8 | 4 | 0 |
| 9 | 4 | 13 |
| 10 | 4 | 95 |
| 11 | 4 | 139 |
| 12 | 5 | 140 |
| 13 | 6 | 64 |
| 14 | 5 | 118 |
| 15 | 6 | 58 |
| 16 | 6 | 39 |
| 17 | 6 | 33 |
| 18 | 6 | 13 |
| 19 | 5 | 12 |
| 20 | 5 | 18 |
| 21 | 5 | 8 |
| 22 | 5 | 4 |
| 23 | 1 | 0 |
| 24 | 1 | 0 |
| 25 | 5 | 0 |
Figure 5.
IMS-TOFMS signals detected and reconstructed in experiments with a 0.5 mg/mL depleted human blood plasma sample. A) 5-bit encoded IMS-TOFMS signal from human blood plasma fraction 14 acquired in technical replicate #4; B) reconstructed IMS-TOFMS signal from the 2D map in A. The inset in B shows one of the identified peptides from the depleted human blood plasma (sequence: IINEPTAAAIAYGLDKK) at a mass measurement accuracy of 0.16 ppm.
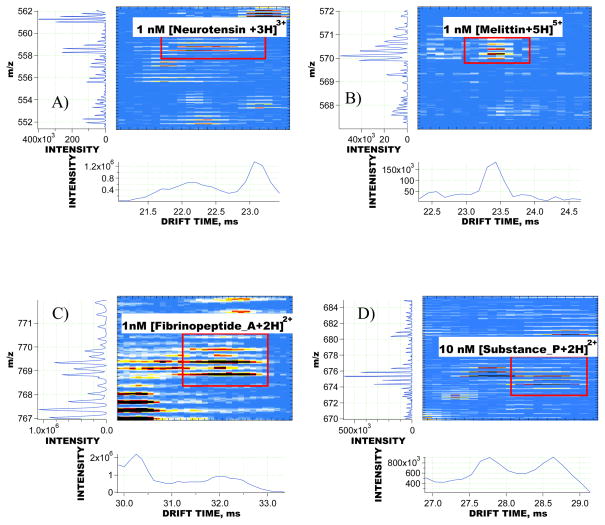
Sensitivity of the MP IMS-TOFMS approach was evaluated based on the signals of model peptides mixed with human blood plasma sample prior to RPLC fractionation. Figure 6a–d shows small regions of the reconstructed IMS-TOFMS 2D maps where signals of honey bee venom melittin, human neurotensin, human fibrinopeptide A and substance P were detected. Melittin, neurotensin and fibrinopeptide A were introduced into the depleted human blood plasma sample at a concentration of 1 nM, while substance P was mixed at a concentration of 10 nM. All the peptides in Figure 6 were detected at SNR of greater than 5 in the presence of human blood plasma species. Figure 6c–d demonstrates the utility of IMS separation for enhanced detection and identification of low concentration peptides from complex samples. As seen in both 2D maps in Figure 6c–d, the spiked peptides overlapped in m/z domain with the endogenous human blood plasma species. If no IMS separation were employed, peptide signal recognition and deisotoping would have represented a challenging task. The IMS instrument enabled spatial separation of the isobaric peptides, resulting in reliable detection and quantitation of the peptides of interest. Detailed analysis of signals from all 14 peptides spiked into the plasma sample showed that the dynamic range of MP IMS-TOFMS spans more than three orders of magnitude and the instrument is capable of detecting analytes at concentrations of less than 1 nM in a highly complex matrix.
Figure 6.
Regions of the reconstructed IMS-TOFMS 2D maps encompassing signals from low concentration peptides. The peptides were mixed with a 0.5 mg/mL depleted human blood plasma sample prior to RP fractionation. A) 1 nM [Neurotensin +3H]3+, detected in fraction 15, signal was encoded with a 6-bit PRS; B) 1 nM [Melittin+5H]5+, fraction 18, 6-bit encoded; C) 10 nM [Substance_P +2H]2+, fraction 12, 5-bit encoded; D) 1 nM [Fibrinopeptide_A +2H]2+, fraction 11, 4-bit encoded. Note that IMS-TOFMS signals of the spiked peptides in C and D overlap in m/z domain with those of endogenous human plasma peptides, and therefore could only be resolved and then successfully “deisotoped” due to IMS separation. All spiked peptides were detected at a mass measurement accuracy of < 5 ppm.
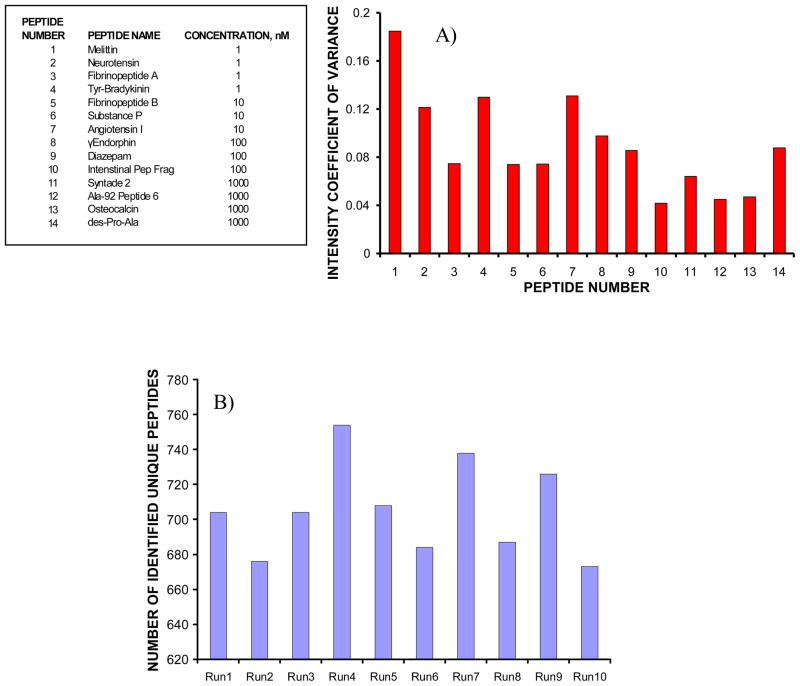
Reproducibility of MP IMS-TOFMS system was investigated using signals of the same spiked peptides throughout all 10 experiments (i.e., technical replicates) and the results are summarized in Figure 7a. Each data point in Figure 7a represents intensity coefficient of variance (CV) for a given peptide. Intensity CVs were derived by integrating peptide signals in the IMS domain. A general trend in Figure 7a indicates that intensity CVs increase at lower peptide concentrations, with an average intensity CV of ~9 %. The overall performance of the MP IMS-TOFMS system was evaluated based on identifications of unique peptides from depleted human blood plasma sample in 10 experiments. Figure 7b shows the number of identified unique human plasma peptides as a function of the experiment number. The average number of the identified unique peptides per run was found to be 705, with a standard deviation of 27. This corresponds to ~15 % variation in the number of identifications between technical replicates. Reproducibility in IDs of the matched peptides was greater than 60 %. The false discovery rate (FDR) in peptide identifications was estimated by shifting the human plasma peptide database by 11 Da34 and determining the matches of the detected IMS-TOFMS features against entries in the shifted database at a mass measurement accuracy of 5 ppm and retention time tolerance of 4 %. Without invoking ion mobility information, the average FDR was ~ 7.5 %. A potential FDR decrease due to the use of ion mobility information was roughly estimated in experiments with 150 nM solutions of BSA and PHB tryptic digests using the following procedure. First, experimental datasets of BSA and PHB tryptic digests were “deisotoped” and matched against the corresponding in-silico protein digests at mass error tolerances of 20, 50 and 100 ppm. Both monoisotopic masses and drift times of the matched IMS-TOFMS features were determined. Second, the matched IMS-TOFMS features were searched against in-silico digest of the other protein within the same mass error tolerances (i.e., the matched BSA experimental features were searched against PHB in-silico digest and visa versa). No matches were found, indicating that the PHB in-silico digest could be used as a decoy database for the BSA experimental dataset. The same was found to be valid when examining the PHB experimental dataset with respect to the BSA in-silico digest. Therefore, an assumption was made that all the matches of e.g. the BSA experimental dataset against the PHB in-silico digest would be false positive identifications. Third, the matches of e.g. the BSA experimental dataset against the PHB in-silico digest were examined with and without the use of ion mobility information. Identifications were considered to be false if the difference in drift times between the IMS-TOFMS matches from the BSA experimental dataset (i.e., incorrect matches) and that from the PHB experimental dataset (i.e., putative correct matches) exceeded 2 ms. As a result of using drift time information, the number of false positive identifications was reduced by a factor of 2. This improvement is limited by partial orthogonally between IMS and MS dimensions and the IMS resolving power, and can potentially be further increased by incorporation of peptide sequence information from tandem MS experiments. Extrapolating to the human blood plasma experiments, one would expect to attain an FDR of < 4 % when using ion mobility information for peptide identifications from this initial analysis. More rigorous evaluation of the ion mobility effect will be performed by generating an extended AMT tag database that incorporates peptide drift times, and additional improvements in the FDR are anticipated. This work is in progress and will be reported in a separate publication.
Figure 7.
A) Intensity coefficients of variance (CVs) for the peptides mixed with a 0.5 mg/mL human blood plasma sample at concentrations ranging from 1 nM to 1 μM. Mixing was performed prior to RP fractionations. Intensity CVs were calculated by integrating signals in the IMS domain. B) Number of identified unique peptides from human blood plasma as a function of the MP IMS-TOFMS experiment number. The peptides were matched against the detected IMS-TOF MS features at a mass measurement accuracy of 5 ppm and a normalized retention time tolerance of 4 %. The projected false discovery rate was estimated as ~ 4%.
Several important features of the developed IMS-TOFMS instrument make it highly attractive for candidate biomarker discovery experiments with complex biochemical samples. First, the use of efficient ion accumulation in combination with multiplexing ensures the improved ion utilization efficiency at a duty cycle of 50 %. For lower concentration samples, the duty cycle of dynamically multiplexed IMS-TOFMS experiments approaches 100 %, resulting in ultra-high sensitivity. Since ion counts in TOFMS measurements exhibit statistical fluctuations described by the Poisson probability distribution, the measurement precision is inversely proportional to the square root of the number of ion counts per peak. Therefore, higher sensitivity due to a combination of ion trapping and multiplexing also contributes to improvements in mass measurement accuracy. Second, the coupling of the TOF MS detector to an ADC-based data acquisition system enhances the dynamic range of the IMS-TOFMS instrument by two orders of magnitude as compared to the TDC-based analyzer and concurrently provides improvement in the mass measurement accuracy due to more precise determination of the peak centroid. The summed mass spectra of complex proteolytic digests were routinely calibrated at a mean square root error of < 2 ppm that constitutes a 5-fold increase in the mass measurement accuracy as compared to the TDC-based data acquisition system. Third, the developed platform has a 10-fold higher throughput than the conventional proteomics platform employing 2-hour RPLC separation and Fourier Transform MS detection. The coupling of brighter ESI sources35 to the multiplexed IMS-TOFMS is expected to further increase the speed of analysis. Fourth, IMS separations are generally more reproducible than RPLC, with an average standard deviation in drift times of less than 1 % under carefully controlled conditions.36 This provides a basis for using IMS information as additional parameter for confident peptide identifications. Furthermore, IMS stage spatially separates ion species based on their mobility, thus reducing the impact of a dense ion cloud on the detector as compared to that observed with an ion trap-TOFMS and, thereby, further expands the instrument dynamic range. Spatial separation of the isotopic distributions overlapping in m/z domain also contributes to improved quantitation for these species. When combined, the above characteristics position MP IMS-TOFMS technology in the mainstream of proteomics research and arguably make it a viable alternative to the currently used LC-MS approaches.
Conclusions
A novel dynamic multiplexing approach with an IMS-TOFMS instrument has been developed and rigorously evaluated in analysis of reverse-phase fractions of depleted human blood plasma. IMS-TOFMS has been coupled to a data acquisition system based an analog-to-digital converter (ADC). In combination with efficient ion accumulation and ADC detection, multiplexing resulted in two orders of magnitude improvement in IMS-TOFMS dynamic range. The multiplexed IMS-TOFMS instrument enabled peptide identifications at concentrations of 1 nM at a signal-to-noise ratio of up to 50 in the presence of highly complex matrix, such as human blood plasma depleted of high abundance proteins. Reproducibility of the system was investigated based on the signals of model peptides introduced into the depleted human blood plasma sample prior to reverse-phase fractionation, and the average intensity coefficient of variance was found to be ~ 9 %. Per single experiment, the average number of identified unique human plasma peptides was ~ 700 at a false discovery rate (FDR) of 7.5 %. When accounting for ion mobility information, a projected FDR of ~ 4% was estimated. The developed multiplexed IMS-TOFMS instrument represents an ultra-high sensitivity high-through platform that can potentially be employed in applications such as candidate biomarker discovery experiments.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Yehia Ibrahim for insightful discussions. Portions of this research were supported by the NIH National Center for Research Resources (RR18522), Science Applications International Corporation-Frederick (25XS118), the National Cancer Institute (R21 CA12619101), and the W.R. Wiley Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory). Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RLO-1830.
References
- 1.Gillig KJ, Ruotolo B, Stone EG, Russell DH, Fuhrer K, Conin M, Schultz AJ. Anal Chem. 2000;72:3965–3971. doi: 10.1021/ac0005619. [DOI] [PubMed] [Google Scholar]
- 2.McLean JA, Russell DH. J Proteome Res. 2003;2:427–430. doi: 10.1021/pr034004p. [DOI] [PubMed] [Google Scholar]
- 3.Wittmer D, Chen YH, Luckenbill BK, Hill HH. Anal Chem. 1994;66:2348–2355. [Google Scholar]
- 4.Hoaglund CS, Valentine SJ, Clemmer DE. Anal Chem. 1997;69:4156–4161. [Google Scholar]
- 5.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Proteomics. 2003;3:422–432. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 7.Huang HL, Stasyk T, Morandell S, Mogg M, Schreiber M, Feuerstein I, Huck CW, Stecher G, Bonn GK, Huber LA. Electrophoresis. 2005;26:2843–2849. doi: 10.1002/elps.200500167. [DOI] [PubMed] [Google Scholar]
- 8.Guerrier L, Lomas L, Boschetti E. J Chromatogr A. 2005;1073:25–33. doi: 10.1016/j.chroma.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Righetti PG, Castagna A, Antonioli P, Boschetti E. Electrophoresis. 2005;26:2843–2849. doi: 10.1002/elps.200406189. [DOI] [PubMed] [Google Scholar]
- 10.Merrell K, Southwick K, Graves SW, Esplin MS, Lewis NE, Thulin CD. J Biomol Tech. 2004;15:238–248. [PMC free article] [PubMed] [Google Scholar]
- 11.Issaeva T, Kourganov A, Unger K. J Chromatogr A. 1999;846:13–23. [Google Scholar]
- 12.Shen Y, Smith RD, Unger KK, Kumar D, Lubda D. Anal Chem. 2005;77:6692–6701. doi: 10.1021/ac050876u. [DOI] [PubMed] [Google Scholar]
- 13.Woods AS, Ugarov M, Egan T, Koomen J, Gillig KJ, Fuhrer K, Gonin M, Schultz JA. Anal Chem. 2004;76:2187–2195. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- 14.Revercomb HE, Mason EA. Anal Chem. 1975;47:970–983. [Google Scholar]
- 15.Lee YJ, Hoaglund-Hyzer CS, Traska JA, Zientara GA, Counterman AE, Clemmer DE. Anal Chem. 2001;73:3549–3555. doi: 10.1021/ac010295e. [DOI] [PubMed] [Google Scholar]
- 16.Tang K, Shvartsburg AA, Lee HN, Prior DC, Buschbach MA, Li F, Tolmachev AV, Anderson GA, Smith RD. Anal Chem. 2005;77:3330–3339. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myung S, Lee YJ, Moon MH, Taraska J, Sowell R, Koeniger S, Hilderbrand AE, Valentine SJ, Cherbas L, Cherbas P, Kaufmann TC, Miller DF, Mechref Y, Novotny MY, Ewing MA, Sporleder CR, Clemmer DE. Anal Chem. 2003;75:5137–5145. doi: 10.1021/ac030107f. [DOI] [PubMed] [Google Scholar]
- 18.Ibrahim YM, Belov ME, Tolmachev AV, Prior DC, Smith RD. Anal Chem. 2007;79:7845–7852. doi: 10.1021/ac071091m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clowers BH, Ibrahim YM, Prior DC, Danielson WF, III, Belov ME, Smith RD. Anal Chem. 2008;80:612–623. doi: 10.1021/ac701648p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwit M, Sloane NJ. Hadamard Transform Optics. Academic Press; New York: 1979. [Google Scholar]
- 21.Clowers BH, Siems WF, Hill HH, Massick SM. Anal Chem. 2006;78:44–51. doi: 10.1021/ac050615k. [DOI] [PubMed] [Google Scholar]
- 22.Szumlas AW, Ray SJ, Hieftje GM. Anal Chem. 2006;78:474–4471. doi: 10.1021/ac051743b. [DOI] [PubMed] [Google Scholar]
- 23.Belov ME, Buschbach MA, Prior DC, Tang K, Smith RD. Anal Chem. 2007;79:2451–2462. doi: 10.1021/ac0617316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clowers BH, Belov ME, Prior DC, Danielson WF, Ibrahim YM, Smith RD. Anal Chem. 2008;80:2464–2473. doi: 10.1021/ac7022712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz JC, Zhou XG, Bier ME. 5,572,022. US Patent. 1996 November 5;
- 26.Belov ME, Nikolaev EN, Harkewicz R, Masselon CD, Alving K, Smith RD. Int J Mass Spectrom. 2001;208:205–225. [Google Scholar]
- 27.Kinter MM, Sherman NE. Protein Sequencing and Identification Using Tandem Mass Spectrometry. Wiley Inter Science; New York: 2000. [Google Scholar]
- 28.http://www.genwaybio.com
- 29.Conrads TP, Anderson GA, Veenstra TD, Pasa-Tolic L, Smith RD. Anal Chem. 2000;72:3349–3354. doi: 10.1021/ac0002386. [DOI] [PubMed] [Google Scholar]
- 30.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen YF, Conrads TP, Veenstra TD, Udseth HR. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 31.http://ncrr.pnl.gov/software
- 32.Zimmer JSD, Monroe ME, Qian WJ, Smith RD. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merenbloom SI, Koeniger SL, Valentine SJ, Plasencia MD, Clemmer DE. Anal Chem. 2006;78:2802–2809. doi: 10.1021/ac052208e. [DOI] [PubMed] [Google Scholar]
- 34.Petyuk VA, Qian WJ, Chin MH, Wang HX, Livesay EA, Monroe ME, Adkins JN, Jaitly N, Anderson DJ, Camp DG, Smith DJ, Smith RD. Genome Res. 2007;17:328–336. doi: 10.1101/gr.5799207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly RT, Page JS, Zhao R, Qian WJ, Mottaz HM, Tang K, Smith RD. Anal Chem. 2008;80:143–149. doi: 10.1021/ac701647s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eiceman GA, Nazarov EG, Rodriguez JE, Stone JA. Rev Sci Instrum. 2001;72:3610–3621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.