Abstract
During transcription of protein-coding genes, bacterial RNA polymerase (RNAP) is closely followed by a ribosome that is engaged in translation of the newly synthesized transcript. Here we show that as a result of translation-transcription coupling the overall elongation rate of transcription is tightly controlled by translation. Acceleration and deceleration of a ribosome results in corresponding changes in the speed of RNAP. Consistently, we found an inverse correlation between the number of rare codons in a gene, which delay ribosome progression, and the rate of transcription. We further show that the stimulating effect of a ribosome on RNAP is achieved by preventing RNAP from adopting non-productive states. The moving ribosome inhibits spontaneous backtracking of RNAP, thereby enhancing its pace and also facilitating read-through of roadblocks in vivo. Such a cooperative mechanism ensures the two gene expression machineries match precisely each other rates, so that the transcriptional yield is always adjusted to translational needs at different genes and under various growth conditions.
In contrast to eukaryotes, where translation and transcription take place in different cellular compartments, in bacteria the two principal events of gene expression are coupled in time and space. The majority of bacterial genes initiate translation soon after the ribosome-binding site (RBS) has emerged from the RNA exit channel of RNAP. Although translation-transcription coupling has been known for decades, the only functional manifestation of this phenomenon was limited to specific cases of transcription attenuation and polarity. (1). When a ribosome slows down due to, for example, amino acid deficiency, the growing gap between moving RNAP and lagging ribosome provides the termination factor Rho with access to the nascent RNA, resulting in premature transcription termination (2). The ribosome can also be responsible for early transcription termination (attenuation) at certain metabolic operones by allowing the formation of an intrinsic termination hairpin in the leader RNA sequences (3). Notably, both attenuation and polarity occur when the ribosome and RNAP are physically separated. In each case the effect of a ribosome on RNAP is indirect; it is mediated by either Rho factor or RNA secondary structures. In contrast, we found that for the most part of a coding region, the first trailing ribosome directly assists RNAP during elongation. Such cooperation between the two macromolecules, in which the ribosome plays the leading role, explains the precise match of translational and transcriptional rates under various growth conditions (Table 1). Ribosome-RNAP cooperation thus represents the fundamental mechanism that coordinates the two major events of bacterial gene expression.
Table 1.
Overall transcription elongation rates match that of translation under various growth conditions.
| Strain | Growth condition | Rates of transcription (nt/s) | Rates of translation (aa/s) | (nt/aa) |
|---|---|---|---|---|
| MG1655 | 42 | 14 | 3.0 | |
| MG1655 | + Cm | 27 | 9 | 3.0 |
| MG1655 | Stationary | 21 | 7 | 3.0 |
| MG1655 | Glycerol | 31 | 10 | 3.1 |
| MG1655 | αMG | 23 | 8 | 2.9 |
| CH184 | + Sm | 31 | 10 | 3.1 |
| CH184 | 19 | 6 | 3.2 | |
|
| ||||
| CH184 | Stationary + Sm | 22 | 7 | 3.1 |
|
| ||||
| CH184 | Stationary | 12 | 4 | 3.0 |
MG1655 (wild type) and CH184 (rpsL[SmP]) were grown in LB to the exponential phase (OD600~0.4) prior to IPTG induction, unless indicated otherwise. “Stationary phase” – IPTG was added at OD600~2.5. Alternative carbon sources: “Glycerol” – cells were grown in M9 minimal media supplemented with glycerol (0.5%) and Casamino Acids (0.2%); “aMG” – cells were grown in glucose minimal media in the presence of α-methyl-glucoside at the 15× to glucose ratio. IPTG was added at OD600~0.4. Numbers represent averaged values from three independent experiments (p<0.05), including those described in Fig. 1, fig. S2, S5, S6, and S7. The standard error for each value in each individual experiment was less than 10%.
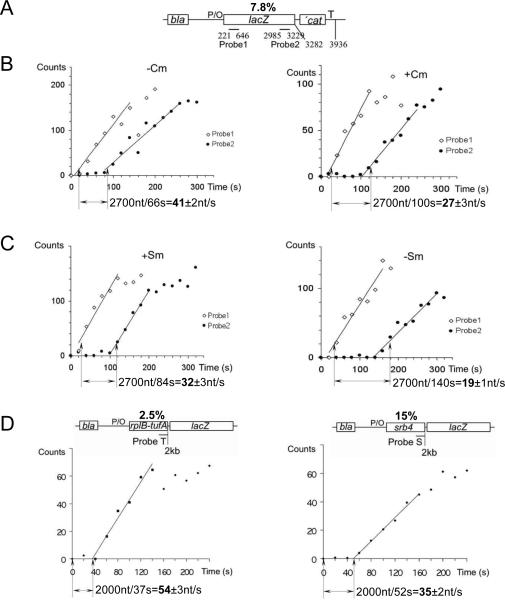
To measure the overall transcription elongation rate in vivo we utilized a plasmid carrying the IPTG controllable version of the A1 promoter of bacteriophage T7 fused to the lacZ reporter gene (Fig. 1A). To calculate the elongation rate, the E.coli culture was induced with IPTG and the time elapsing between the appearance of a specific hybridization signal from probes complementary to the 5' and 3' segments of the lacZ transcript was determined by dot blot hybridization (Fig. 1; fig. S1) (4, 5). Although the absolute values of the hybridization signal may vary depending on factors such as Rho termination, RNA stability, or rate of transcription initiation, the timing for the linear signal increase between the two probes depends only on RNAP speed. This setup therefore allows for the accurate and unbiased determination of the elongation rate in vivo (4, 5). In parallel we measured the translation elongation rate by monitoring the induction lag for b-galactosidase synthesis (fig. S2) (6). We first investigated the effect of the antibiotic chloramphenicol (Cm), a specific inhibitor of translation elongation, on the transcription elongation rate. Cm was added to exponentially growing cells at 1 μg/ml – a concentration that had only a mild inhibitory effect on bacterial growth. In the absence of Cm the transcription elongation rate was determined to be 42 nt/s under specified growth conditions, matching a translation elongation rate of 14 aa/s (Table 1; fig. S2). However, Cm reduced the transcription elongation rate to 27 nt/s (Fig. 1B), which matched a reduced translation rate of 9 aa/s (Table 1; fig. S2). This result suggests that it is the ribosome which controls the rate of RNAP propagation.
Fig. 1. The overall transcription elongation rate depends on the rate of translation.
(A) Schematics of the test plasmid pUV12 used in (B) and (C) (4, 5). Open bars show genes. P/O indicates the T7A104/03 promoter/operator site; T - terminator sequence. Lines indicate probes complementary to the lacZ transcript at specified positions relative to the 5' end. The percentage of rare codons in the open reading frame (%) is indicated on top. (B) Effect of chloramphenicol (Cm) on transcription elongation rate. Panels display representative induction curves used to calculate the elongation rate. At time zero, IPTG was added. RNA was extracted at the indicated times after induction and used for parallel dot blot hybridization with the early (probe 1) and late (probe 2) lac probes (fig. S1A). The distance between probes divided by the time of the linear increase in the hybridization signal between probes 1 and 2 (shown by arrows) gives the elongation rate. Radioactivity at time 0 (before IPTG induction) was taken as background and subtracted from all values. (C) Effect of the slow ribosome mutant (SmP) and streptomycin (Sm) on transcription elongation rate. Panels display representative induction curves used to calculate the elongation rate. Corresponding dot blots of the early (probe 1) and late (probe 2) probes are shown in fig. S1B. (D) Rate of transcription elongation as a function of codon usage. Schematics of the test plasmids with the percentage of rare codons in the open reading frame (%) are shown on top. Two representative induction curves used to calculate the elongation rate are displayed. The amount of radioactivity in each probe was divided by the corresponding radioactivity in the bla probe, and the ratio is plotted against sampling time. Corresponding dot blots are shown in fig. S1C. The distance between the probe and the start of transcription divided by the time of linear increase in the hybridization signal (shown by arrows) gives the elongation rate.
To provide further support for this conclusion and to rule out any potential indirect effect of Cm on transcription, we took advantage of a bacterial strain carrying a chromosomal mutation in the rpsL gene that renders the ribosome inherently slow (7). Importantly, the slow translation phenotype of this mutant (CH184 cells) can be partially recovered by the antibiotic streptomycin (Sm), which also restores the growth rate of the mutant strain (7). Indeed, our b-gal measurements indicate that without Sm the translation elongation rate of CH184 was 6 aa/s, and in the presence of 100 μg/ml Sm - 10 aa/s (Table 1; fig. S2), consistent with a previous study (7). Remarkably, the speed of transcription elongation in CH184 was very slow (only 19 nt/s), but accelerated to 30 nt/s upon addition of Sm (Fig. 1C), perfectly matching the translation elongation rates with and without Sm (Table 1). The opposite effect of translation antibiotics (Cm and Sm) on transcription elongation demonstrates a strong reliance of RNAP speed on that of a ribosome.
Codon usage affects the rate of translation in bacteria and other organisms (8–10). Rare codons, which pair to less abundant tRNA isoacceptors, delay the progression of a ribosome and often compromise protein expression (11–13). If the transcriptional rate relies so heavily on translation, the codon-reading program encoded by each individual gene should determine the speed of RNAP. To test this hypothesis, we compared the rates of transcription elongation at several genes with different rare codon frequency. The E.coli genes rplB and tufA have a relatively small number of rare codons (Table 2; fig. S3). In contrast, a foreign gene (srb4), which encodes a subunit of yeast Mediator, is characterized by a high frequency of rare codons (Table 2; fig. S3). Open reading frames of the rplB-tufA fusion and srb4, which are the same in length (2 kb), were inserted upstream of lacZ in the same test vector utilized in our previous experiments (Fig. 1D). Remarkably, transcription of rplB-tufA was more than 1.5 fold faster than that of srb4 (Fig. 2D, Table 2), matching its fast translation rate (Table 2). Moreover, genes carrying the intermediate amount of rare codons (lacZ and infB) displayed predictably intermediary rates of transcription (Table 2): slower than rplB-tufA, but faster than srb4. These results implicate codon usage as a key determinant of transcription rate.
Table 2.
Inverse correlation between rare codon frequency and transcription elongation rates at individual genes.
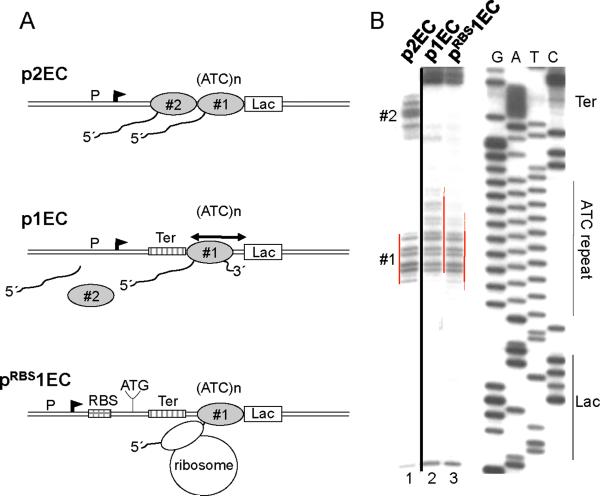
Fig. 2. The trailing ribosome inhibits RNAP backtracking.
(A) Diagram describing p2EC, p1EC and pRBS1EC constructs and footprinting results of (B). The leading EC1 is halted by the lac repressor within the ATC repeated sequence (ATC)n. The trpT terminator (Ter) is positioned so that the trailing EC (EC2) stalled at the termination site dissociates completely. (B) Primer extension analyses of in situ CAA modifications of the non-template strand in p2EC (line 1), p1EC (line 2), and pRBS1EC (line3). The locations of the Lac-operator (lac), ATC repeat, terminator (Ter), EC1, and EC2 are indicated. Red lines show the dynamic position of the EC1 transcription bubble. The presence of EC2 or trailing ribosome shifts the backtracked EC1 forward. The space between the RBS and blocked EC1 allows for only one ribosome to be engaged in translation.
Previously we showed that the rate of transcription elongation depends on the efficiency of initiation, i.e. promoter strength (4, 14). The mechanism underlying this phenomenon was explained by the cumulative anti-backtracking effect of trailing RNAPs on leading elongation complexes (EC) (fig. S4). Backtracking or spontaneous reverse sliding of the EC along DNA and RNA occurs frequently in vitro and usually determines the overall elongation rate (4, 14). During backtracking the 3' end of RNA is disengaged from the RNAP catalytic site, thereby causing temporal (pausing) or permanent EC inactivation (15, 16). Reactivation of the EC in vitro can be promoted by trailing ECs that rescue leading backtracked ECs by pushing them forward (4, 14). The trailing ribosome could act in a similar way, by “pushing” backtracked ECs forward, thereby controlling the rate of transcription (fig. S4).
To test this hypothesis we developed an assay in which the effect of a ribosome on RNAP backtracking could be directly monitored in vivo. We constructed a set of plasmids in which RNAP initiated from a constitutive promoter is halted at a downstream site by the lac repressor bound to its operator motif (Fig. 2A). Previously we showed that RNAP backtracks upon collision with the repressor in vitro and in vivo, which restricts its ability to read through the roadblock (14). To monitor the changes in positioning of the blocked EC in response to translation, we utilized in situ footprinting of the transcription bubble with the single-strand-specific probe, chloroacetaldehyde (CAA) (Fig. 2B) (14). The plasmids were designed so that the repressor halts either two ECs transcribing in tandem (p2EC) or only one isolated EC (p1EC) (Fig. 2). In the latter case, the trp terminator was placed between the promoter and the repressor-binding site to completely terminate the trailing EC upon stalling (14). A derivative of p1EC (pRBS1EC) was also made, which contained the strong T7 g10 RBS 120 nt upstream of the repressor-binding site (Fig. 2A). Analysis of CAA modifications on the non-template strand of p2EC and p1EC revealed two ECs (#1 and #2) and one EC (#1) between the promoter and the repressor, respectively (Fig. 2B). CAA signals from #1 and #2 ECs disappear upon IPTG addition, demonstrating that in each case the lac repressor blocked the ECs (14). CAA footprinting revealed a significant difference in positioning of EC#1 on each plasmid. Clearly, isolated EC#1 in p1EC backtracks over a longer distance than EC#1 in pRBS1EC (or in p2EC), since new CAA reactive sites are detected upstream and the reactivity of the downstream margin of the footprint is strongly decreased (Fig. 2B, compare lanes 2 and 3). We conclude that similar to the situation with trailing ECs (lane 1) (14), the trailing ribosome prevents backtracking of the leading EC.
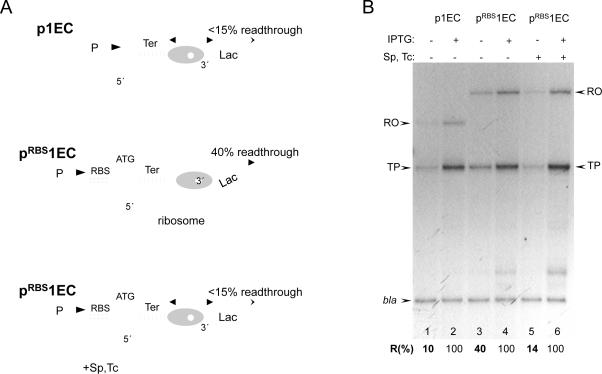
Because trailing ECs help leading ECs traverse the roadblock by inhibiting backtracking (14), the inhibition of backtracking by a ribosome should also facilitate transcriptional read-through. This premise was tested directly by comparing the amounts of downstream cat mRNA generated from pRBS1EC and p1EC plasmids and also with and without the translation inhibitors spectinomycin (Sp) and tetracycline (Tc) (Fig. 3A). Cat message accumulation was measured by reverse transcriptase primer extension and normalized to the plasmid-encoded b-lactamase transcript (bla) (Fig. 3B). Without RBS, only a small fraction (~10%) of ECs transcribed beyond the roadblock (lane 1). However, the level of read-through increased considerably (to 40%) in the case of the RBS-containing plasmid (lanes 3). Moreover, the ribosome-targeting antibiotics reversed the stimulating effect of RBS on transcriptional read-through (lane 5). Because the space between the blocked RNAP and RBS can accommodate only one ribosome at a time, this analysis revealed direct in vivo cooperation between a single moving ribosome and a single moving RNAP in overcoming the transcriptional roadblock.
Fig. 3. Cooperation between the ribosome and leading RNAP in overcoming a transcriptional roadblock in vivo.
(A) Diagram describing the cooperative mechanism and the results of (B). The ribosome reactivates the blocked EC (which is predominantly backtracked) by pushing it forward. The activated EC is now able to traverse the lac repressor as soon as the latter dissociates. Inhibition of translation by antibiotics (Sp and Tc) disrupts cooperation, thereby inhibiting RNAP readthrough. (B) Stimulating effect of translation on transcriptional readthrough of the lac repressor in vivo. Autoradiogram shows the RT extension products obtained from 32P primers hybridized to either cat or bla transcripts. p1EC and pRBS1EC template plasmids and experimental conditions were as in Fig. 2. The ribosome inhibitors Sp and Tc were added as indicated (lanes 5, 6). RO and TP stand for the full size (runoff) reverse transcription product and the product interrupted at the base of the trp terminator hairpin, respectively. The efficiency of transcriptional readthrough (%R) was calculated as a fraction of extension products (RO+TP) normalized to 100% readthrough in the presence of IPTG. The results are the mean from three independent experiments (p<0.05).
RNAP and ribosomes are two molecular motors with fundamentally different working cycles. Although both enzymes utilize Brownian ratchet principles in converting thermal energy into movement (17–19), translocation of a ribosome is virtually irreversible, whereas RNAP oscillates back and forth along RNA and DNA at numerous sites in vitro (15, 16, 19, 20). Such an equilibrium between productive and non-productive (pre-translocated or backtracking) states determines the pace of transcription elongation (19). Elongation factors, such as NusG or RfaH, shift the equilibrium towards the productive (post-translocated) state. They accelerate transcription by changing the intrinsic translocation properties of RNAP (19, 21). In contrast, a moving ribosome appears to control transcription “mechanically” by physically blocking backtracking (Fig. 2; fig. S4) (22). Such cooperation between RNAP and a ribosome is reminiscent of the situation with queuing ECs. Trailing ECs help leading ECs to overcome backtracking-type elongation blocks in vitro and in vivo (4, 14, 23). The cooperation effect in this case is proportional to the promoter strength (4), hence it works particularly well at highly expressed genes, such as ribosomal and other stable RNA genes. The majority of genes, however, have relatively weak promoters which compromise RNAP cooperation at those operons. The backtracking-prone elongation by RNAP, therefore, provides a means for precise adjustment to the rate of translation by a mechanical coupling with a moving ribosome.
Indeed, we observed a perfect match between translation and transcription rates under various growth conditions including variations in carbon sources and growth phase transition (Table 1; fig. S5–S7). In the stationary phase transcription and translation rates decelerate synchronously in “wild type” (MG1655) or “slow ribosome” (CH184) cells (fig. S6 and S7, Table 1). Moreover, Sm accelerated both transcription and translation in the stationary growing CH184 to the levels that of the stationary growing MG1655 (fig. S7, Table 1). Thus, the transcription elongation rate remains under tight translational control throughout bacterial growth. To coordinate translation and transcription rates so precisely at individual genes (each with a unique combination of rare codons, RNA secondary structure, and other parameters affecting translation) is a great challenge, unless RNAP progression relies on a ribosome itself. Not only does this cooperation save energy by limiting any excessive transcripts that cannot be translated in a timely manner, but it also prevents premature Rho termination by ensuring continuous coupling between transcription and translation.
In summary, the moving ribosome directly controls the rate of transcription by preventing RNAP from spontaneous backtracking. Because of such cooperation, the rate of transcription is determined by codon usage and nutrient availability sensed by the ribosome. As a result, there is a precise adjustment of transcriptional yield to translational needs under various growth conditions. Macromolecule trafficking and cooperation (fig. S4) is thus the fundamental mechanism of bacterial gene regulation and adaptation to environmental changes.
Supplementary Material
Acknowledgments
We thank Dr. Diarmaid Hughes for the CH184 (SmP) strain. This work was supported by grants from the NIH and Dynasty Foundation (E.N.).
References and Notes
- 1.Adhya S, Gottesman M. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- 2.Richardson JP. Cell. 1991;64:1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- 3.Yanofsky C. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 4.Epshtein V, Nudler E. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 5.Vogel U, Jensen KF. J Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohman K, Ruusala T, Jelenc PC, Kurland CG. Mol. Gen. Genet. 1984;198:90–99. doi: 10.1007/BF00328706. [DOI] [PubMed] [Google Scholar]
- 7.Ruusala T, Andersson D, Ehrenberg M, Kurland CG. EMBO J. 1984;3:2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong H, Nilsson L, Kurland CG. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 9.Lavner Y, Kotlar D. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 10.Elf J, Nilsson D, Tenson T, Ehrenberg M. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 11.Jansen R, Bussemaker HJ, Gerstein M. Nucleic Acids Res. 2003;31:2242–2251. doi: 10.1093/nar/gkg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angov E, Hillier CJ, Kincaid RL, Lyon JA. PLoS One. 2008;3:e2189. doi: 10.1371/journal.pone.0002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, Ignatova Z. PLoS One. 2009;4:e5036. doi: 10.1371/journal.pone.0005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 16.Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 17.Ramakrishnan V. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 18.Steitz TA. Nat Rev Mol Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 19.Bar-Nahum G, Epshtein V, Ruckenstein AE, Mustaev A, Nudler E. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 20.Nudler E. Annu Rev Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Nucleic Acids Res. 2007;35:5694–705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.In addition to preventing backtracking-type pauses, the ribosome can also suppress pausing induced by RNA secondary structures. However, the hairpin-mediated pauses are rare compared to ubiquitous backtracking-type pauses (25). They have been detected only within the leader sequences of a few metabolic genes (3, 26) and require specific sequences upstream and downstream of the pause hairpin (27). Usually, hairpins do not induce pausing. For example, strong termination hairpins do not pause RNAP (28) (fig. S8). Instead, it is a poly-T stretch of the terminator that induces backtracking-type pauses at the site of termination (28). Also, factors that suppress RNAP backtracking (e.g. NusG and RfaH) accelerate the overall elongation rate (19, 21) (fig. S8). Therefore, it is unlikely that random secondary structures affect the overall elongation rate to the same extent as ubiquitous backtracking-type pauses in vivo.
- 23.Saeki H, Svejstrup J. Mol Cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varenne S, Buc J, Lloubes R, Lazdunski C. J Mol Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 25.Depken M, et al. Biophys J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henkin T, Yanofsky C. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 27.Chan C, Landick R. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 28.Gusarov I, Nudler E. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.