Abstract
To better understand the structure and function of Z lines, we used sarcomeric isoforms of α-actinin and γ-filamin to screen a human skeletal muscle cDNA library for interacting proteins by using the yeast two-hybrid system. Here we describe myozenin (MYOZ), an α-actinin- and γ-filamin-binding Z line protein expressed predominantly in skeletal muscle. Myozenin is predicted to be a 32-kDa, globular protein with a central glycine-rich domain flanked by α-helical regions with no strong homologies to any known genes. The MYOZ gene has six exons and maps to human chromosome 10q22.1-q22.2. Northern blot analysis demonstrated that this transcript is expressed primarily in skeletal muscle with significantly lower levels of expression in several other tissues. Antimyozenin antisera stain skeletal muscle in a sarcomeric pattern indistinguishable from that seen by using antibodies for α-actinin, and immunogold electron microscopy confirms localization specifically to Z lines. Thus, myozenin is a skeletal muscle Z line protein that may be a good candidate gene for limb-girdle muscular dystrophy or other neuromuscular disorders.
The α-actinins are a family of actin-binding proteins evolutionarily related to dystrophin and the spectrins. Family members are characterized by an N-terminal actin-binding domain, a central rod region composed of spectrin-like triple-α-helical repeated units, and an EF hand calcium-binding domain that is functional in nonmuscle α-actinin but degenerate in the spectrins, muscle α-actinins, and dystrophin (1, 2). Biochemically, the primary function of α-actinin dimers is F-actin binding and crosslinking. In skeletal and cardiac muscle, α-actinin isoforms encoded by the ACTN2 and ACTN3 genes (3) are major components of the Z lines and intercalated discs where they function to anchor the actin/nebulin-containing thin filaments in a constitutive manner.
The filamins represent another family of cytoskeletal proteins whose actin-binding domains are related evolutionarily to those of the α-actinins, spectrins, and dystrophin (4). In skeletal muscle, the FLNC gene (previously γFLN, ABP-L, FLN2) encodes γ-filamin that is localized to Z lines and I bands immediately adjacent to the Z lines (5, 6). Cytoskeletal (nonmuscle) isoforms of filamin dimerize to crosslink actin filaments in a loosely spaced network of roughly 50-nm periodicity. In skeletal muscle, γ-filamin may play a similar role in regulating the spacing of F-actin-containing thin filaments.
Although it is clear that α-actinin and γ-filamin are important structural components, the composition of Z lines is complex and still not well known (reviewed in ref. 7). Recently, several new Z line proteins, including telethonin (8), myotilin (9), cypher (10), ZASP (11), Nspl1 (12), and the actinin-associated LIM proteins (13, 14), have been described. Although α-actinin is known to interact directly with many of these, less is known about γ-filamin at the Z line, and it is highly likely that additional Z line components remain to be identified.
To further understand Z line structure and function, and to identify additional candidate genes for human neuromuscular disease, we have conducted a yeast two-hybrid screen of human skeletal muscle cDNAs by using portions of α-actinin-2 and -3 as well as γ-filamin as “bait.” Here we report the identification and characterization of a novel α-actinin- and γ-filamin-binding Z line protein named “myozenin.” While this manuscript was in preparation, another group reported this same protein, using the name “FATZ” (15).
Methods
Yeast Two-Hybrid Assays.
All amino acid numbering is based on the following GenBank reference sequences: α-actinin-2, M86406; γ-filamin, NM_001458; and myozenin, AF240633. Subclones of the human ACTN2, ACTN3, and FLNC genes (3, 5) were constructed in pGBT9 and used in yeast GAL4 two-hybrid assays to screen a human skeletal muscle MATCHMAKER-1 library cloned into pGAD10 (16). HF7c yeast were transformed sequentially with α-actinin-containing “bait” clones followed by the cDNA library. Assays for GAL4-HIS3 and GAL4-lacZ activity followed standard company protocols (CLONTECH). Selection in synthetic minimal media (SD) lacking Trp, Leu, and His for double transformants that also activated their GAL4-HIS3 genes was done in the presence of 25 mM 3-amino-1,2,4-triazole (Sigma). To confirm positive interactions, α-actinin sequences were subcloned into pGAD424, library-derived “prey” inserts were subcloned into pGBT9, and interactions were assayed by cotransformation into SFY526 yeast followed by assay for HIS3 and lacZ activity as above.
Myozenin cDNA and Gene Characterization.
All DNA sequences were analyzed on ABI 373 or ABI PRISM 377 automated sequencer by using dideoxy-fluorescent dye terminator chemistry (Applied Biosystems). Sequences were assembled by using SEQUENCHER 3.1.1 software (Gene Code, Ann Arbor, MI). Subsequent amino acid prediction and sequence analyses were performed by using MACVECTOR 6.5.3 software (Oxford Molecular Group, Oxford, U.K.). All positive pGAD10 clones were sequenced partially from both ends by using pGAD10 sequencing primers (CLONTECH). The largest (full length) myozenin cDNA clone (3c8) was sequenced in its entirety on both strands by using custom gene-specific primers. To identify additional sequences, 5′ RACE was performed on “Marathon-Ready” human skeletal muscle cDNA by following the company protocol (CLONTECH). Database searches used blast and fasta programs available through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the DNA Database of Japan (http://www.ddbj.nig.ac.jp/Welcome-e.html). Human myozenin genomic organization was determined by long-range genomic PCR across introns by using cDNA-based primers and LA Taq polymerase (Takara Shuzo, Kyoto) for long accurate genomic PCR, followed by DNA sequencing to determine intron–exon boundaries. Multiple-tissue Northern blots (CLONTECH) were hybridized according to the manufacturer's protocol with the full-length 3c8-insert labeled by using the random priming method.
Antibodies.
Rabbit antimyozenin antisera, produced by Research Genetics (Huntsville, AL), were raised against synthetic peptides corresponding to the N-terminal 17 amino acids of human myozenin (MPLSGTPAPNKKRKSSK) synthesized by the multiple antigenic peptide method (ref. 17; 3c8Na) and to the myozenin C-terminal 18 amino acids (VDYNVDIGIPLDGETEEL) conjugated to keyhole limpet hemocyanin (3c8Ca). Each immunogen was injected into two rabbits. All animals generated strong antipeptide responses as judged by ELISA, and both pairs of replicate antisera exhibited similar patterns of reactivity (data not shown). Isoform-specific anti-α-actinin-2 and -3 and γ-filamin Abs have been described (5, 18–20).
In Vitro Transcription and Translation (IVTT) and SDS/PAGE.
The proteins α-actinin-2 or -3 and myozenin were produced by IVTT of the complete coding sequences cloned into the expression vectors pMGT1 and pFHR3 (21) by using the TNT T7-coupled reticulocyte lysate system (Promega) in 50-μl reactions per the manufacturer's protocol. The C-terminal region (residues 2,191–2,705) of γ-filamin was expressed from clone 2–14, and partial dystrophin fragments were as described (5, 22). pFHR3 introduces an N-terminal FLAG nonapeptide (MDYKDDDDK) epitope-tag that was recognized by using M2 Abs (Eastman Kodak). When indicated, some reactions were carried out in a reaction buffer in which 40 μCi of l-(methyl-35S) Met (>1,000 Ci/mmol; Amersham Pharmacia) was added to a 50-μl reaction mix lacking Met. SDS/PAGE analysis used 10–20% (wt/vol) gradient gels (Invitrogen). Gels were fixed and dried before autoradiography on storage phosphor plates for several days. Plates were scanned by PhosphorImager and analyzed with imagequant software (Molecular Dynamics).
Coimmunoprecipitation (CoIP) Assays.
CoIP assays were performed on 35S-labeled proteins taken directly from the above IVTT reactions. Ten-microliter total IVTT products (equal amounts of each protein) were incubated with 20 μl of TBST buffer containing 10 mM Tris-buffered saline (pH 8.0), 0.1% Tween-20, and 150 mM NaCl. After a 2-hour incubation on ice, 2 μl of precipitating Ab [anti-α-actinin-2 or anti-α-actinin-3 specific polyclonal Ab (18), antifilamin “FLN2-A1” (5), antimyozenin “3c8Ca”, or anti-FLAG “M2” monoclonal Ab] was added with 20 μl of TBST. After a 1-hour incubation on ice, 50 μl of 50% (vol/vol) suspension of protein G-Sepharose (Sigma) was added and incubated for 30 min. Beads were centrifuged, and the supernatant was removed. Beads were washed three times with 1 ml of TBST and analyzed by SDS/PAGE and autoradiography as above.
Tissue Extracts and Blot-Overlay Assays.
Human tissue extracts were prepared by cryosectioning multiple 25-μm sections of frozen-unfixed tissue and mixing with sample buffer [62.5 mM Tris-buffered saline (pH 6.8)/2.5% (vol/vol) SDS/10% (vol/vol) glycerol/50 mM DTT/10 mM EDTA/1.5 μg/ml aprotinin/1.25 μg/ml leupeptin/40 μg/ml PMSF/1 μg/ml pepstatin/3 μl/ml saturated bromophenol blue/2% (vol/vol) β-mercaptoethanol], quantified by the amido black method (23), and boiled immediately before loading. Nonradioactively labeled IVTT products or tissue extracts (2–15 μg per lane) were subjected to SDS/PAGE and blotted onto PROTRAN nitrocellulose membrane (Schleicher & Schuell). After overnight treatment with blocking buffer [0.1% gelatin/5% (vol/vol) BSA/0.1% Tween-20 in 1 × PBS] at 4°C, membranes were incubated with 35S-Met-labeled IVTT protein products in blot-overlay buffer [150 mM NaCl/20 mM Hepes/2 mM MgCl2/5% (vol/vol) BSA] at a concentration of 10 μl of IVTT mixture per milliliter of blot-overlay buffer for 4 h at 4°C. Membranes were washed with 0.4–0.6% Tween-20/150 mM NaCl/20 mM Hepes/2 mM MgCl2/5% (vol/vol) BSA. Dried blots were exposed to PhosphorImager plates for various time periods, and individual bands were quantified by using imagequant software (Molecular Dynamics).
Immunohistochemistry, Confocal, and Immunoelectron Microscopy.
Confocal and immunogold electron microscopy of normal human skeletal muscle was performed essentially as described (5). Primary culture of human myoblasts (24) and indirect immunofluorescence microscopy of these and human nemaline myopathy skeletal muscle were as described (19, 25). Rabbit anti3c8Ca and anti3c8Na antisera were diluted 1:500. Mouse monoclonal antifast myosin (My-32, Sigma M4276) and antisarcomeric tropomyosin (Sigma T9283) were diluted 1:200 and 1:100. Cy3, Texas red, and fluorescein-labeled secondary Abs (Jackson ImmunoResearch) were diluted 1:200.
Results
Identification and Cloning of a Gene for a Skeletal Muscle α-Actinin and γ-Filamin-Binding Protein.
To identify α-actinin-binding proteins in skeletal muscle, we constructed a series of pGBT9 subclones containing portions of the rod or C-terminal EF hand domains of α-actinin-2 and -3 (3). Larger GAL4-DNA-binding domain fusion clones encoding the rod and EF hand domains of both proteins all selfactivated the GAL4-controlled reporter genes in the absence of GAL4 activation domain-containing plasmids, as did any clones containing the α-actinin-2 EF hand or the α-actinin-3 rod domains. However, a pGBT9 subclone encoding the α-actinin-2 rod domain (clone pGBT9–3c, residues 274–757) did not self activate and was used to screen an adult human skeletal muscle cDNA library cloned into pGAD10. A screen of 8.5 × 105 pGBT9–3c and pGAD10 cDNA library double transformants yielded 50 HIS3-positive colonies, of which 11 also strongly expressed lacZ. Inserts from each interacting clone were sequenced partially, and all were found multiple times in human expressed sequence tag (EST) databases. One of the pGBT9–3c-positive clones contained a cDNA fragment of α-actinin-2, confirming that the assay was working properly as α-actinin is known to form both homo- and heterodimers (18). In addition, this screen identified 10 clones representing three potentially interacting partners designated 3c2 (1 clone), 3c8 (8 clones), and 3c41 (1 clone). None of these clones selfactivated in the absence of the actinin-containing pGBT9 clones, nor did they interact with a control plasmid, pVA3, containing part of murine p53 in pGBT9. Preliminary DNA sequence analysis revealed that 3c41 encoded a portion of synaptopodin, an actin-binding protein at synapses and glomerular podocytes (26), whereas 3c2 represented myopodin/genethonin 2, a related actin-binding protein of skeletal muscle (27). Transcript 3c8 represented a gene with no existing database entries except for ESTs.
In a parallel yeast two-hybrid screen, a C-terminal portion of the γ-filamin gene, FLNC (clone 2–14; ref. 5), cloned into pGBT9, was used to screen the same skeletal muscle cDNA library. Of 5 × 105 double transformants, 13 were HIS3- and lacZ-positive. DNA sequence analysis revealed that six of these potential γ-filamin-interacting clones also contained portions of the 3c8 transcript.
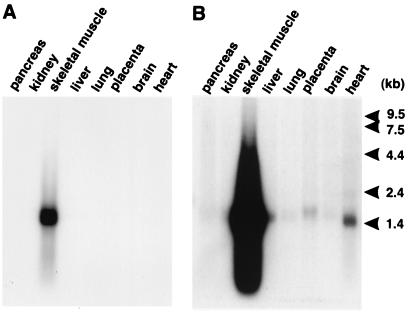
Thus, independent screens by using ACTN2 rod domain and FLNC C terminus bait clones identified eight and six clones of transcript 3c8, respectively, as potential binding partners for these two sarcomeric proteins. Sequence analysis of the largest clones and 5′ RACE products revealed a 1,266-bp 3c8 transcript containing an ORF bound by a potential Kozak consensus initiation codon (ATG) at nucleotides 68–70 and a termination codon (TGA) at nucleotides 965–967. This mRNA is predicted to encode a 32-kDa protein, hereafter referred to as “myozenin” (gene symbol “MYOZ,” GenBank accession no. AF240633; LocusLink ID 58529; www.ncbi.nlm.nih.gov/LocusLink/index.html). A homologous EST in the public database (HSPD03874) has been radiation hybrid mapped to chromosome 10q22.1, approximately 8.56 centirays from WI-2997, making this the likely locus for MYOZ (ref. 28; http://grup.bio.unipd.it/muscle/). Myozenin contains 299 amino acids with no significant homology to any previously described proteins or domains on blast searches of GenBank and ProDom databases (29). Secondary-structure predictions suggest the presence of two α-helical domains consisting of amino acid residues 1–72 and 187–244 (approximately) with a flexible glycine-rich region in between (32 of 81 residues between positions 95 and 175), and a C-terminal tail of approximately 55 amino acids (Fig. 1A). A predictprotein (http://dodo.cpmc.columbia.edu/predictprotein/) estimate of globularity suggests that myozenin is a compact, globular protein without significant coiled-coil regions (30). Northern blot analysis identified a single skeletal muscle-specific transcript of apparently 1.5 kb; however, long exposures detected this, or related, mRNAs in several other tissues, including cardiac muscle and placenta, where the size appeared to be slightly larger (Fig. 2).
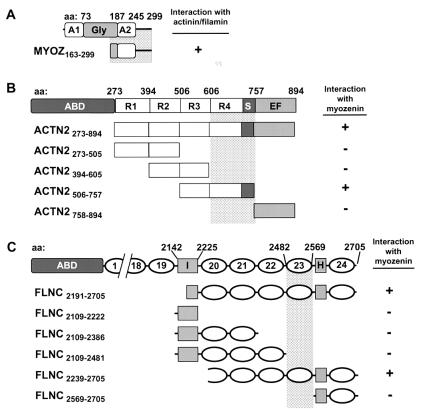
Figure 1.
Schematic diagrams (Left) and results of yeast two-hybrid analysis with deletion mutants (Right). Amino acid (aa) numbers at domain junctions are shown above schematics and amino acid residues contained in each deletion construct are indicated in clone name (Lower Left). Minimum critical regions necessary for interaction with binding partners are indicated by shaded bar. (A) Myozenin structure illustrating two putative α-helical domains (A1 and A2) and central glycine-rich domain (Gly). (B) α-Actinin-2 structure showing actin-binding domain (ABD), central repeats (R1–R4), spacer (S), and EF hand domain (EF). (C) γ-Filamin schematic illustrating ABD, repeats (numbered), repeats 19 and 20 interdomain insertion (I), and hinge region (H).
Figure 2.
Northern blot analysis of MYOZ illustrating predominant expression in skeletal muscle on short exposure (A) and lower-level expression (and/or cross-hybridization to the related locus) in heart, placenta, and other tissues on long exposure (B).
The genomic structure of the MYOZ gene was determined by long-range genomic PCR across exons and sequencing of PCR products to determine intron–exon borders. Additional 5′ end sequences were identified in two genomic sequences (GenBank accession nos. AQ343349 and AC073389), deposited as part of the Human Genome Project. Accession no. AC073389 maps to chromosome 10q22.2 in the September 2000 draft version of the human genome (http://genome.ucsc.edu/goldenPath/hgTracks.html). The MYOZ gene contains six exons ranging in size from under 100 to 531 base pairs and spanning approximately 10 kb (GenBank accession nos. AF243386–AF243390) (Table 1). The ORF begins 18 base pairs into exon 2 and extends to the middle of exon six.
Table 1.
Exon-intron boundary sequence* of human MYZN gene
| Exon no. | Exon length, bp | Splice acceptor | Splice donor | Intron no. | Intron length† |
|---|---|---|---|---|---|
| 1 | 49 | TCCAAAGCTG | CGAGCAGCCGgtaagtctccagctc | 1 | 1.8 kb |
| 2 | 91 | tcccaccctgtccagGCTGAATCCA | CTCACTGGAGgtaggacatgtttgc | 2 | 3.0 kb |
| 3 | 179 | ttgcctccctgaaagGTGGACAGGA | CAGCTCAATGgtgagtctggaactt | 3 | 3.5 kb |
| 4 | 250 | ttgtcttcttttcagGATCACTTCC | ACAGGATCAGgcaagtacactcacc | 4 | 418 bp |
| 5 | 166 | tcccatggctaccagGAGACCAGGC | CCTTCAACAGgtaggggtggtgagg | 5 | 2.5 kb |
| 6 | 509(−1245) | gtgtccccttctcagGACGGCAATG | GAAAATAAAG |
Lowercase letters refer to intron sequences, and uppercase letters refer to exon sequences.
Intron sizes were estimated by gel analysis and/or direct sequencing of genomic interexon PCR products.
blast homology searches of all available databases identified myozenin orthologs in human, bovine, porcine, murine, and rat genomes but not in any nonmammalian species. Furthermore, an apparently paralogous human gene (UniGene cluster Hs.42346, http://www.ncbi.nlm/UniGene/) was identified as having a 29% identity and, of the remaining residues, a 20% similarity, to myozenin with its greatest homologies at the N- and C-terminal α-helical regions.
Localization of Binding Sites in Myozenin, α-Actinin, and γ-Filamin.
Each of the original yeast two-hybrid assays by using α-actinin and γ-filamin baits pulled out both full-length and partial cDNA clones of myozenin, all of which encoded at least the C terminus of myozenin. The smallest clones from each screen encoded amino acids 163–299, suggesting that this 15-kDa C-terminal region includes the likely α-actinin- and γ-filamin-binding site or sites (Fig. 1A). To confirm and localize the binding sites for myozenin interactions with α-actinin and γ-filamin in the yeast two-hybrid system, a series of pGAD424 α-actinin and γ-filamin subclones was generated, and the 3c8 insert was “swapped” from pGAD10 to pGBT9. Both interactions between larger clones were reproducible, confirming that myozenin binds both α-actinin and γ-filamin in the yeast two-hybrid system. The deletion analysis suggests that sequences necessary for myozenin binding to α-actinin-2 reside in the fourth repeat of the central rod domain and/or the spacer region before the EF hands, whereas sequences in repeat 23 of γ-filamin are essential for myozenin binding to that protein (Fig. 1).
Myozenin Competitively Binds Both Skeletal Muscle Isoforms of α-Actinin as Well as γ-Filamin in Vitro.
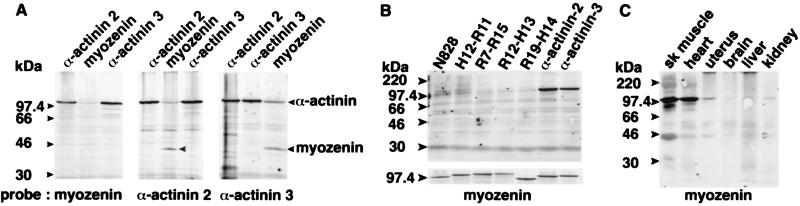
To extend and further confirm these in vivo data, portions of myozenin, α-actinin-2, and γ-filamin were cloned into expression vectors for production of recombinant proteins by IVTT. Blot-overlay assays by using radiolabeled α-actinin-2 and -3 showed that these proteins bound to themselves, to each other, and to myozenin (Fig. 3A). In contrast, the myozenin probe bound to both α-actinin-2 and -3 but not to itself or to portions of the dystrophin rod domain, which has evolutionary homology to the α-actinins (Fig. 3 A and B). When radiolabeled myozenin was used to probe total tissue lysates, it predominantly bound a ≈100-kDa protein in skeletal and cardiac muscle (likely α-actinin-2 and -3) and weakly labeled a similarly sized protein (probably α-actinin-1 and -4) in uterus (smooth muscle), as well as other tissues to a lesser extent (Fig. 3C). Binding to the diffuse band at ≈43 kDa likely represents α-actin and may be specific or simply a mass effect reflecting the large amounts of this protein in skeletal muscle. No obvious binding to 265-kDa γ-filamin likely reflects the much smaller quantity of this protein in skeletal muscle (5).
Figure 3.
Blot-overlay analysis of myozenin binding to α-actinins. 35S-labeled probes are indicated below and nonradioactive IVTT products (A and B) or whole-tissue homogenates (C) loaded in each lane are indicated above A–C. (A) Myozenin binds to both α-actinin-2 and -3 but not to itself (Left), whereas α-actinin-2 and -3 both bind to myozenin as well as to themselves (Center and Right). Binding to α-actinin in myozenin lanes likely represents binding to α-actinin-1 and -4 produced from contaminating ACTN1 and ACTN4 mRNA in reticulocyte lysates. (B) Control experiment showing lack of myozenin binding to various partial dystrophin fragments: N828, N-terminal nos. 1–828; H12–R11, hinge no. 2–repeat no. 11; R7–R15, repeat nos. 7–15; R12–H13, repeat no. 12–hinge no. 3; and R19–H14, repeat no. 19–hinge no. 4. Parallel radiolabeled IVTT products of all clones are shown below to confirm production of expected size products. (C) Detection of myozenin-binding partners in human tissues by blot-overlay assay. Predominant binding is to a ≈100-kDa protein at expected size for α-actinin.
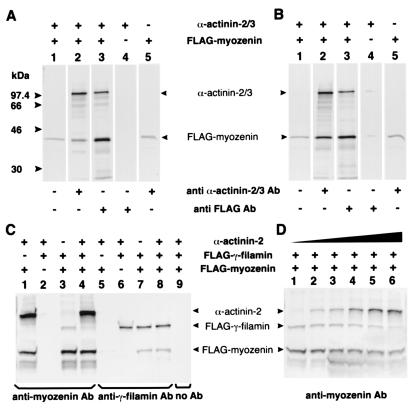
To further characterize these interactions, radiolabeled IVTT products of α-actinin, γ-filamin, and FLAG-tagged myozenin were used in CoIP assays. In separate experiments, myozenin coimmunoprecipitated with full-length products of both α-actinin-2 and -3 as well as with the C terminus of γ-filamin, and these results were reproducible regardless of which precipitating Ab was used (Fig. 4 A–C). Because the yeast two-hybrid data suggested that the same region of myozenin was involved in binding both α-actinin and γ-filamin, a competitive CoIP assay was performed where the amounts of myozenin and either γ-filamin or α-actinin were held constant whereas the third protein's concentration was varied. At low levels of α-actinin, antimyozenin antiserum coimmunoprecipitated both γ-filamin and α-actinin; however, CoIP of γ-filamin was related inversely to the amount of α-actinin (Fig. 4D), as was CoIP of α-actinin when γ-filamin was varied (data not shown), demonstrating a competitive interaction between these two proteins for binding to myozenin.
Figure 4.
CoIP of IVTT myozenin, α-actinin, and γ-filamin. Presence (+) or absence (−) of 35S-labeled IVTT products in each reaction is shown above and precipitating Abs are indicated below. Identity of each precipitated protein is indicated (Middle). A and B illustrate CoIP of full-length α-actinins-2 and -3, respectively, with FLAG-tagged full-length myozenin by using either anti-α-actinin or anti-FLAG Abs (lanes 2 and 3). Each Ab is specific (lanes 4 and 5); however, there is some degree of aggregation and nonspecific precipitation of IVTT myozenin (lanes 1 and 5). (C) CoIP of myozenin, α-actinin, and γ-filamin (FLNC2191–2705) by using specific antimyozenin and antifilamin Abs. Myozenin coimmunoprecipitates both α-actinin-2 (lane 1) and γ-filamin (lane 3). When equal volumes of all three IVTT products are combined, α-actinin CoIP predominates (lane 4). Use of anti-γ-filamin-precipitating Abs results in CoIP of myozenin but not α-actinin (lanes 5–8). Similarly, use of anti-α-actinin Abs coimmunoprecipitated myozenin but not γ-filamin (not shown), indicating a lack of direct interaction between α-actinin and γ-filamin. (D) Competitive CoIP of α-actinin and γ-filamin (FLNC2191–2705) by using specific antimyozenin Abs. Increasing relative amounts of α-actinin (lanes 1–6 contain 0-, 1-, 2-, 4-, 10-, and 20-fold levels, respectively) competitively decreased CoIP of γ-filamin. Similarly, increasing amounts of γ-filamin reduced CoIP of α-actinin (not shown).
Myozenin Colocalizes with α-Actinin and γ-Filamin at Skeletal Muscle Z Lines and on Nemaline Rods.
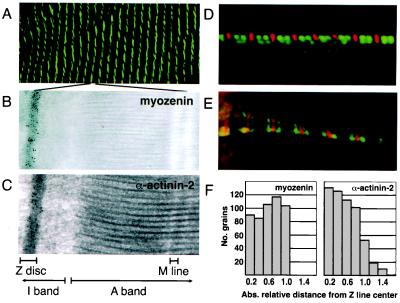
To study the subcellular distribution of myozenin in skeletal muscle, specific antimyozenin antisera were raised in rabbits immunized with either N- or C-terminal peptides. Two rabbits were immunized with each peptide, and all four resulting antisera gave similar results (data not shown). Indirect immunofluorescence assays on transverse sections of human skeletal muscle revealed that myozenin was expressed in the interior of all muscle fibers. On longitudinal sections, confocal microscopy revealed localization in a striped pattern characteristic of sarcomeric proteins (Fig. 5A). Double-label immunofluorescence microscopy for myozenin and either myosin or tropomyosin shows that this staining was restricted to the middle of the I bands, likely at the Z lines (Fig. 5 D and E).
Figure 5.
Subcellular localization of myozenin in human skeletal muscle. (A) Confocal indirect immunofluorescence microscopy of human skeletal muscle by using antimyozenin antisera showing striated staining pattern characteristic of sarcomeric proteins. (B and C) Immunogold transmission electron microscopy for myozenin (B) and α-actinin-2 (C) in human skeletal muscle showing predominant labeling at Z lines. (D and E) Double-label indirect immunofluorescence microscopy for myozenin (red) and myosin (D, green) or tropomyosin (E, green) in cultured human myotubes. (F) Quantitative grain measurements showing distribution of α-actinin-2 centered in middle of Z line (Right) and myozenin slightly skewed toward edges of Z lines (Left).
Immunogold transmission electron microscopy studies confirmed that myozenin is localized exclusively to Z lines in a pattern similar to that seen for α-actinin (Fig. 5 B and C). However, although both proteins appear localized throughout the entire width of the Z lines, quantitative grain measurements revealed that α-actinin is distributed with the majority of grains centered on the Z line, whereas myozenin distribution appears to be centered closer to the periphery of Z lines (Fig. 5F).
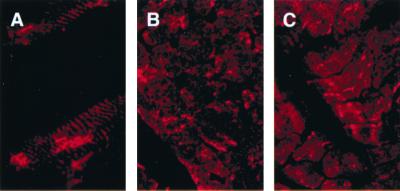
Nemaline myopathy is a congenital myopathy characterized by the presence of nemaline rods that contain large amounts of α-actinin and seem to be abnormal extensions of Z lines (31). To learn whether myozenin expression is perturbed similarly in nemaline myopathy, indirect immunofluorescence microscopy was performed by using anti-α-actinin-2 (19) and antimyozenin antisera. In a skeletal muscle biopsy from a patient with “typical” nemaline myopathy and nemaline rods in the majority of fibers, myozenin antisera labeled the rods in a pattern indistinguishable from that of α-actinin (Fig. 6).
Figure 6.
Indirect immunofluorescence analysis of α-actinin and myozenin in nemaline rods. Longitudinal (A) and cross sections (B) stained with antimyozenin Ab. (C) Cross section stained with anti-α-actinin-2 Ab. All Abs stain both Z lines as well as nemaline rods.
Discussion
Z line structure is complex, likely with many as-yet-uncharacterized protein constituents and interprotein interactions. Here we report the identification of myozenin, an α-actinin- and γ-filamin-binding protein of skeletal muscle. These interactions were identified by yeast two-hybrid assay and confirmed by blot overlay and CoIP experiments. Given that these three proteins also colocalize at or near Z lines, it is highly likely that they undergo biologically significant interactions in vivo.
Both α-actinin and filamin are capable of numerous interactions with other proteins, some of which involve overlapping binding sites (5, 9–11, 13, 14, 32, 33). For instance, the third and fourth spectrin-like repeats of α-actinin are involved in binding actinin-associated LIM protein (14), vinculin (34), and myotilin (9), although not all interactions may occur simultaneously. Here we have shown that myozenin also binds α-actinin in this region, likely somewhere within residues 606–757. This region has a 73% identity and, of the remaining residues, a 17% similarity, between α-actinin-2 and -3, supporting the observation that myozenin binds both isoforms effectively.
The apparent myozenin-binding site on γ-filamin resides within repeat 23 near the C terminus of the protein. This site is near, but distinct from, the muscle-specific repeat 20 interdomain insertion that binds myotilin and is important for localization of γ-filamin at Z lines (32). It is interesting to note that filamin repeat 24 contains sequences responsible for dimerization. Thus, one speculative role for myozenin may be to modulate both filamin and α-actinin dimerization, as the central repeats of the α-actinins are also the site of dimerization for those proteins (18).
Although it is important to bear in mind that the competitive CoIP experiments used only the C-terminal fifth of γ-filamin, it is not surprising that α-actinin and γ-filamin binding to myozenin appeared to be competitive, because both proteins interact with the C-terminal 137 amino acids of myozenin. This observation suggests that myozenin may be multifunctional, serving analogous but distinct roles in binding either α-actinin or γ-filamin, but not both simultaneously. All three proteins colocalize at or near Z lines; however, α-actinin is present throughout the Z line, whereas γ-filamin is distributed preferentially toward the periphery and within the I bands near the Z lines (5, 6). Although significant labeling for myozenin was not detected in the I bands, the distribution was shifted subtly toward the periphery of the Z lines, perhaps reflecting a γ-filamin-bound subpopulation at the edges of the Z lines.
Based on both the high frequency with which myozenin was isolated in two independent yeast two-hybrid assays and the results of Northern blot analysis, it appears to be an abundant transcript in skeletal muscle with no strong homologies to any known proteins except a single, uncharacterized paralogous muscle gene represented by UniGene cluster Hs.42346. Many muscle-specific proteins, such as actin, filamin, or α-actinin, are evolutionary adaptations of preexisting nonmuscle proteins involved in the control of cell shape or movement. However, myozenin is atypical in this regard, as it represents a class of truly muscle-specific proteins without known nonmuscle paralogs, suggesting that its function is unique to Z lines and discs in skeletal and cardiac muscle. The presence of numerous myozenin ESTs in most mammalian databases, but not in the Drosophila genome or existing EST databases of any other nonmammalian species, further argues that this protein represents a relatively recent evolutionary event, possibly providing unique functional characteristics to mammalian muscle.
The function of myozenin is unclear. It may simply act as an adapter molecule for binding of α-actinin or γ-filamin to telethonin or other Z line protein(s) (8, 15). Alternatively, its binding sites on α-actinin and γ-filamin suggest that it may influence dimerization of these proteins, perhaps modulating the spacing between monomers and hence spacing of actin-thin filaments. The possible direct interaction between myozenin and actin also requires further study.
The Z line localization and protein-binding characteristics of myozenin have parallels with telethonin and in particular myotilin, both of which have been implicated in the pathogenesis of limb-girdle muscular dystrophies (35, 36). Thus, myozenin is a particularly attractive candidate gene for human neuromuscular disease. Although all three of these proteins are present in nemaline rods, there is no direct evidence that mutations of any of these cause nemaline myopathy. Indeed, a myozenin mutation search of 50 patients with nemaline myopathy was negative (F.T. and A.H.B., unpublished data). Nevertheless, myozenin remains an important candidate gene for other myopathies and dystrophies.
Acknowledgments
We thank I. Dalkilic for many helpful interactions, Dr. E. Gussoni (Children's Hospital, Boston, MA) for a gift of primary human myoblasts, M. Sullivan for assistance with electron microscopy, Dr. Y. Chan for advice, and Dr. I. Nonaka for helpful discussions. This publication was made possible through generous support from the Joshua Frase Foundation and the Gimbel Foundation, by grants from the Muscular Dystrophy Association (to A.H.B., T.G.T, and S.C.W.), and by National Institutes of Health Grants R01 AR44345 and K02 AR02026 (to A.H.B.). The Core DNA sequencing facility was supported by National Institutes of Health Grant P30 HD18655. L.M.K. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- IVTT
in vitro transcription and translation
- CoIP
coimmunoprecipitation
- EST
expressed sequence tag
Note Added in Proof.
Myozenin is identical to calsarcin-2 (37).
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF240633 and AF243386–AF243390).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041609698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041609698
References
- 1.Blanchard A, Ohanian V, Critchley D R. J Muscle Res Cell Motil. 1989;10:280–289. doi: 10.1007/BF01758424. [DOI] [PubMed] [Google Scholar]
- 2.Hartwig J H. Protein Profile. 1994;1:706–778. [PubMed] [Google Scholar]
- 3.Beggs A H, Byers T J, Knoll J H, Boyce F M, Bruns G A, Kunkel L M. J Biol Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- 4.Noegel A A, Rapp S, Lottspeich F, Schleicher M, Stewart M. J Cell Biol. 1989;109:607–618. doi: 10.1083/jcb.109.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson T G, Chan Y M, Hack A A, Brosius M, Rajala M, Lidov H G, McNally E M, Watkins S, Kunkel L M. J Cell Biol. 2000;148:115–126. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Ven P F, Obermann W M, Lemke B, Gautel M, Weber K, Furst D O. Cell Motil Cytoskeleton. 2000;45:149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Stromer M H. Microsc Res Tech. 1995;31:95–105. doi: 10.1002/jemt.1070310202. [DOI] [PubMed] [Google Scholar]
- 8.Valle G, Faulkner G, De Antoni A, Pacchioni B, Pallavicini A, Pandolfo D, Tiso N, Toppo S, Trevisan S, Lanfranchi G. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- 9.Salmikangas P, Mykkanen O M, Gronholm M, Heiska L, Kere J, Carpen O. Hum Mol Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Ruiz-Lozano P, Martone M E, Chen J. J Biol Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
- 11.Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, et al. J Cell Biol. 1999;146:465–745. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisler J G, Palmer R J, Stubbs L J, Mucenski M L. J Muscle Res Cell Motil. 1999;20:661–668. doi: 10.1023/a:1005533013926. [DOI] [PubMed] [Google Scholar]
- 13.Pomies P, Macalma T, Beckerle M C. J Biol Chem. 1999;274:29242–29250. doi: 10.1074/jbc.274.41.29242. [DOI] [PubMed] [Google Scholar]
- 14.Xia H, Winokur S T, Kuo W L, Altherr M R, Bredt D S. J Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faulkner G, Pallavicini A, Comelli A, Salamon M, Bortoletto G, Ievolella C, Trevisan S, Kojic S, Dalla Vecchia F, Laveder P, et al. J Biol Chem. 2000;275:41234–41242. doi: 10.1074/jbc.M007493200. [DOI] [PubMed] [Google Scholar]
- 16.Chien C-T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam J P. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan Y, Tong H Q, Beggs A H, Kunkel L M. Biochem Biophys Res Commun. 1998;248:134–139. doi: 10.1006/bbrc.1998.8920. [DOI] [PubMed] [Google Scholar]
- 19.North K N, Beggs A H. Neuromuscul Disord. 1996;6:229–235. doi: 10.1016/0960-8966(96)00361-6. [DOI] [PubMed] [Google Scholar]
- 20.Wyszynski M, Lin J, Rao A, Nigh E, Beggs A H, Craig A M, Sheng M. Nature (London) 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 21.Ahn A H, Kunkel L M. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan Y, Kunkel L M. FEBS Lett. 1997;410:153–159. doi: 10.1016/s0014-5793(97)00454-7. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura K, Tanaka T, Kuwahara A, Takeo K. Anal Biochem. 1985;148:311–319. doi: 10.1016/0003-2697(85)90234-9. [DOI] [PubMed] [Google Scholar]
- 24.Rando T A, Blau H M. J Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beggs A H, Miner J H, Scangos G A. J Gen Virol. 1990;71:151–164. doi: 10.1099/0022-1317-71-1-151. [DOI] [PubMed] [Google Scholar]
- 26.Mundel P, Heid H W, Mundel T M, Kruger M, Reiser J, Kriz W. J Cell Biol. 1997;139:193–204. doi: 10.1083/jcb.139.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leinweber B D, Fredricksen R S, Hoffman D R, Chalovich J M. J Muscle Res Cell Motil. 1999;20:539–545. doi: 10.1023/a:1005597306671. [DOI] [PubMed] [Google Scholar]
- 28.Pallavicini A, Zimbello R, Tiso N, Muraro T, Rampoldi L, Bortoluzzi S, Valle G, Lanfranchi G, Danieli G A. Hum Mol Genet. 1997;6:1445–1450. doi: 10.1093/hmg/6.9.1445. [DOI] [PubMed] [Google Scholar]
- 29.Sonnhammer E L, Kahn D. Protein Sci. 1994;3:482–492. doi: 10.1002/pro.5560030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 31.North K N, Laing N G, Wallgren-Pettersson C. J Med Genet. 1997;34:705–713. doi: 10.1136/jmg.34.9.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Der Ven P F, Wiesner S, Salmikangas P, Auerbach D, Himmel M, Kempa S, Hayess K, Pacholsky D, Taivainen A, Schroder R, et al. J Cell Biol. 2000;151:235–248. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahlhut M, van Deurs B. Mol Biol Cell. 2000;11:325–337. doi: 10.1091/mbc.11.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGregor A, Blanchard A D, Rowe A J, Critchley D R. Biochem J. 1994;301:225–233. doi: 10.1042/bj3010225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauser M A, Horrigan S K, Salmikangas P, Torian U M, Viles K D, Dancel R, Tim R W, Taivainen A, Bartoloni L, Gilchrist J M, et al. Hum Mol Genet. 2000;9:2141–2147. doi: 10.1093/hmg/9.14.2141. [DOI] [PubMed] [Google Scholar]
- 36.Moreira E S, Wiltshire T J, Faulkner G, Nilforoushan A, Vainzof M, Suzuki O T, Valle G, Reeves R, Zatz M, Passos-Bueno M R, et al. Nat Genet. 2000;24:163–166. doi: 10.1038/72822. [DOI] [PubMed] [Google Scholar]
- 37.Frey N, Richardson J A, Olson E N. Proc Natl Acad Sci USA. 2000;97:14632–14637. doi: 10.1073/pnas.260501097. . (First Published December 12, 2000; 10.1073/pnas.260501097) [DOI] [PMC free article] [PubMed] [Google Scholar]