Abstract
This paper addresses a family of issues surrounding the biological phenomenon of resistance and its representation in realist ontologies. The treatments of resistance terms in various existing ontologies are examined and found to be either overly narrow, internally inconsistent, or otherwise problematic. We propose a more coherent characterization of resistance in terms of what we shall call blocking dispositions, which are collections of mutually coordinated dispositions which are of such a sort that they cannot undergo simultaneous realization within a single bearer. A definition of ‘protective resistance’ is proposed for use in the Infectious Disease Ontology (IDO) and we show how this definition can be used to characterize the antibiotic resistance in Methicillin-Resistant Staphylococcus aureus (MRSa). The ontological relations between entities in our MRSa case study are used alongside a series of logical inference rules to illustrate logical reasoning about resistance. A description logic representation of blocking dispositions is also provided. We demonstrate that our characterization of resistance is sufficiently general to cover two other cases of resistance in the infectious disease domain involving HIV and malaria.
Keywords: Infectious Disease Ontology, Basic Formal Ontology, MRSa
1. Introduction: IDO, SaIDO, and MRSa
The phenomenon of resistance is an important feature of biological reality, encompassing diverse phenomena such as: the resistance of an individual to specific diseases, the herd immunity of an organism population to certain populations of infectious organisms, the resistance of disorders (for example, tumors) to specific treatments, and the resistance of certain pathogens to certain drugs. Treatment decisions and public health policies often hinge on correctly identifying types of resistance [1]. As such, resistance is a phenomenon that needs to be captured in biomedical ontologies in a consistent, coherent, and sufficiently general way.
Our primary goal in this communication is to characterize resistance in the infectious disease domain. The Infectious Disease Ontology (IDO) consortium is developing a set of interoperable ontologies that together are intended to provide progressively expanding coverage of the infectious disease domain. Central to this set is the IDO Core ontology, which provides a representation of entities, drawn from both the biomedical and the clinical domains, that are relevant to infectious diseases in general. Domain-specific extensions (e.g., pathogen-specific extensions) of the IDO Core complete the set by providing ontology coverage for the types of entities relevant to specific sub-domains of the infectious disease field.
IDO is itself an extension of the Basic Formal Ontology (BFO)1 and links to other ontologies constructed according to the principles of the Open Biomedical Ontologies (OBO) Foundry.2 IDO takes its treatment of disease from the Ontology for General Medical Science (OGMS).3, which distinguishes between:
a disease,
its diagnosis,
its signs and symptoms,
its realization in the series of processes we call a disease course, and
the underlying disorder(s) on the side of the patient in which the disease is rooted [2].
We mention these distinctions here because the conflation of entities of any of types listed can lead to confusion and error in reasoning about complex biomedical phenomena in general and about resistant entities in particular.
The Staphylococcus aureus Infectious Disease Ontology (SaIDO) is an extension of IDO concerning Staph aureus (Sa) infection. Sa can be partitioned into two subtypes:4 Methicillin-Susceptible Sa (MSSa) and Methicillin-Resistant Sa (MRSa). The latter subtype is a defined class that is distinguished by its resistance to methicillin (and other β-lactam antibiotics). Due to its rapid evolution in the face of antibiotic selective pressures, MRSa has become the paradigm of resistance (a so-called “superbug”), and has drawn significant attention from NIAID/NIH5, CDC6, and biomedical researchers throughout the developed world.
Subtypes of Sa can also be specified by assigning bacterial strains to clonal complexes based on genotypic differences. Variants can differ in their degree of resistance and in the types of drug to which they are resistant, forming a continuum, in terms of which Sa can be (and is) categorized. This provides one powerful reason to produce an ontologically correct representation of resistance in the infectious disease domain, and there are several others:
A classification of Sa that would allow inference of resistance profile.
IT tools for monitoring the number of infections from resistant strains observed in each hospital, thus allowing for: early detection of increases, anticipation of outbreaks, and tracking the spread of resistant strains.
Tools to guide in the writing of prescriptions.
In this paper, we consider the issues arising from the representation of resistance in realist ontologies and specifically, in IDO. In section 2 we list a set of desirable features for such a representation. In section 3 we survey some problems with resistance representations in other ontologies and devise a definition of ‘protective resistance’ for IDO with our desiderata in mind. We then focus our attention on the antibiotic resistance of MRSa to methicillin as a detailed case-study in section 4. We characterize our representation in terms of blocking dispositions (section 5), and show how our definition is general enough to apply to other important cases of resistance in the infectious disease domain (section 6).
2. Desiderata for an Ontological Representation of Resistance
Before reviewing how ‘resistance’ is defined in other ontologies and providingour own definition, it will be useful to list the desiderata for a good definition. We implicitly append to this list the desiderata for all good ontological definitions, such as non-circularity, Aristotelian form, and providing necessary and sufficient conditions.
2.1. Positivity Principle
An important principle for realist ontology development is to avoid as far as possible the use of negative differentia (e.g., ‘nonphysical’, ‘not part of the heart’, ‘not otherwise specified’) in formulating definitions. This “positivity design principle” enforces the use of terms which capture information about the entities represented in the ontology rather than information about the state of our knowledge at some given time [3].
At some level, however, resistance seems to require a negative aspect for its description. After all, a continuant is resistant precisely when something does not happen. John’s resistance to marriage entails a host of processes that do not happen (for example, John does not buy an engagement ring, does not get a marriage license, and so forth). In the case of MRSa, resistance to methicillin entails that a process of cell wall formation is not interfered with. The key is that the implicit negativity of resistance is only a semantic feature of the description at some level. The biological phenomenon of resistance is manifested at various levels of biological reality: genes, cells and their parts, organs, organisms, and populations. Negative descriptions at a macro-scale here mask the positive and active aspects of resistance at the micro-scale. A comprehensive ontological treatment must, accordingly, consider resistance at different levels of granularity.
In BFO-based ontologies, the lacks relation can be used to capture negative findings at one scale of biological description while avoiding the problems of using negative predicates or characteristics [4]. In describing resistance, we will need to say that independent continuants of a certain type do not exhibit a dependent continuant of a certain type. As we will see below, this amounts to an independent continuant lacking a certain disposition.
2.2. Doing Justice to Multiple Disciplinary Perspectives
Along with the various granularities at which we want to talk about resistance, we also must acknowledge that resistance is referred to by several disciplines: epidemiologists describe the spread of resistance in a population, the medical community speaks of patient resistance to disease and of pathogen resistance to drugs. Geneticists make reference to the genes that confer resistance when certain alleles are present. Incrementally, the IDO suite of ontologies must capture all of these discipline-specific aspects of resistance and the relations between them.
2.3. Nonproliferation of New Relations and Terms
The terms used in our representation will be derived from IDO, the Gene Ontology (GO), and the Protein Ontology (PRO). The relations used are drawn from the OBO Relation Ontology (RO) and its extensions 7. Naïvely, we could introduce a new relation resistant_to and use it to describe every instance of a resistance phenomenon. However, this would hide the complexity of the mechanisms of resistance working at smaller scales and eliminate many important inferences about resistance. Also, it is important to avoid a proliferation of relations in the OBO Foundry, since restriction to a small set of relations promotes reuse and interoperability of the constituent ontologies.
2.4. Explanatory Value
An assertion of resistance of X to Y should not be tautological or otherwise trivial. Appeals to resistance should be explanatory, which means that resistance itself should be represented in such a way as to provide some explanation of why certain processes unfold the way they do. The assertion of resistance of X to Y should be a useful response to a query; it should be a proposition on which to base further reasoning [5].
2.5. Formalizability
Finally, the definition should be expressible using the ontological tools of the trade. IDO supports machine-readable representations using both OWL and OBO formats. For use in OWL, resistance should be expressible using description logic restrictions.
In summary, we have five desiderata for a representation of resistance:
Positivity Principle: What is the active mechanism producing resistance and what is the associated physical basis?
Multiscale and Multiperspective: What are the relations between what has resistance and what confers resistance?
Nonproliferation of New Relations and Terms: What OBO Foundry relations and terms can be utilized?
Explanatory Value: Is resistance characterized so as to be a suitable result of an inference or a response to a query?
Formalizability: Can resistance be inferred from a formal representation of the relevant domain using first order logic? Can it be expressed using description logic?
3. Resistance in Existing Ontologies
We surveyed the treatment of resistance in existing ontologies.
3.1. Gene Ontology (GO)
The treatment of resistance is, strictly speaking, outside the scope of the GO, as resistance is not a biological process, molecular function, or cellular component. Within the sub-ontology of biological processes, however, GO contains the term ‘response to drug’, with putative synonyms drug resistance’ and ‘drug susceptibility/resistance’ (although ‘drug resistance’ has been obsoleted from GO, it still remains a synonym for the term ‘response to drug’)
[GO:0042493] Response to Drug: A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a drug stimulus.
It is of course incorrect to view the narrower term ‘drug resistance’ as a synonym of the broader term ‘response to drug’. Drug resistance arises spontaneously as the result of genetic diversification. The presence of the drug provides an environment in which those individuals (cells or viral particles) that have the resistance conferring gene or mutation have a fitness advantage, thus they outcompete the susceptible individuals. The resistance is not a direct response to the drug stimulus, although the manifestation of resistance may be a consequence of prior exposure to the drug. A response to a drug is a process, whereas resistance is a continuant, and thus ‘response to drug’ should not be a synonym of ‘drug resistance’. The GO definition defines resistance at the scale of cell or organism, but would not apply to molecules or populations. Finally, the definition seems to hinge on a ‘change in state’, but cells which do not change state are manifesting a ‘response to a drug’ just as much as are those which do, and in fact, resistant cells may not change state at all.
3.2. NCI Thesaurus
The NCI Thesaurus has the following entry for ‘resistance’:
[C19391] Resistance: Natural or acquired mechanisms, functions, activities, or processes exhibited by an organism to maintain immunity to, or to resist the effects of, an antagonistic agent, e.g., pathogenic microorganism, toxin, drug.
The primary problems with this treatment of resistance are that:
the definition is circular, since it uses ‘resist’ in defining ‘resistance’, and
the definition applies at the scale of the organism, ignoring the scale of the cell, molecule, or population
the term ‘resistance’ is a child of “resistance process”, making resistance a process and excluding many types of resistance, because the definition of ‘resistance process’ is biased towards multicellular organism resistance mediated by host defense mechanisms.
3.3. SNOMED-CT
SNOMED-CT contains the entry ‘drug resistance (disorder)’ with two defining relationships:
Drug Resistance Is a Drug-Related Disorder
Drug Resistance has Causative Agent (Attribute)
Drug or Medicament.
The former, which assigns to ‘drug resistance’ the parent term ‘drug-related disorder’, is formulated from the perspective of the patient. From the perspective of the pathogen or tumor, in contrast, drug resistance is not a disorder, but rather a benefit. That SNOMED adopts this patient-based perspective is not surprising. SNOMED, specifies that drug resistance is caused by a drug, but drug resistance is caused by the presence of a gene or mutation. It is only the manifestation of such resistance that results from the presence of the drug. Finally, as with other terms in SNOMED, only necessary but not sufficient conditions for drug resistance are provided. Good definitions should spell out both.
3.4. Infectious Disease Ontology (IDO)
In IDO, resistance is represented as a BFO disposition
Disposition =def A disposition is a realizable entity8 which is such that, if it ceases to exist, then its bearer is physically changed, and whose realization occurs in virtue of the bearer’s physical make-up when this bearer is in some special circumstances.
In English, the word ‘resistance’ is polysemous and can be used to refer to either a disposition or a quality (i.e., a categorical property). When referring to a quality, ‘resistance’ is roughly synonymous with ‘low susceptibility’. If we think of degree of susceptibility as a continuum, then the quality of resistance is the region of this continuum beneath a certain threshold. As a disposition, resistance is possessed in virtue of the internal physical arrangement of its bearer, is not always manifested when borne, and is realized in active processes at some physical scale. It is this realizable sense of ‘resistance’ that we want to represent: resistance is the capability (and in some cases the function) to resist under certain conditions.
IDO includes the term ‘protective resistance’, the definition of which attempts to address some of the problems described above:
Protective resistance is a disposition that inheres in a material entity (x) by virtue of the fact that the entity has a part (e.g. a gene product), which itself has a disposition 1) to ensure a physiologic response of a certain degree to an entity of type Y with the capability to damage x, or 2) to prevent the completion of some process caused by an entity of type Y with the capability to damage x. The realization of the disposition protects x from or mitigates the damaging effects of Y. The protective resistance disposition is realized in a biological process.
Here we write lowercase x to indicate an instance, and capital Y to indicate a type.
4. Towards a More Robust Ontological Treatment of Protective Resistance
To better understand the representational demands posed by resistance (and to expose the problems raised by this and similar phenomena from an ontological point of view), it will be useful to go through a detailed example. We choose drug resistance for a single combination of pathogen, antibiotic, and resistance-mechanism types. In this section we sketch the outlines of a formal representation of the resistance of MRSa to methicillin as conferred by PBP2a, a penicillin binding protein (PBP) and a product of the gene mecA. Both methicillin and penicillin are β-lactam antibiotics and, for the purposes of our formalization, a PBP can be considered to be a methicillin binding protein. Chambers gives a concise description of the form of resistance involved:
[M]ethicillin resistance in staphylococci is due to expression of PBP2a, a novel, low-affinity PBP for which there is no homologue in methicillin-susceptible strains[7].
We formalize this information as a set of triples expressing the relevant ontological relationships. We also include a series of inference rules that would lead a logic-driven reasoner to deduce from the triples that MRSa is resistant to methicillin. Such inference rules will one day be used by automated reasoners to compute antibiotic resistance from logical formalizations of such domains. Using ontologies as predictive tools will help to guide treatment decisions and support automated drug discovery.
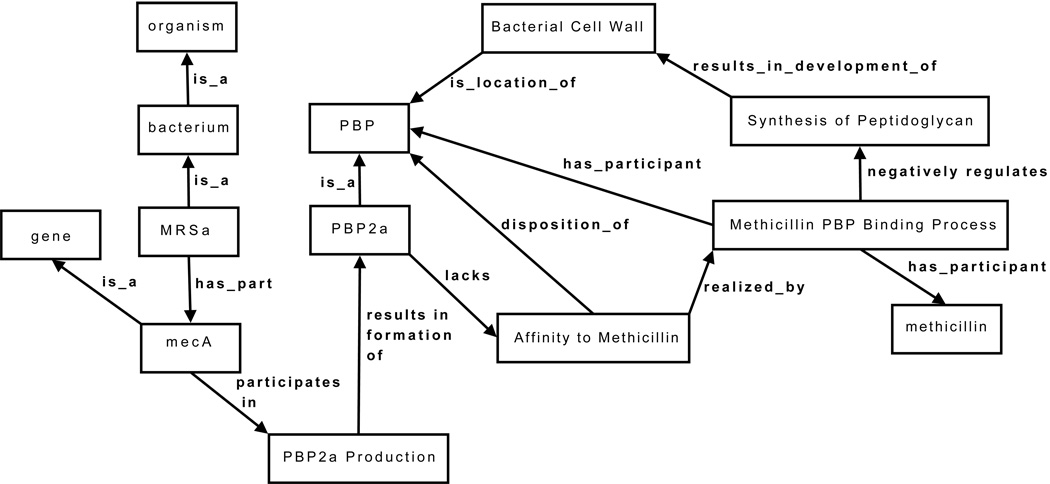
A faithful representation of the MRSa domain requires at least the following components (where is_a and has_part are used for relations between both continuant and occurrent universals):
bacterium is_a organism
MRSa is_a bacterium
synthesis_of_peptidoglycan is_a process and has_participant Penicillin_Binding_Protein (PBP)9
PBP has_function_realized_as_process synthesis_of_peptidoglycan
Bacterial_cell_wall is_location_of PBP
Canonically, synthesis_of_peptidoglycan results_in_development_of bacterial_cell_wall
formation_of_bacterial_cell_wall is_a process
PBP2a is_a PBP
methicillin_PBP_binding_process is_a binding process that has_participants methicillin and PBP
affinity_to_methicillin disposition_of some PBP to undergo a methicillin_PBP_binding_process that is realized in the presence of a methicillin.
methicillin_PBP_binding_process negatively_regulates synthesis_of_peptidoglycan.
PBP2a lacks affinity_to_methicillin
mecA is_a gene
MRSa has_part mecA
mecA participates_in PBP2a_production
PBP2a_production results_in_formation_of PBP2a
A subset of these triples is depicted graphically in Figure 1. Resistance to methicillin should be inferred from such representations in a logical manner. Such inferencing should be explanatory: it should tell us why MRSa bears such resistance.
Figure.
These triples will be used along with several rules of inference and derived facts (labeled IRn and Dn respectively in what follows). For readability, all variables are italicized and initial universal quantifier symbols are suppressed. First, we specify that is_a and has_part (for both continuants and occurrents) are transitive, allowing us to derive some basic taxonomic facts about the domain:
(IR1) x is_a y ∧ y is_a z → x is_a z
(IR2) x has_part y ∧ y has_part z → x has_part z
(D1) MRSa is_a organism
The parts of an organism are the products of the organism’s expressed genes, and these products are located in the appropriate places:
(IR3)o is_a organism ∧ g is_a gene ∧ o has_part g∧
g participates_in proc ∧ proc results_in_formation_of prod
∧ o has_part locp ∧ locp is_location_of prod →
o has_part prod located_in locp
(D2)MRSa has_part PBP2a located_in bacterial_cell_wall
The inference rule (IR3) makes a few simplifying assumptions. Since not all genes are expressed, we are only modeling the situation in which g is an expressed gene. We also assume that the process proc leading to prod is active, and that the single gene g participates in proc (rather than a set of genes).
If a continuant lacks a disposition to undergo a process in some situation, and that process negatively regulates a second process which has the continuant as a participant, then the continuant participates in the second process in that situation:
(IR4)p lacks disposition to undergo proc1 realized in situation s∧
proc1 negatively_regulates proc2 ∧ proc2 has_participant p →
In situation s, p participates_in proc2
(D3)In the presence of methicillin,
PBP2a participates_in synthesis_of_peptidoglycan
This lack of a disposition (i.e., the affinity to methicillin) has a categorical basis in the fact that methicillin binds to PBPs and prevents them from carrying out their function. However, PBP2a lacks this affinity, so the presence of methicillin does not prevent the essential sub-processes of cell-wall construction in MRSa.
If an organism has a continuant as a part and that part participates in a process in some situation, then the process unfolds in the organism in that situation.
(IR5)In situation s, p1 participates_in proc∧
p1 located_in p2 ∧ o has_part p2 →
proc unfolds_in o in situation s
(D4)synthesis_of_peptidoglycan unfolds_in MRSa
in the presence of methicillin
Finally, if a process unfolds in an organism in some situation and the process results in the development of a continuant which (canonically) is a part of the organism, then the organism has the continuant as a part in that situation.
(IR6)In situation s, proc unfolds_in o∧
Canonically, proc results_in_development_of p →
p part_of o in situation s
(D5)Bacterial_cell_wall part_of MRSa in the presence of methicillin
The canonical cell wall is a rigid configuration of peptidoglycan. The canonical cell wall is a healthy one for MRSa. The assertion (D5) captures the active, and thus positive, microphysical side of the resistance coin.
However the chain of reasoning here presents a puzzle. What does the lack of a disposition in (IR4) amount to? Consider the following pair:
Continuant C lacks disposition D to undergo process P in situation S
Continuant C undergoes P in a situation S.
Both (A) and (B) can be true at the same time. In fact the conjunction of (A) and (B) implies that (B) happens for a non-dispositional reason (i.e., (B) is not, in the corresponding case, a manifestation of the disposition D). Even if John lacks the disposition to feel hungry when in the presence of sushi, he may still feel hungry in such a situation because he has been fasting for three days. We need a way to express the fact that PBP2a necessarily lacks affinity to methicillin, and that this is what allows for the relevant cell-wall formation to take place. In order to frame the necessary lack as a positive and explanatory account, we will need the framework of blocking dispositions described below.
An important aspect of the chain of reasoning is the use of canonicity in domain triple 6:
Canonically, synthesis_of_peptidoglycan results_in development_of
bacterial_cell_wall.
Here ‘canonically … ’ works to specify a baseline of what is held to be true according to some canonical ontological reference (e.g., the anatomy of a model organism). Although this construction resembles the syntax of the modal operator ‘Necessarily,… ’, it does not yield a substantial ontological claim, but rather just says what is true relative to the particular reference. This form of reasoning with canonicity is consistent with its use in anatomical reasoning [8, 9]. In our case, we assume that a reference ontology of the cell specifies that synthesis of peptidoglycan results in the development of a cell wall. Such assumptions are axiomatic for IDO because they are beyond the scope of the core ontology and they provide a useful constraint for a reasoner. The chain of reasoning relies on such axioms and we expect that they will grow in number to accommodate the reasoning needs of IDO extensions.
5. Resistance as Blocking Disposition 10
An explanatory positive account for PBP2a lacking an affinity to methicillin can be given if we consider what prevents the manifestation of this disposition. Often what prevents the realization of a disposition is the manifestation of another disposition. We call the latter a blocking disposition and the former a blocked disposition. Dispositions are often said to manifest given certain background conditions, contexts, or circumstances [11, 12]. Blocking dispositions emphasize the ontological interactions in the background. In general, if D1 is a disposition and D2 is a blocking disposition for D1, then it must be the case that the realization of D2 prevents the realization of D1. A blocking disposition might be understood in different ways:
Incompatible occurrents: The realization of D1 and the realization of D2 are somehow incompatible occurrents, meaning either that they cannot co-occur or that one negatively regulates the other.
Incompatible qualities: The realization of D2 results in a quality of a continuant that is incompatible with the quality of the same continuant that would have resulted from the realization of D1. That is, we have two qualities that cannot be simultaneously exhibited (e.g., a square circular object).
By giving resistance a positive characterization, in which we describe what dispositions are actively realized, resistance can play a more explanatory role. We can describe this resistance without reference to blocking dispositions by noting the lack of affinity to methicillin (a disposition) in the relevant portion of the penicillin-binding protein of MRSa (PBP2a). As an explanation of why MRSa is resistant, however, invoking the lack of affinity to methicillin seems to be begging the question; MRSa is resistant to methicillin because one of its parts lacks an affinity for it. The same situation can be described in a positive (active) way by considering the disposition of PBP2a to synthesize peptidoglycan (an essential component of the bacterial cell wall) as a blocking disposition for the disposition of methicillin to bind to penicillin-binding proteins.11 In this way, protective resistance can be redescribed as an active response to methicillin.
In this situation, we can argue for incompatible occurrents: the process of cell wall construction (as a realization of the typical disposition of PBP) is incompatible with the process of methicillin binding (which is the realization of affinity to methicillin that PBP2a lacks). We could also argue for incompatible qualities: for a particular peptidoglycan molecule being bound by methicillin is incompatible with being bound to peptidoglycan peptide subunits. As a result, the molecular structure of a well-formed bacterial cell wall (i.e., a peptidoglycan lattice) is incompatible with the molecular structure of a compound sufficiently bound to methicillin. Cell wall construction is something a bacterium will participate in when no methicillin is present. In order to see this canonical process as an active response, we need the machinery of blocking dispositions. Protective resistance to methicillin is exhibited by MRSa in the process of cell wall construction by blocking the disposition of methicillin to bind to PBP.
In order for a theory of blocking dispositions to be useful in computational inference, it should be expressible in a formal language. For this task, we prefer description logic because: (1) it is the logic underlying OWL-DL, and (2) description logic is relatively inexpressive, so if we can capture blocking dispositions in description logic, we should be able to represent blocking dispositions in a more expressive formalism. It is easiest to formulate a blocking disposition as a description logic restriction by using the RO_Proposed relation negatively_regulates
D2_blocking_disposition_of_D1 ≡
∃realized_by (∃negatively_regulates ∃realizes D1 ∏ ∃realizes D2)
But we may also describe the inability for D1 and D2 to co-occur using a cardinality restriction:12
∃realizes D1 ∏ ∃realizes D2 ∏ ∃occurs_at T = ∅
Description logic does not provide schema variables in the way we have used them in D2 blocking_disposition_of D1, so each such disposition must be fleshed out in concrete terms by the IDO extension ontologies.
Such an analysis is not without its problems. One minor concern is that calling something a blocking disposition may be considered too perspectival, biasing the ontological term towards D1 being blocked by rather than blocking D2. A more serious problem is how can we empirically distinguish between something not happening to a specific continuant as the result of (1) an external blocking disposition or (2) as the result of its own internal makeup.
A further worry involves the identity criteria for blocking dispositions. Storm-resistant walls on a particular house are most likely also lemonade-resistant as well, but in virtue of the same underlying structure (i.e., categorical properties). So is the particular lemonade resistance inhering in those walls identical to the particular water resistance inhering in those walls? It seems counterintuitive to say so, but if we say these are not identical are we not opening the door to a combinatorial explosion of resistance dispositions? Similarly, penicillin binding protein has an affinity to penicillin (as its name suggests) which is conferred by the same qualities that yield methicillin resistance, but we do not want to say that these forms of resistance are identical because some staph aureus may be susceptible to methicillin but resistant to penicillin. The standard answer to such worries from the realist ontology camp is that terms are included in an ontology in reflection not of what is combinatorially possible but rather of the actual needs of biologists who are describing real biological phenomena. Whether dispositions referred to by such terms are or are not identical will need to be decided on a case-by-case basis, but such a decision is then not in principle more problematic for dispositions than for entities of other sorts.
6. Other Examples
The blocking disposition characterization presented above can be applied to different types of resistance. In this section, we present two examples involving infectious disease: (1) CCR5 mutation confers protective resistance against certain strains of HIV, and (2) the sickle cell trait confers protective resistance against malaria. Like the case of MRSa, we will see that the simple macroscale characterization of resistance gives way to a network of related entities at the microscale.
6.1. CCR5- Δ32 and HIV
Certain strains of HIV have a disposition to bind to CCR5 (complemented, like a lock and key, with the disposition of CCR5 to bind to HIV) and thus enter cells. CCR5-Δ32 is a deletion mutation of the CCR5 gene resulting in cells which lack a functioning CCR5 receptor on their surfaces[13]. Note that HIV does not lose the disposition to bind to CCR5, the disposition simply goes unmanifested.
Similar to the case of antibiotic resistance in MRSa, we are dealing here with a part of an organism lacking a continuant; in this case, however, what is missing is an independent continuant (i.e., a portion of canonical CCR5). To characterize something as lacking a disposition is to provide a negative characterization that stands in need of further explanation — as contrasted with characterizing something as lacking a part, which is a positive characterization. The fact that a cell lacks CCR5 receptors on its surface is a quality of the cell.
This case of resistance to HIV is covered by the clause of the IDO definition for protective resistance in which the process caused by a potentially damaging entity is prevented from completing. In terms of blocking dispositions, this situation can be described in terms of incompatible qualities. If d1 is the disposition of HIV to bind to a CCR5 molecule, and d2 is the disposition of individuals with the CCR5-Δ32 mutation to develop cells without CCR5 on their surface, then we have d2 blocking d1 because the realization of both would require the same continuant (i.e., a T cell or macrophage) to exhibit incompatible qualities by simultaneously having and lacking CCR5 on its surface.
6.2. The Sickle-Cell Trait and Malaria
There are many hypothesized mechanisms by which the sickle cell hemoglobin gene (HbS) confers resistance to Plasmodium falciparum, one of the infectious organisms that causes malaria. One such mechanism is through the impact of HbS on red blood cell hydration and density [14]. In individuals with HbS, red blood cells are disposed to dehydration and a consequent increase in density.
Plasmodium falciparum merozoites have a disposition to spread through host red blood cells. This spreading process consists of four subprocesses: plasmodium replication inside a single red blood cell, red blood cell lysis, release of plasmodium merozoites from the lysed cell, and entry of released merozoites into a new red blood cell. Merozoite invasion of dense, dehydrated red blood cells is reduced. Thus, an essential subprocess of the spread of plasmodium in the host is reduced (the process is negatively regulated), thus conferring protective resistance against malaria.
Using blocking dispositions, we can again characterize this situation in terms of incompatible qualities. If d1 is the disposition of certain red blood cells to become dehydrated, and d2 is the disposition of plasmodium to spread through red blood cells, then the realization of d1 results in a hydration quality inhering in the red blood cells, thereby negatively regulating the realization of d2, which requires hydrated red blood cells. The qualities are incompatible because a red blood cell cannot simultaneously be hydrated and dehydrated.
7. Mereological Issues
If we take resistance to be a specifically dependant continuant that inheres in an independent continuant, then we must still answer some mereological questions: Is the resistance of the relevant portion of PBP2a (i.e., of a part) identical to the resistance of the cell (i.e., of the including whole)? Furthermore, is cell resistance identical to the resistance of a portion of tissue in which the cell resides or the containing host organism or, for that matter, of the containing population? The ontology of resistance must address which scales of biological reality resistant continuants occupy, and the identity of resistance across scales.
We can begin to address this question for our case study by considering a biochemical explanation of why methicillin does not bind to PBP2a. Indeed, a structural explanation involving the biochemistry of PBP2a and β-lactam antibiotics is known [15]. This explanation involves facts about chemical structure of the peptide links between glycan chains in peptidoglycan and the chemical structure of methicillin. The configuration of such structures, in turn, hinges on facts about how proteins fold and the basic chemical rules governing how bonds between carbon, oxygen, hydrogen, and nitrogen form. There are several ontological resources to represent such chemical structures (e.g., ChEBI13), but at this molecular scale we are only reasoning about structures (qualities in BFO) that are the physical basis for the resistance disposition, not about the disposition itself. Simply put, the resistance of MRSa to methicillin is not identical to an aggregate of chemical structures, but it is borne in virtue of such structures. Importantly, we also want to be able to talk about resistance at the physiological and population levels when we can talk about the consequences at those levels of the chemical structures that confer resistance, and also when when we have incomplete microscale information about such structures. This is another reason an ontology like IDO should provide terms for the entities existing at both scales.
A related issue that should be addressed at different scales of biological reality is the way in which facts at each scale are used to explain the phenomenon of resistance. At the genetic scale, MRSa having mecA and MSSa lacking mecA are explanatory. At the cellular level, inference (D5) is explanatory. To help link scales of biological reality, the proposed IDO definition for protective resistance given above specifies that a material entity is resistant in virtue of one of its parts (or one of its members in the case of a population), but further work of specification of parts must be done in the respective IDO extension ontologies in order for us to be able to exploit multiscale reasoning. As stated above, structural facts (BFO qualities) alone may not be explanatory because they do not include information of how entities with different structures interact. Interactions (BFO processes) alone are not explanatory because they do not include information about what it is in the interacting participants that enables these interactions to happen. Dispositions are the explanatory glue between structures and interactions.
8. Conclusion
We have attempted to provide a definition of protective resistance that is general enough to cover the varied types of resistance in the infectious disease domain, specifies the components of resistance at multiple scales and across ontological types (along with how those components relate to each other), and is capable of being extended to cover specific forms of resistance in this domain. We have seen that resistance is an important multi-scale phenomenon, often with a one-to-many relationship between a resistant organism and the underlying mechanisms of resistance. Several desiderata for an ontological representation were found lacking in existing ontologies. Our preliminary formal representation of resistance honors a positivity design principle, by providing an analysis for its negative characterizations (e.g., the lack of a disposition). It also conforms to a principle of non-proliferation of relations by reusing existing RO relations. Using the formalism of blocking dispositions, we are able to analyze the multiscale interactions that give rise to resistance. Such an analysis adds explanatory value to an inferred fact about the resistance of MRSa to methicillin. Certain assumptions of canonicity are needed, namely the assumptions that all entities (e.g., anatomical and cellular component entities) are canonical and that no exogenous factors are present unless explicitly stated (e.g., methicillin). This canonical representation of resistance is, however, able to cover several different types of resistance. Our definition of protective resistance was shown to be sufficiently general to cover resistance phenomena involving malaria and HIV. In these cases, as in the case of MRSa, we demonstrated how resistance involves the prevention of the completion of an essential subprocess by the potentially harmful entity.
Some issues remain (e.g., in providing a systematic account for the lack of a disposition that overcomes some of the issues we have identified), but we are confident that further study of resistance will have great benefits for biomedical ontologies. For example, this work might be extended by considering the logical rules necessary to infer novel forms of resistance from a known resistance type, potentially across related drugs and species.
Acknowledgements
This work was funded by the National Institutes of Health through Grant R01 AI 77706-01. Smith’s contributions were also funded through the NIH Roadmap for Medical Research, Grant 1 U 54 HG004028 (National Center for Biomedical Ontology). Cowell’s contributions were also funded by a Burroughs Wellcome Fund Career Award at the Scientific Interface. We would like to thank Wacek Kusnierczyk and two anonymous reviewers for their feedback on earlier drafts of this work.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Further refinement of SaIDO may involve the creation of MRSaIDO and MSSaIDO sub-ontologies which overlap in SaIDO.
For more on BFO realizable entities, see [6].
Here we chain together two triples for brevity.
This section is adapted from a conference submission by the authors to Formal Ontology in Information Systems [10].
Note that since we are dealing with the impossibility of co-occurrence, we could also take the disposition to bind to PBP as a blocking disposition for the disposition to synthesize peptidoglycan.
By our notational convention, ∅ denotes a cardinality of 0
References
- 1.Knobler SL, Lemon SM, Najafi M, B T, editors. The Resistance Phenomenon in Microbes and Infectious Disease Vectors. Washington DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- 2.Scheuermann R, Ceusters W, Smith B. Toward an ontological treatment of disease and diagnosis. Proceedings of the 2009 AMIA Summit on Translational Bioinformatics. 2009:116–120. [PMC free article] [PubMed] [Google Scholar]
- 3.Spear A. Ontology for the twenty-first century: An introduction with recommendations. 2006 http://www.ifomis.org/bfo/manual.pdf.
- 4.Ceusters W, Elkin P, Smith B. Negative findings in electronic health records and biomedical ontologies: A realist approach. International Journal of Medical Informatics. 2007;76(3):s326–s333. doi: 10.1016/j.ijmedinf.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sober E. Dispositions and subjunctive conditionals, or dormative virtues are no laughing matter. The Philosophical Review. 1982;91(4):591–596. [Google Scholar]
- 6.Arp R, Smith B. Function, role, and disposition in basic formal ontology. Proceedings of Bio-Ontologies Workshop (ISMB2008) 2008:45–48. [Google Scholar]
- 7.Chambers H. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. Journal of Infectious Disease. 1999;179(2):S353–S359. doi: 10.1086/513854. [DOI] [PubMed] [Google Scholar]
- 8.Neuhaus F, Smith B. Modeling principles and methodologies—relations in anatomical ontologies. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. New York: Springer; 2007. pp. 289–306. [Google Scholar]
- 9.Goldfain A. Canonicity and disease ontologies. Biomedical Computation Review. 2009;5(3):33. [Google Scholar]
- 10.Goldfain A, Smith B, Cowell LG. Dispositions and the Infectious Disease Ontology. forthcoming in Proceedings of FOIS2010. 2010 [Google Scholar]
- 11.Mumford S. Dispositions. Oxford: Oxford University Press; 1998. [Google Scholar]
- 12.Bird A. Nature’s Metaphysics: Laws and Properties. Oxford: Clarendon Press; 2007. [Google Scholar]
- 13.McNicholl JM, Smith DK, Qari SH, Hodge T. Host genes and hiv: The role of the chemokine receptor gene CCR5 and its allele (Δ32 CCR5) Emerging Infectious Diseases. 1997;3(3):261–271. doi: 10.3201/eid0303.970302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiffert T, Lew VL, Ginsubrg H, Krugliak M, Croisille L, Mohnadas N. The hydration state of human red blood cells and their susceptibility invasion by Plasmodium falciparum. Blood. 2005;105(12):4853–4860. doi: 10.1182/blood-2004-12-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim D, Strynadka NCJ. Structural basis for the β-lactam resistance of pbp2a from methicillin-resistant staphylococcus aureus. Nature Structural Biology. 2002;9(11):870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]