Abstract
The maintenance of the fetus during pregnancy has been attributed to the absence of major histocompatibility complex (MHC) class II antigens on fetal trophoblastic cells that make contact with the maternal immune system. However, the mechanism(s) by which class II genes are regulated in trophoblast cells is unclear. We have identified a negative regulatory element (IAαNRE) in the promoter of the mouse class II gene IAα that represses IAα transcription in trophoblast cells. IAαNRE, located from −839 to −828, binds transacting factors from rat, mouse and human trophoblast cells, but not from 18 other cell lines tested. These results indicate that IAαNRE binding proteins (IAαNRE BPs) are conserved in species with hemochordial placentas, and suggest that IAαNRE binding activity is restricted primarily to trophoblast cells. Interestingly, the IAαNRE BPs bind to the IAαNRE antisense strand in a sequence-specific manner. IAαNRE represses transcription from the IAα promoter in a position-dependent manner, and has a minor down-regulatory effect on the activity of the SV40 promoter/enhancer. Our results demonstrate that MHC class II gene transcription is repressed in fetal trophoblast cells by sequence-specific, single-stranded DNA binding proteins, and suggest a possible mechanism by which the conceptus is protected from immune rejection during pregnancy.
Keywords: trophoblast, MHC class II genes, transcription, negative regulatory element
INTRODUCTION
The survival of the semiallogeneic fetus during pregnancy suggests that the maternal immune system lacks the ability to mount an effective allograft rejection reaction against the fetus. Since the same tissues transplanted at other sites are rejected, the pregnant uterus has been termed an “immunological sanctuary.” The mechanism(s) by which the developing fetus evades the maternal immune system are unclear, but it has been proposed that the lack of major histocompatibility complex (MHC) class II antigens on trophoblast cells may play a role in preventing rejection of the fetal allograft (Medawar, 1953; Billingham, 1975). Trophoblast cells form the fetal component of the maternal-fetal interface, and in species with hemichordial placentas they are the only fetal cells continuously exposed to the maternal bloodstream. Trophoblast cells do not constitutively express class II antigens, and expression cannot be induced by IFN-γ or TNF-α (Chatterjee-Hasrouni and Lala, 1981; Hunt et al., 1990a,b; Peyman and Hammond, 1992; Peyman et al., 1992; Salter-Cid and Flajnik, 1995). The importance of class II gene regulation in fetal maintenance is suggested by studies demonstrating that aberrant expression of class II antigens on mouse or human placental tissue strongly correlated with abortion (Athanassakis-Vassiliadis et al., 1990, 1995).
MHC class II antigen expression is controlled primarily at the level of transcription (reviewed in Benoist and Mathis, 1990; Glimcher and Kara, 1992). The proximal promoters of all class II genes contain several highly conserved cis-acting sequences that are required for both constitutive and IFN-γ inducible transcription in B cells and other cell types (reviewed in Benoist and Mathis, 1990; Glimcher and Kara, 1992). The function of these cis-acting sequences, termed the W (also named H, S or Z), X and Y boxes, is mediated by the binding of transacting factors (Glimcher and Kara, 1992; Abdulkadir and Ono, 1995). Positive regulatory sequences have also been defined in the distal promoters of class II genes; these include the B cell response elements (BRE-1 and BRE-2; Boothby et al., 1988) and the lipopolysaccharide element (LRE; Gravellese et al., 1989). Recently, the gene encoding class II transactivator (CIITA) was isolated and shown to be essential for both constitutive and IFN-γ-inducible class II gene transcription (Steimle et al., 1993, 1994; Chang et al., 1994; Chin et al., 1994). CIITA does not interact directly with DNA but appears to mediate its activating function through protein:protein interactions (Steimle et al., 1994; Riley et al., 1995).
In contrast to the characterization of class II gene-activating elements and factors, there are relatively few reports of negative regulation of class II gene expression. The class II-negative cell types that have been examined contain the same transacting factors that bind to the proximal promoter as do class II-positive and inducible cells, but genomic footprinting demonstrated that the promoters are unoccupied in vivo (Boothby et al., 1989; Kara and Glimcher, 1991). At least two negative regulatory regions have been defined in the HLA-DQβ promoter that function in human fibroblasts (Boss and Strominger, 1986). The region of the mouse IEα promoter between −873 and −353 contains negative cis-acting elements that decrease basal promoter activity in a xeroderma cell line that is class II-inducible (Thanos et al., 1988). Albert et al. (1994) identified a negative element in the IAβ promoter that represses transcription in renal epithelial cells, T cells and fibroblasts. Recently, the DNA binding protein YB-1 was shown to repress class II antigen expression in U937 cells by competing for binding to the X box (MacDonald et al., 1995).
The mechanisms for repression of class II genes described above were all defined in cells that constitutively or inducibly express these genes, but few studies have examined regulatory mechanisms in cells such as trophoblasts that cannot be induced to express class II antigens. We have identified a novel negative regulatory element (IAαNRE) in the promoter of the mouse class II gene IAα that functions to repress IAα transcription in trophoblast cells. This negative regulatory element functions in a location-dependent manner in the IAα promoter, and has a minor repressive effect on transcription from the SV40 promoter/enhancer complex. IAαNRE binds proteins from multiple different trophoblast cell lines derived from rat, mouse and human, but not from 18 other cell lines tested, suggesting that the activity of IAαNRE binding proteins is restricted primarily to trophoblasts, and is conserved in species with a hemochordial placenta. These results suggest that class II antigen expression is repressed in trophoblast cells at the level of transcription by novel transacting factors that bind to the IAαNRE.
MATERIALS AND METHODS
Cell Culture
The mouse B cell lymphoma line A20 was obtained from the American Type Culture Collection (Rockville, MD) and was maintained in RPMI-1640 (GIBCO/BRL, Gaithersburg, MD) supplemented with L-glutamine (Sigma, St. Louis, MO), 50 µM β-ME (Sigma) and 10% fetal bovine serum (FBS, Hyclone, Logan, UT). The rat trophoblast cell lines Rcho-1 (Faria and Soares, 1991) R8RP.3, HRP-1 and LRP-2 (Hunt et al., 1989) and the mouse trophoblast cell line SM-9 (J.S. Hunt, unpublished) were maintained in RPMI-1640 supplemented with L-glutamine, 1 mM sodium pyruvate, 50 µM β-ME and 10% FBS.
RNA Isolation and Northern Analysis
RNA was isolated and Northern blot analysis performed as previously described (Pazmany et al., 1995). MHC class II gene mRNAs were detected using cDNA probes that correspond to the conserved α2 and β2 domains of the IAα (Davis et al., 1984), IAβ (Robinson et al., 1982), IEα (McNichols et al., 1982), and IEβ (Steinmetz et al., 1982) genes, respectively.
Plasmid Construction, Transfection Procedures and Chloramphenicol Acetyl Transferase (CAT) Assays
The IAαproCAT plasmid constructs were generated by making 5′ deletions of a 1.9-kb IAα promoter fragment extending from 1.9 kb upstream to within IAα exon I (subcloned in pUC19, [pUC(IAα1.9)]) using restriction sites present in the upstream region and the BamHI site at +1 relative to the transcription start site. The fragment staggered ends were filled in with Klenow enzyme (Pharmacia, Gaithersburg, MD) and subcloned into the blunted Pst I site of the pCAT basic vector (Promega, Madison, WI). These constructs [pIAα(393)CAT, pIAα(449)CAT, pIAα(483)CAT, pIAα(501)CAT, pIAα(541)-CAT, pIAα(631)CAT, pIAα(890)CAT and pIAα(1200)CAT] are diagrammed in Figure 2. The pIAα (864)CAT and pIAα(810)CAT constructs were generated by ligating blunt-ended restriction fragments into the filled-in HindIII site of pCATbasic. The pIAα(780)CAT construct, also shown in Figure 2, was generated by polymerase chain reaction (PCR) amplification using the IAα(−21/−1) AS primer (see below) and a primer corresponding to −780 bp to −750 bp of the IAα promoter. PCR was done as described below and the resulting amplification product was also subcloned into pCAT basic. Plasmids used in CAT assays were purified twice by cesium chloride centrifugation and analyzed by agarose gel electrophoresis for nicked DNA prior to transfection.
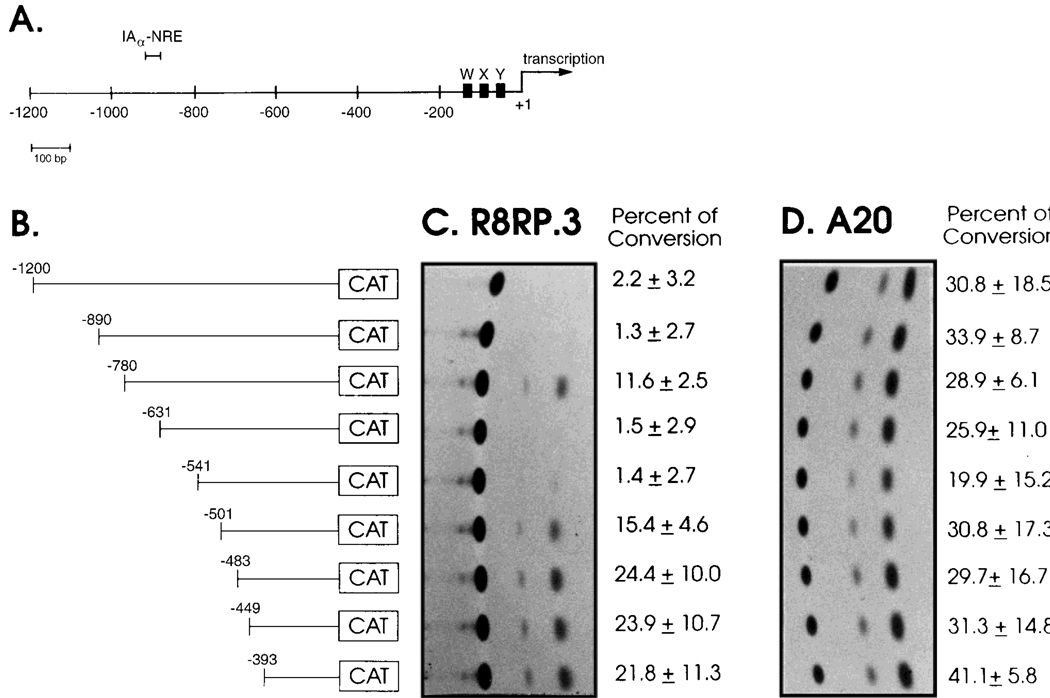
Fig. 2.
Identification of negative regulatory regions in the IAα promoter that function in trophoblast cells. A: Schematic of the 1.20-kb IAα promoter fragment that was used to create 5′ deletions. The locations of the X, Y and W boxes and the transcriptional start site are shown. B: Schematic of the IAαpro CAT constructs used in the promoter analysis. The resulting constructs were transfected into: C: R8RP.3 cells or: D: A20 cells. Cell extracts were prepared and assayed for CAT activity. Representative experiments are shown; the numbers to the right of each autoradiograph represent the average percent conversion of the nonacetylated to the acetylated form of chloramphenicol and the standard error calculated from four experiments.
Selective deletions of the 890-bp IAα promoter were generated by two-step PCR. The pIAα(Δ541/492)CAT plasmid was created by a primary amplification of two fragments, A and B, on either side of the IAα −541 to −492 sequence. The primary reaction mixture consisted of 100 ng of template DNA [pUC(IAα 1.9)], 200 µM dNTPs, 2.5 µM of each primer, 1 U of Taq polymerase (Boehringer Mannheim, Indianapolis, IN) and the supplied buffer. The primers used for primary amplification were:
Fragment A:
primer IAα (−556 to −543/ −494 to −478)S:
5′-AAGAACCTCACCAATGGGGATACCTTGT-3′
primer IAα (−21 to −1)AS:
5′-GTGTCTAGACAGCCAAAGAGATCACACAC-3′
Fragment B:
primer IAα (−556 to −543/ −494 to −478)AS:
5′-ACCCAAGGTATCCCCATTGGTGAGGTTCTT-3′
primer IAα (−900 to −880)S:
5′-TGCAAGCTTCTCAGTCTTCTATAAGCAAG-3′
The outermost primers contain restriction enzyme sites, Xbal and HindIII, which are underlined and were used for subcloning. The PCR program consisted of five cycles containing an annealing step at 37°C for 1 min, an amplification step at 72°C for 2 min and a denaturation step at 95°C for 45 sec, followed by 30 cycles containing an annealing step at 60°C for 1 min, an amplification step at 72°C for 2 min and a denaturation step at 95°C for 45 sec. All oligonucleotides used in this study were generated in the Roswell Park Biopolymer Facility. Following the primary amplification, the products were agarose gel-purified and ethanol-precipitated. Equal concentrations of the individual fragments were combined and the PCR reaction was repeated using the IAα(−21 to −1)AS and IAα(−900 to −880)S primers. The secondary PCR product was digested with Xbal and HindIII, agarose gel-purified and subcloned into the pCAT basic vector. All PCR-generated constructs were sequenced by dideoxy sequencing using Sequenase (U.S.B.).
The pIAα(Δ840/810)CAT plasmid was generated as above. The primary PCR reaction was done with the following primers:
Fragment A:
primer IAα (−860 to −842/−808 to −788)S:
5′-TCACATAAGGGGGCAGTTTTCTAAGTTAG-3′
and the IAα( −21 to −1)AS primer.
Fragment B:
primer IAα( −860 to −842/ −808 to −788)AS:
5′-CTAACTTAGAAAAGCTGCCCCCTTATGTGA-3′
and the IAα( −900 to −880)S primer.
The secondary PCR was done with equal molar amounts of fragments A and B and the IAα(−900 to −880)S and IAα(−21 to −1)AS primers.
The pIAα[(NRE)(393)]CAT construct was generated by cloning double-stranded oligonucleotides (sense strand: 5′-CTAAAGCTTAGCAAATTTGGAAAAGCATGCCGA-3′) containing HindIII and SphI restriction sites (underlined) and the IAα promoter sequence corresponding to −842 to −828 into the HindIII-SphI sites of pIAα(393)CAT.
The p(NREU)4[SV40]CAT construct was generated by inserting two copies of a double-stranded oligonucleotide containing a tandem repeat of the IAα sequence from −842 to −824 upstream of the SV40 promoter in the pCAT control vector (Promega). The same oligonucleotide was inserted downstream of the SV40 enhancer in the pCAT control vector to generate p(NRED)4[SV40]CAT.
Cells (4 × 106) were transiently transfected by the electroporation technique (Potter et al., 1984) using 16 µg of pIAαproCAT DNA and 4 µg of tk-luciferase plasmid as an internal control for transfection efficiency. Cells were cultured for 2 days following transfection, harvested and disrupted. Protein concentrations were determined by BioRad protein analysis and luciferase assays were performed using a Promega luciferase kit according to the manufacturer’s instructions. Samples for CAT assays were normalized according to their luciferase activity and used in CAT assays as described (Titus, 1991). Quantitation was performed by Image-Quant laser densitometry (Molecular Dynamics, Sunnyvale, CA) and presented as indicated in the figure legends. The data are representative of at least three independent transfections for a given recombinant, and include at least two independent preparations of each plasmid DNA. Identical results were obtained in CAT assays with SM9 and R8RP.3 cells. However, SM9 cells were observed to be more refractory to transfection than R8RP.3 cells; therefore, only the data for R8RP.3 cells is shown.
Nuclear Extract Preparation and Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared by modifications of the method of Stein et al. (1989). Briefly, 2–3 150-mm dishes of subconfluent cells were washed 2× with ice cold phosphate-buffered saline (PBS), scraped off the plates, pelleted, and resuspended in 4× packed cell volumes (PCV) lysis buffer [10 mM HEPES pH 7.9/1 mM EDTA/60 mM KCl/1 mM dithiothreitol (DTT)/0.2% NP-40 (0.075% NP-40 for B cells)/5 µg/ml each of pepstatin, aprotinin and leupeptin]. After 5 min on ice, nuclei were pelleted for 6 min at 2,000× g and 4°C, washed with lysis buffer lacking NP-40, and recentrifuged. Purified nuclei were resuspended in 2× nuclear volume of nuclear lysis buffer [250 mM Tris:HCl pH 7.5/60 mM KCl/1 mM DTT/protease inhibitors], and lysed by three cycles of freezing and thawing in dry ice-methanol and a 37°C water bath. Nuclear extracts were clarified by centrifugation at 8,500× g for 10 min at 4°C, and aliquotts stored at −80°C. Protein concentrations were determined by the Bradford Assay using Biorad reagents. A minimum of three independent nuclear extracts from each cell line was tested in EMSA.
Oligonucleotides corresponding to the IAαNRE sequences that were used in EMSA are described in Tables 1 and 2. One hundred ng of each oligonucleotide was end-labeled with T4 DNA kinase and separated from unincorporated radioactivity using Select-D G-25 columns (5-Prime-3 Prime, Inc., Boulder, CO). Octamer-1 (Oct 1) consensus oligonucleotides were purchased from Promega and are described elsewhere (Parslow et al., 1984).
TABLE 1.
The Results of EMSA Using R8RP.3 Nuclear Extracts and Oligonucleotides Corresponding to IAαNRE Sequences
| DSa | SSa | ASa | |
|---|---|---|---|
 |
− − | − − | − −b |
| − − | − − | + +b | |
| − − | − − | + + | |
| − − | − − | + + | |
| − − | − − | + + | |
| − − | − − | + + | |
| − − | − − | − − | |
| − − | − − | − − |
DS, double-stranded oligos; SS, sense strand oligos; AS, antisense strand oligos.
(+) indicates that sequence specific DNA:protein complexes were detected; (−) no specific complexes were observed.
TABLE 2.
IAαNRE BP Binding Specificity as Defined by Electrophoretic Mobility Shift Competition Analysis*
| Oligonucleotide | Sequence | Competition |
|---|---|---|
| IAαNRE antisense strand |
5′-TTT TCC AAA TTT-3′ | + + |
| IAαNRE sense strand | 5′-AAA TTT GGA AAA-3′ | − − |
| m1NRE | 5′-TTT ctC cAA TTc-3′ | + + |
| m2NRE | 5′-TTT ctgtgA TTc-3′ | + + |
| m3NRE | 5′-TTT cCa ttA TTa-3′ | + + |
| m4NRE | 5′-TTT TtC gAA TTT-3′ | + + |
| m5NRE | 5′-TTT TCC AAA cTg-3′ | − − |
| m6NRE | 5′-TTT TCC AAA ggg-3′ | − − |
| m7NRE | 5′-ggg TCC AAA TTT-3′ | − − |
| m8NRE | 5′-gTT TCC AAA TTT-3′ | + + |
| m9NRE | 5′-TgT TCC AAA TTT-3′ | + + |
| m10NRE | 5′-TTg TCC AAA TTT-3′ | + + |
| m11NRE | 5′-Tgg TCC AAA TTT-3′ | + + |
| m12NRE | 5′-TTT TCC AAA gTT-3′ | + + |
| m13NRE | 5′-TTT TCC AAA TgT-3′ | + + |
R8RP.3 nuclear extracts were preincubated with 200-fold excess of the unlabeled oligonucleotides shown above, followed by the binding reaction with the labeled IAαNRE antisense strand oligonucleotide. DNA:protein complexes were resolved on 4% acrylamide gels. The mNRE oligonucleotides correspond to mutations of the wildtype IAαNRE antisense sequence. Competition for binding to the IAαNRE oligo is denoted by the (++). Competitions were performed with each oligonucleotide a minimum of three times.
EMSA was performed as follows: nuclear extracts (4–5 µg) were incubated with 0.2–1.0 ng 32P-labeled oligonucleotides for 20 min at room temperature in 20 µl binding reactions containing: 10 mM Tris:HCl (pH 7.5)/1 mM MgCl2/50 mM NaCl/0.5 mM DTT/0.5 mM EDTA/4% glycerol (vol/vol)/ and 0.5 µg poly (dI:dC; Pharmacia). The specificity of protein:DNA complexes was examined by incubating the nuclear extracts with a 100- to 300-fold molar excess of unlabeled competitor oligonucleotides for 15 min at room temperature, followed by the binding reaction with the labeled oligonucleotides. Protein:DNA complexes were resolved by electrophoresis on 4% nondenaturing polyacrylamide (acrylamide:bis 80:1)/0.5× tris:borate:EDTA(TBE) gels at 10 V/cm at room temperature. Gels were subsequently dried and exposed to Kodak X-Omat film at −70°C with the use of an intensifying screen.
RESULTS
MHC Class II mRNA Is Not Expressed in Trophoblast Cells
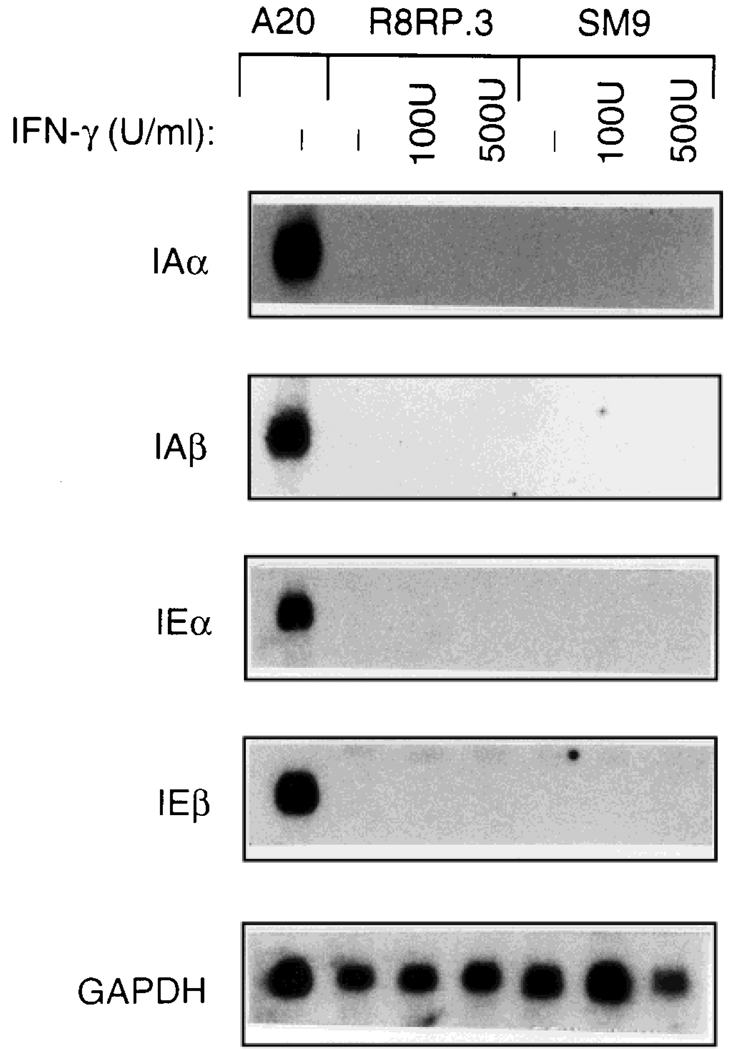
Previous studies demonstrated that MHC class II antigens are not expressed on trophoblast cells, even after treatment with IFN-γ (Chatterjee-Hasrouni and Lala, 1981; Hunt et al., 1990a,b; Peyman and Hammond, 1992; Peyman et al., 1992; Salter-Cid and Flajnik, 1995). In order to examine the mechanism(s) by which class II antigen expression regulated in trophoblast cells, we initiated studies utilizing trophoblast cell lines derived from rat (R8RP.3) and mouse (SM9). Northern blot analysis was performed using total RNA isolated from R8RP.3 and SM9 cells cultured in the presence or absence of IFN-γ for 24 hr to determine whether the mRNA encoding class II antigens is expressed in these trophoblast cell lines. Figure 1 demonstrates that none of the probes corresponding to MHC class II genes (IAα, IAβ, IEα or IEβ) hybridized to either R8RP.3 or SM9 cell RNA, even from cells exposed to IFN-γ. Rehybridization of the blots with a probe for the housekeeping gene glyceraldehyde phosphate dehydrogenase (GAPDH) demonstrated that the lack of detectable class II mRNA in the trophoblast samples was not due to RNA degradation or variations in RNA loading. Thus, the absence of class II antigen expression in trophoblast cell lines is mediated at the level of transcription and/or RNA stability. Furthermore, class II mRNA expression appears to be coordinately regulated in trophoblast cells, as observed previously for class II-positive cells (Boothby et al., 1989). These results are consistent with previous studies using in situ hybridization to examine class II mRNA expression in trophoblasts at different stages of pregnancy (Peyman and Hammond, 1992; Lata et al., 1992).
Fig. 1.
MHC class II mRNA is not expressed in trophoblast cells. Total RNA was isolated from rat R8RP.3 and mouse SM9 cells grown for 24 hr in the absence or presence of 100 or 500 U/ml IFN-γ (rat for R8RP.3 and murine for SM9) and subjected to Northern blot analysis using cDNA probes for the mouse class II genes IAα, IAβ, IEα and IEβ. These probes contain the conserved α2 and β2 domains of the genes that crossreact with the corresponding rat mRNAs. RNA from mouse A20 B cells was included as a positive control for class II mRNA expression. Blots were rehybridized with the labeled cDNA encoding the housekeeping gene glyceraldehyde-6-phosphate dehydrogenase (GAPDH) as a control for RNA loading and integrity.
Identification of Negative Regulatory Regions Within the IAα Promoter That Function in Trophoblast Cells
To determine whether the lack of MHC class II mRNA expression in trophoblast cells is mediated at the level of transcription, transient transfection assays were performed using vectors containing the bacterial CAT gene under the control of promoter fragments from the mouse IAα gene. Our initial studies suggested that transcription from the IAα promoter is repressed in R8RP.3 and SM9 trophoblast cells by negative regulatory element(s) present between −1,200 and −393 (data not shown). In order to more precisely localize the putative negative regulatory region(s), a series of plasmids containing sequential 5′ deletions of the IAα promoter linked to the CAT gene was constructed and used in CAT assays (Fig. 2). No CAT activity was observed from R8RP.3 cells transfected with CAT plasmids containing 890 bp or more of upstream IAα promoter sequence [pIAα(1,200)CAT or pIAα(890)CAT; Fig. 2]; however, deletion to −780 [pIAα(780)CAT] resulted in clearly detectable activity. These results suggest that a negative regulatory element that functions to repress IAα gene transcription in trophoblast cells is localized between −890 and −780 of the IAα promoter. More detailed analysis of this region of the IAα promoter localized the negative regulatory region to between −864 and −810, since CAT activity was detected from R8RP.3 cells transfected with CAT plasmids containing 810 bp of IAα sequence, but not 864 bp (data not shown). Extracts from R8RP.3 cells transfected with CAT constructs containing IAα sequences from −631 or −541 to +1 had no detectable CAT activity (Fig. 2), suggesting that the sequences between −780 and −631 may contain a positive regulatory region. Further deletion to −501 resulted in restoration of IAα promoter activity (Fig. 2), suggesting that the region between −541 and −501 may also have a negative regulatory function in trophoblast cells. The results also demonstrate that the 393 bp IAα promoter fragment [pIAα(393)CAT], which contains the conserved W, X, and Y elements found in all class II promoters, is functional in trophoblast cells. Similar results were observed in CAT assays with SM9 trophoblast cells (data not shown).
The mouse B cell line A20, which constitutively expresses class II antigens, was included as a positive control for IAα promoter activity. Comparable CAT activity is observed in A20 cells transfected with pIAα(890)CAT and pIAα(780)CAT (Fig. 2), suggesting that the negative regulatory element located between −864 and −810 functions in class II-negative trophoblast cells but not class II-positive B cells. Deletion of the IAα promoter to −541 results in a minor decrease in CAT activity in A20 cells, but the levels increase after further deletion to −501. These results suggest that the IAα promoter region between −541 and −501 may function as a weak negative regulatory region in both R8RP.3 and A20 cells. However, this region (−541 to −501) does not appear to function in the context of the full length IAα promoter (see below).
Sequence-Specific, Single-Stranded DNA Binding Proteins From Trophoblast Cells Bind to the IAα-Negative Regulatory Region
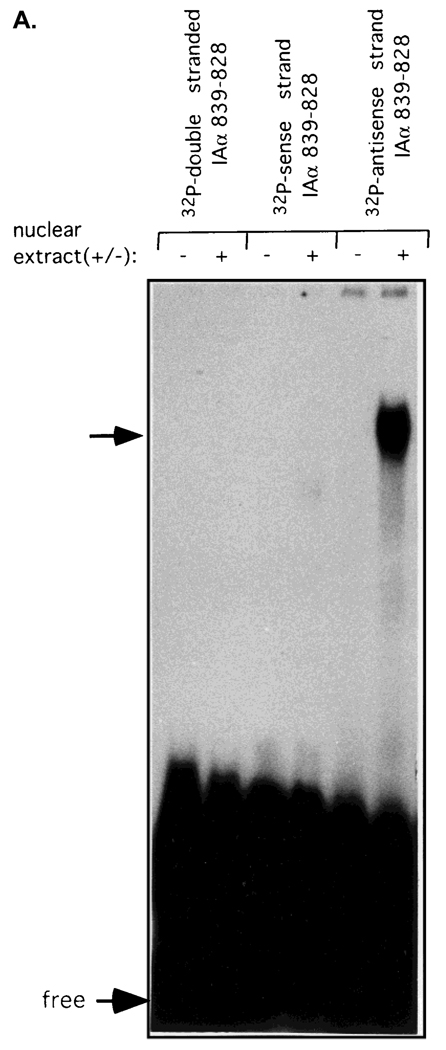
Since the upstream negative regulatory region (−864 to −810) appeared to be sufficient for repressing IAα promoter activity in trophoblast cells (see below), we concentrated on examining potential protein:DNA interactions between transcription factors and this IAα sequence. A series of oligonucleotides was synthesized that correspond to this region (see Table 1), and used in electrophoretic mobility shift analysis (EMSA) with nuclear extracts isolated from R8RP.3 trophoblast cells. No DNA:protein complexes were detected when either double-stranded IAα oligonucleotides or single-stranded oligonucleotides corresponding to the IAα sense strand sequences were utilized in EMSA (Fig. 3A; Table 1). However, DNA:protein complexes were observed with the single-stranded oligonucleotides that contained the sequences corresponding to −839 to −828 (5′-TTTTCCAAATTT-3′) of the IAα antisense strand (Fig. 3A; Table 1). The binding of proteins from R8RP.3 trophoblasts to the IAα antisense strand from −839 to −828 appeared to be specific, since preincubation of the R8RP.3 nuclear extracts with excess unlabeled oligonucleotides corresponding to the sense strand of IAα, or a series of unrelated oligonucleotides, had no effect on complex formation, while excess antisense strand oligonucleotide competed for binding (Fig. 3B; data not shown). The IAα sequence between −839 to −828 will hereafter be referred to as the IAα-negative regulatory element (IAαNRE).
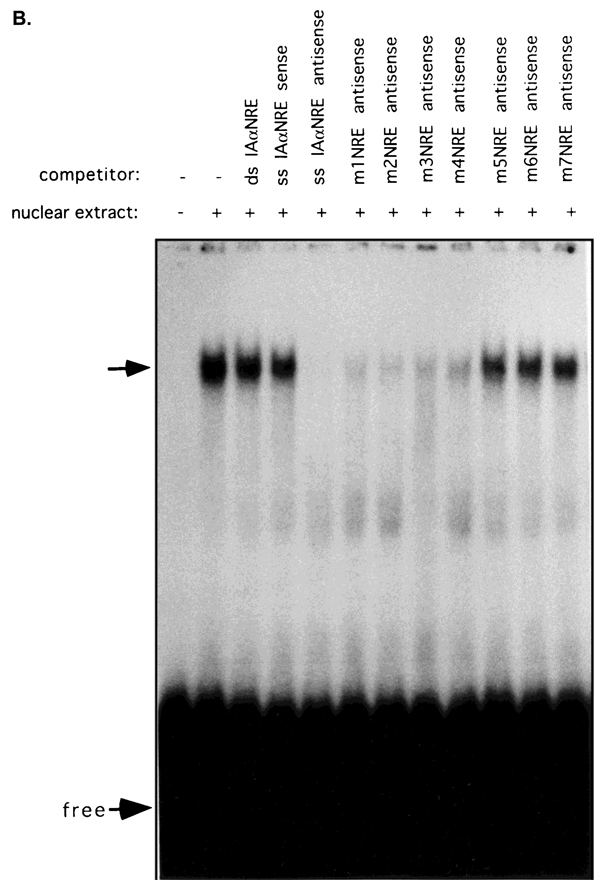
Fig. 3.
Transacting factors from trophoblast cells bind specifically to the antisense strand of the IAα promoter sequence from −839 to −828. A: EMSA was performed using nuclear extracts from R8RP.3 cells and double- or single-stranded oligonucleotides corresponding to the IAα sequence from −839 to −828. B: Competition analysis of IAαNRE BP binding. R8RP.3 nuclear extracts were preincubated with a 200-fold molar excess of unlabeled competitor oligonucleotides for 15 min prior to the binding reaction with the radiolabeled IAαNRE oligonucleotide. The sequences of the competitor oligonucleotides are shown in Table 2. The top arrows indicate the specific IAαNRE DNA:protein complexes. “Free” refers to the unbound oligonucleotides.
Competition analysis was subsequently performed using a series of oligonucleotides corresponding to mutated versions of IAαNRE in order to examine the specific nucleotide bases that are essential for factor binding (Fig. 3B; Table 2). Oligonucleotides containing mutations of single Ts within either of the T triplets (m8NRE; m9; m12; and m13) successfully competed for binding of the wildtype IAαNRE sequence; however, oligonucleotides in which all three Ts within either triplet (m6NRE; m7NRE) were mutated did not compete. None of the mutations of internal bases had an effect on competition for binding to the wild-type sequence. These results suggest that IAαNRE binding proteins (IAαNRE BPs) are relatively promiscuous in their sequence requirements for binding DNA, but that both of the T triplets are necessary for recognition.
Cell Type Specificity of IAαNRE Binding Activity
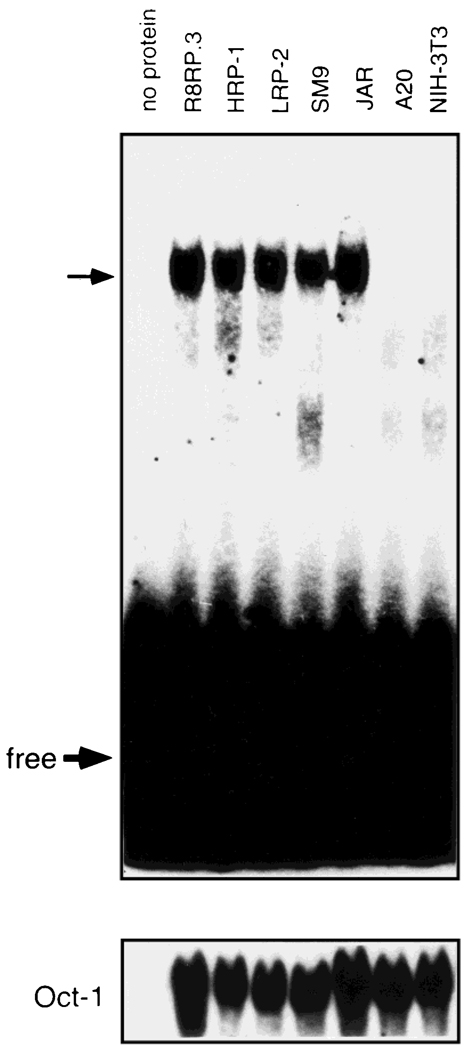
Nuclear extracts from a total of 24 cell lines (Table 3) were used in EMSA to examine the cell and tissue-type distribution of IAαNRE BPs. IAαNRE DNA:protein complexes of identical mobility were observed in nuclear extracts from one mouse (SM9) and all four rat (R8RP.3; HRP-1; LRP-2; Rcho-1) trophoblast cell lines that were tested (Fig. 4; Table 3). In addition, IAαNRE binding activity was detected in the human choriocarcinoma cell line JAR. The IAαNRE BPs from all six trophoblast cell lines shared similar profiles in competition analysis (data not shown). These results indicate that IAαNRE BPs are conserved in species with hemochordial placentas. With the exception of a Chinese hamster ovary (CHO) cell line, IAαNRE binding activity was not detected in any of the other cell lines tested, including A20 B cells and NIH-3T3 fibroblasts (Fig. 4), and F9 and P19 EC cells (Table 3). These results suggest that the activity of the IAαNRE BPs is restricted primarily to trophoblast cells.
TABLE 3.
IAαNRE Binding Activity Is Restricted Primarily to Trophoblasts*
| Cell line | Species | Cell/tissue type |
IAαNREBP (+/−) |
|---|---|---|---|
| HRP-1 | rat | trophoblast | + + |
| LRP-2 | rat | trophoblast | + + |
| Rcho-1 | rat | trophoblast | + + |
| R8RP.3 | rat | trophoblast | + + |
| SM9 | mouse | trophoblast | + + |
| JAR | human | choriocarcinoma | + + |
| NIH-3T3 | mouse | fibroblast | − − |
| C3H 10T1/2 | mouse | fibroblast | − − |
| F9 | mouse | EC | − − |
| P19 | mouse | EC | − − |
| MC3T3.E1 | mouse | osteoblast | − − |
| UMR-106 | rat | osteosarcoma | − − |
| BRL | rat | liver | − − |
| HeLa | human | cervical carcinoma | − − |
| CHO | hamster | ovary | + + |
| SK-OV-3 | human | ovarian carcinoma | − − |
| A20 | mouse | B lymphoma | − − |
| Raji | human | B lymphoma | − − |
| L1210 | mouse | B lymphoma | − − |
| SCC | human | squamous carcinoma | − − |
| Sa1 | mouse | sarcoma | − − |
| CV-1 | monkey | kidney | − − |
| WEHI | mouse | B cell | − − |
| Hs294T | human | melanoma | − − |
EMSA was performed using oligonucleotides corresponding to the IAαNRE antisense strand sequence (839−828) and nuclear extracts from the cells listed. At least three independent extracts were examined for each cell type.
Fig. 4.
IAαNRE binding activity is restricted primarily to trophoblast cells. EMSA was performed as described in Figure 3 using the oligonucleotide corresponding to the antisense strand of the IAα sequence from −839 to −828 and nuclear extracts from rat trophoblast cells (R8RP.3; HRP-1 and LRP-2), mouse trophoblast (SM9), human choriocarcinoma (JAR), mouse B cells (A20), and fibroblasts (NIH-3T3). The same nuclear extracts were also used in EMSA with an Octamer-1 (Oct-1) oligonucleotide as a control for protein loading and extract integrity. The top arrow corresponds to the IAαNRE: protein complex. “Free” refers to unbound oligonucleotide.
The IAαNRE Binding Protein Sequence Is Sufficient to Repress IAα Transcription
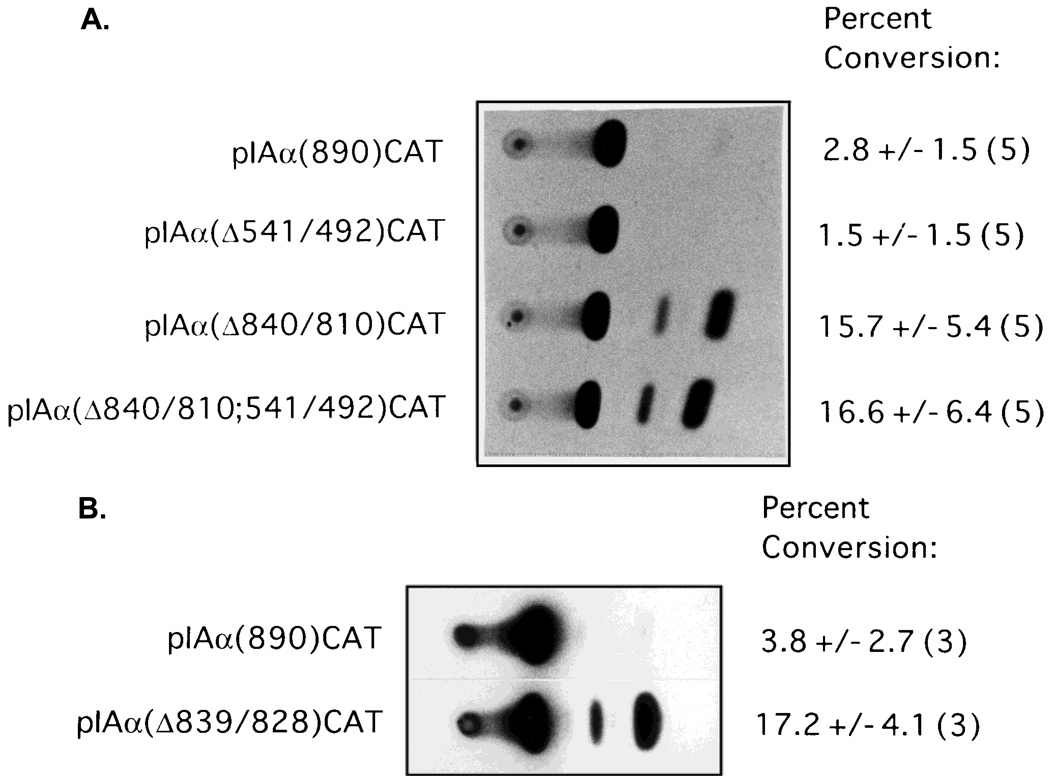
To further characterize the functional role of the putative negative regulatory regions in the repression of IAα promoter activity in R8RP.3 cells, PCR-based mutagenesis was used to selectively delete the regions from −541 to −492 and −840 to −810 from the 890-bp IAα promoter, and the resulting mutants were inserted into pCATbasic to form pIAα(Δ541/492)CAT and pIAα(Δ840/810)CAT, respectively. No CAT activity was observed from R8RP.3 cells transfected with pIAα(Δ541/492)CAT (Fig. 5A). However, selective deletion of the IAα sequences between −840 and −810 alleviated repression of transcription from the 890 bp promoter in R8RP.3 cells (Fig. 5A), while having no significant effect on IAα promoter activity in A20 cells (data not shown). Deletion of both the sequences from −840 to −810 and −541 to −492 resulted in little or no increase in CAT activity relative to deletion of −840 to −810 alone. These results are consistent with the results of the 5′ deletion analysis and indicate that the IAα sequence from −840 to −810 is sufficient to repress the activity of the 890 bp IAα promoter fragment in R8RP.3 cells. Furthermore, the region of the IAα promoter between −541 and −501 does not function as a negative regulatory element in the context of the full length IAα promoter.
Fig. 5.
The minimal sequence required for IAα promoter repression corresponds to transacting factor binding sites. A: CAT assays were performed using extracts of R8RP.3 cells transfected with pIAα(890)CAT and its derivatives containing site-specific deletions of the regions from −541 to −492 (pIAαΔ541/492CAT) and −840 to −810 (pIAαΔ840/810CAT). B: PCR-based mutagenesis was used to selectively delete the sequences between −839 and −828 from the 890 bp IAα promoter, and the resulting mutant promoter was cloned into pCATbasic to form pIAα(Δ839/828)CAT. Shown are representative experiments with the average percent conversion of the nonacetylated to the acetylated form of chloramphenicol and the standard error calculated from the number of experiments indicated in parentheses.
Identical results were obtained in transient transfection assays with mouse SM9 and rat Rcho-1 trophoblast cells (data not shown), indicating that IAαNRE activity is not unique to R8RP.3 cells. In addition, stable R8RP.3 cell lines containing integrated copies of the IAα(890)CAT, pIAα(541/492)CAT, pIAα(Δ840/810)CAT and pIAα(Δ541/492;840/810)CAT plasmids were constructed and tested for CAT activity. The results with the stable cell lines were identical to those from the transient assays, suggesting that the repressive function of IAαNRE is not affected by chromatin conformation (data not shown).
To examine whether the IAαNRE BP binding site corresponds to the minimal sequence required for transcriptional repression, PCR-based mutagenesis was used to selectively delete the region from −839 to −828 from the 890-bp IAα promoter, and the activity of the resulting mutant promoter was tested in transient transfection studies. Deletion of the sequences from −839 to −828 resulted in constitutive IAα promoter activity in R8RP.3 trophoblast cells (Fig. 5B), indicating that the IAαNRE binding protein site is sufficient to repress IAα promoter activity in trophoblast cells.
The Effects of IAαNRE on Homologous and Heterologous Promoters
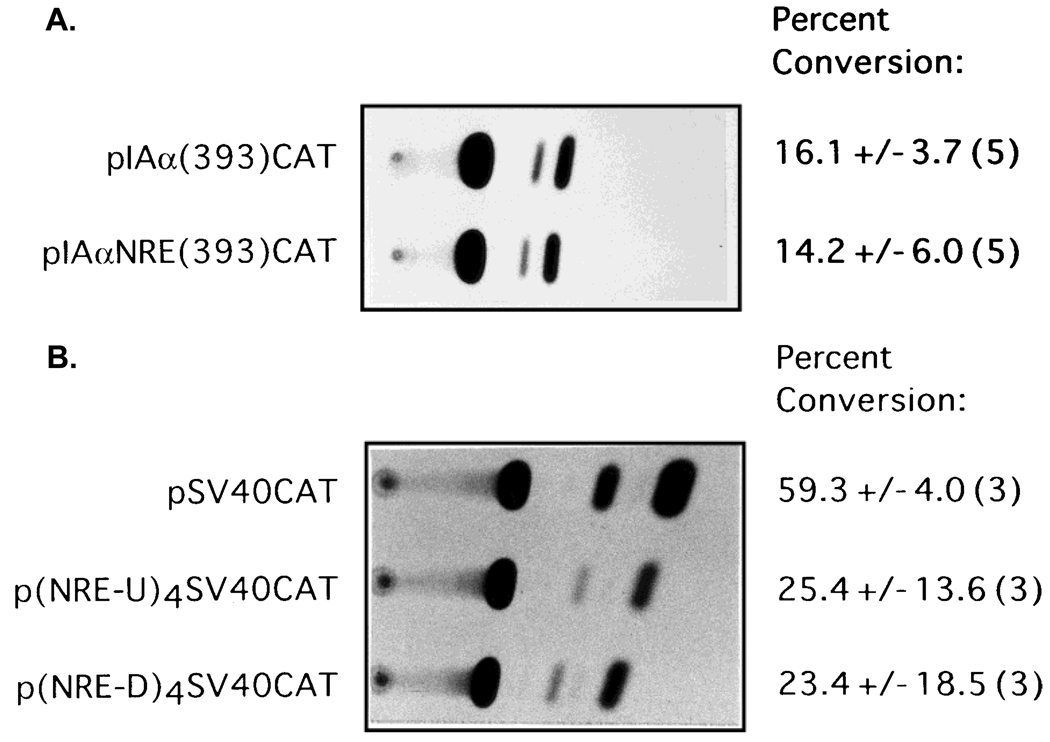
In order to test whether IAαNRE functions in a position- and/or distance-independent manner, the sequences from −839 to −828 were inserted directly upstream of the 393 bp IAα promoter fragment in pIAα(393)CAT, and the resulting vectors were transfected into R8RP.3 cells. The level of CAT activity from R8RP.3 cells transfected with pIAαNRE(393)CAT was indistinguishable from transfectants with pIAα(393)-CAT (Fig. 6A), suggesting that the repressive function of IAαNRE is location-dependent, and that IAαNRE repressive activity may require additional “accessory” sequences within the IAα promoter.
Fig. 6.
The effects of IAαNRE on homologous and heterologous promoters. A: The repressive function of IAαNRE is position-dependent. An oligonucleotide corresponding to the IAα sequence from −839 to −828 was cloned directly upstream of the 393 bp IAα proximal promoter in pIAα(393)CAT to form pIAαNRE(393)CAT. B: The IAαNRE region reduces the activity of a heterologous promoter. Four repeats of IAαNRE were subcloned directly upstream of the SV40 promoter in pCAT control to create p(IAαNRE-4U)[SV40]CAT, or downstream of the SV40 enhancer to create p(IAαNRE-4D)[SV40]CAT. For both A and B, R8RP.3 cells were transfected and assayed for CAT activity. Representative experiments are shown; the numbers to the right of each autoradiograph represent the average percent conversion of the nonacetylated to the acetylated form of chloramphenicol and the standard error calculated from the number of experiments indicated in parentheses.
To examine whether IAαNRE could down-regulate the activity of a heterologous promoter, four repeats of the IAαNRE sequence were inserted either upstream of the SV40 promoter or downstream of the SV40 enhancer in pCATcontrol (Promega) and the resulting recombinants were tested in R8RP.3 cells in transient transfection assays. Insertion of four copies of IAαNRE upstream of the SV40 promoter [p(IAαNREU)4-[SV40]CAT] decreased CAT activity 2-fold in R8RP.3 cells relative to pCATcontrol (Fig. 6B), but had no effect in B cells (data not shown). The presence of IAαNRE downstream of the SV40 enhancer [p(IAαNRED)4[SV40]CAT] resulted in a 2.2-fold reduction of activity relative to pCATcontrol (Fig. 6B). These results suggest that although IAαNRE can downregulate a heterologous promoter, the effect is relatively minor.
DISCUSSION
Two basic mechanisms have been proposed to explain the absence of MHC class II gene transcription in class II-negative cells: positive transacting factors required for class II gene transcription are missing or mutated, or negative transacting factors actively repress class II gene transcription. The absence of positive transacting factors in class II-negative cells has been demonstrated in several studies. For example, the B cells of patients with a severe combined immunodeficiency disease called bare lymphocyte syndrome (BLS) do not express class II antigens (dePravel et al., 1985; Griscelli et al., 1989). The B cells of patients in one complementation group of BLS lack RFX, a transactivating factor that binds to the X box of all class II promoters, and is required for class II expression (Reith et al., 1988, 1989). Another group of BLS patients fail to express class II transactivating factor (CIITA), which does not bind DNA but has been shown to be required for both constitutive and cytokine-inducible class II gene expression (Steimle et al., 1993, 1994; Chang et al., 1994; Chin et al., 1994). Furthermore, the downregulation of MHC class II gene expression during B cell differentiation into plasma cells appears to result from silencing of CIITA expression (Silacci et al., 1994).
Several groups have defined negative regulatory elements (NREs) in class II genes that function in cells which are inducible for class II gene expression by IFN-γ. Boss and Strominger (1986) demonstrated that sequential deletion of the human HLA-DQβ promoter between −2,500 and −82 resulted in stepwise increases in promoter activity in the human fibroblast cell lines M1XP and 143b. The sequences of the HLA-DQβ promoter from −159 to −128 and −107 to −82 were identified as negative regulatory regions, and the region between −2,500 and −159 also contained repressor element(s) that function in human fibroblasts (Boss and Strominger, 1986). Deletion analysis of the murine IEα promoter demonstrated that the region between −873 and −353 contains negative cis-acting element(s) that decreased basal promoter activity in a class II cytokine-inducible xeroderma cell line (Thanos et al., 1988). An NRE has been identified between −552 and −489 of the class II IAβ promoter that functions in low class II-expressing renal tubular epithelial cells, NIH-3T3 fibroblasts and thymoma cells (Albert et al., 1994). This element, termed IAβNRE, represses the activity of both the IAβ promoter and the SV40 early promoter, and functions in a position- and distance-independent but orientation-dependent manner (Albert et al., 1994).
We have identified a negative regulatory element (IAαNRE) in the IAα promoter, located from −839 to −828, that represses IAα promoter activity in three different trophoblast cell lines. The IAαNRE sequence has only limited homology to the IAβNRE described by Albert et al. (1994), implying that these elements are different. The functional properties of these NREs and cell type distribution of the respective binding proteins further suggest that this is the case. While IAβNRE functions in NIH-3T3 cells (Albert et al., 1994), deletion of IAαNRE has no effect on IAα promoter activity in NIH-3T3 cells (data not shown). Furthermore, EMSA analysis indicated that IAαNRE binding protein activity is not present in NIH-3T3 cells, and is restricted primarily to trophoblasts (Fig. 4). Lastly, IAβNRE function is position- and distance-independent while IAαNRE is not (Fig. 6A).
The majority of transcription factors recognize and bind to double-stranded DNA; however, there are a number of reports of single-stranded DNA binding proteins that have been implicated in both positive and negative regulation of gene expression (Pan et al., 1990; O’Neill et al., 1991; Kamada and Miwa, 1992; Altiok and Groner, 1993; Tanuma et al., 1995; Sun et al., 1995). Among these are cell type-specific single-stranded DNA binding proteins present in mammary epithelial cells that regulate β-casein gene transcription (Altiok and Groner, 1993), and factors in myoblasts and fibroblasts that negatively regulate α-actin gene transcription (Sun et al., 1995). The IAαNRE BPs are similar to a subset of these single-stranded DNA binding proteins in that they recognize a sequence that is composed primarily of pyrimidines. The IAαNRE sequence (5′-TTTTCCAAATTT-3′) is 75% pyrimidine, and is located within a 68-bp region that is 69% pyrimidine. Regions of DNA that are pyrimidine-rich have been shown to be important in the regulation of transcription (O’Neill et al., 1991), in some cases by forming triplex DNA structures (Htun and Dahlberg, 1988; Wells et al., 1988). The observation that IAαNRE activity is position- and distance-dependent suggests that it may require additional “accessory” sequences within the IAα promoter in order to function. Perhaps the Py-rich region surrounding the IAαNRE is required to alter the DNA conformation such that IAαNRE becomes single-stranded, which would facilitate binding by the negative transacting factors.
The YB-1 protein, which is expressed in both class II-positive and -negative cells, has been shown to downregulate class II gene transcription by interacting with the sense strand of the X box (MacDonald et al., 1995; Lloberas et al., 1995). The authors proposed that YB-1 binding to the sense strand may result in unwinding or melting of the proximal promoter region, which would effectively compete for or prevent binding by RFX (MacDonald et al., 1995). The precise mechanism by which IAαNRE BPs repress class II gene transcription in trophoblast cells is not yet clear, but it may be similar to YB-1, which inhibits the binding of transacting factors to the X and Y boxes in the proximal promoter. However, since the IAαNRE is over 700 bp from the proximal promoter elements, and the IAαNRE BPs bind the antisense strand, the specific mechanism by which X and Y box factor binding would be repressed by IAαNRE BPs is likely to differ from that of YB-1. Although the factors that bind the X and Y elements are present in class II-negative cells that lack CIITA (such as fibroblasts), genomic footprinting analysis has demonstrated that the W, X and Y boxes in the proximal promoter region are unoccupied in vivo in these cells (Kara and Glimcher, 1991). CIITA is thought to both stabilize the binding of RFX to the X1 box and to form a higher order protein:protein complex that provides a transcriptional activating function (Riley et al., 1995; Zhou and Glimcher, 1995). The expression of MHC class II genes in trophoblasts may therefore be the result of a balance between the activity of IAαNRE and coactivators such as CIITA. Other possibilities include interactions with histones or factors such as HMG that bend DNA (Abdulkadir and Ono, 1995; Kingston et al., 1996). Genomic footprinting of the stable R8RP.3 clones containing integrated copies of the IAαproCAT genes will help address these alternatives.
The Northern blot analysis demonstrating that none of the class II genes are expressed in trophoblast cells (Fig. 1) suggests that the genes are coordinately repressed. Is transcription of all of the class II genes repressed in trophoblasts by IAαNRE BPs, or are multiple regulatory proteins involved? A search of the Genbank database revealed homology between IAαNRE and sequences in the IEα (73%), human HLA-DRα (78%), -DQβ (83%) and the rat RT1.1β (80%) gene promoters, but only low homology with the IAβ or IEβ promoters. These results imply that more than one factor could be involved in repression of class II gene expression in trophoblasts. Precedence for isotype-specific repression of class II gene transcription has been demonstrated in studies on the human HLA-DPA gene (Scholl et al., 1996). However, the competition analysis demonstrating that the IAαNRE BPs are relatively promiscuous in terms of their sequence recognition properties suggest that it may be difficult to identify bonafide IAαNRE homologies that are functionally relevant by computer search. Thus, IAαNRE BPs may regulate all class II genes in trophoblasts by binding poorly conserved versions of the IAαNRE recognition sequence. Cloning of the genes encoding the IAαNRE BPs and generation of recombinant proteins will allow us to directly address this issue.
The conservation of the IAαNRE BPs in species with hemochordial placentas and the observation that their activity appears to be restricted primarily to trophoblast cells raise interesting questions regarding fetal maintenance. Did repression of class II gene transcription in trophoblast cells mediated by IAαNRE BPs arise during evolution to prevent fetal rejection, thereby maintaining reproduction and perpetuation of the species? The importance of class II gene regulation to fetal maintenance is suggested by the studies of Athanassakis-Vassiliadis et al. (1990, 1995). Induction of class II antigen expression on fetal placental cells by 5-azacytidine was shown to correlate with fetal abortion (Athanassakis-Vassiliadis et al., 1989). Moreover, aberrant class II antigen expression was frequently observed in the trophoblast cells of placentas from human miscarriages (Athanassakis-Vassiliadis et al., 1995). Numerous studies have demonstrated that the maternal immune system is capable of recognizing fetally derived paternal antigens and mounting an immune response against the fetus (Teraski et al., 1970; Van Rood et al., 1985). In addition, several groups have demonstrated a T cell proliferative response to antigen presented by “nontraditional” antigen presenting cells (reviewed in Nickoloff and Turka, 1994). Therefore, aberrant expression of class II antigens on trophoblast cells could potentially lead to the presentation of unique fetal antigens (other than paternal class II), which would result in the generation of an immune response and loss of fetal viability. Thus, the repression of class II gene transcription that we observed may play a crucial role in the maintenance of normal pregnancy. Targetted gene knockout of the IAαNRE BP gene may shed light on this issue.
The identification of IAαNRE BPs in trophoblast cells may also have implications in tumor biology. A number of reports have demonstrated an inverse correlation between class II antigen expression and tumorigenicity (Ostrand-Rosenberg et al., 1990; Baskar et al., 1993; Bateman et al., 1991; Fuji and Iribe, 1986). Murine Sa1 sarcoma cells transfected with class II expression vectors were found to be significantly more immunogenic, and less tumorigenic, than the parental cells (Ostrand-Rosenberg et al., 1990; Baskar et al., 1993). Studies in which transformed subclones of C3H 10T1/2 cells were selected for varying levels of IFN-γ-induced class II expression demonstrated that C3H 10T1/2 clones which were noninducible for class II antigen expression were significantly more tumorigenic than their more highly expressing counterparts (Bateman et al., 1991). In addition, class II expression has been shown to be inversely correlated with the tumorigenicity of clones of the L1210 B lymphoma line (Fuji and Iribe, 1986). Preliminary evidence from our laboratory (Murphy et al., unpublished) reveals that a class II-negative, tumorigenic L1210 clone contains a single-stranded IAαNRE binding protein(s) similar to those detected in trophoblast cells, while a class II-expressing variant clone that is nontumorigenic does not contain the factor(s). These data suggest that tumor cells lacking class II antigens may be at a selective advantage for tumor formation and metastasis, and that certain tumors may have subverted the same evolutionary mechanisms developed to prevent fetal rejection in order to maintain growth in an immunocompetent host.
ACKNOWLEDGMENTS
We appreciate the advice in various aspects of this work by Drs. M. Boothby (Vanderbilt University) and J.S. Hunt (Department of Anatomy and Cell Biology, University of Kansas Medical Center, Kansas City, KS). We also thank Drs. M. Boothby and L.H. Glimcher (Harvard Medical School), for the pUC[IAα1.9] and pA10[IAα(0.45)]CAT plasmids, Dr. J.S. Hunt for the rat trophoblast cell lines (R8RP.3, HRP.1, LRP-2) and the mouse trophoblast cell line (SM-9), and Dr. M. Soares (Department of Physiology, University of Kansas Medical Center, Kansas City, KS) for the rat trophoblast cell line Rcho-1. We thank Ms. C. Grande and C. Budzinski for help in the preparation of this manuscript, and Dr. Peter Aplan for critical review.
Contract grant sponsor: NIH; Contract grant number: HD-17013;
Contract grant sponsor: Peter and Elizabeth C. Tower Foundation.
REFERENCES
- Abdulkadir SA, Ono SJ. How are class II MHC genes turned on and off? FASEB J. 1995;9:1429–1435. doi: 10.1096/fasebj.9.14.7589984. [DOI] [PubMed] [Google Scholar]
- Albert SE, Strutz F, Shelton K, Haverty T, Sun MJ, Li S, Denham A, Maki RA, Neilson EG. Characterization of a cis-acting regulatory element which silences expression of the class II-A β gene in epithelium. J Exp Med. 1994;180:233–240. doi: 10.1084/jem.180.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altiok S, Groner B. Interaction of two sequence-specific single-stranded DNA binding proteins with an essential region of the β-casein gene promoter is regulated by lactogenic hormones. Mol Cell Biol. 1993;13:7303–7310. doi: 10.1128/mcb.13.12.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassakis-Vassiliadis I, Aifantis Y, Makrygiannakis A, Koumantakis E, Vassiliadis S. Placental tissue from human miscarriages expresses class II HLA-DR antigens. Amer J Reprod Immunol. 1995;34:281–287. doi: 10.1111/j.1600-0897.1995.tb00954.x. [DOI] [PubMed] [Google Scholar]
- Athanassakis-Vassiliadis I, Galanopoulos VK, Grigoriou M, Papamatheakis J. Induction of class II MHC antigen expression on the murine placenta by 5-azacytidine correlates with fetal abortion. Cell Immunol. 1990;128:438–449. doi: 10.1016/0008-8749(90)90039-t. [DOI] [PubMed] [Google Scholar]
- Baskar S, Ostrand-Rosenberg S, Nabavi N, Nadler LM, Freeman GJ, Glimcher LH. Constitutive expression of B7 restores immunogenicity of tumor cells expressing truncated major histocompatibility complex class II molecules. Proc Natl Acad Sci USA. 1993;90:5687–5690. doi: 10.1073/pnas.90.12.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman WJ, Fiera R, Matthews N, Morris AG. Inducibility of class II major histocompatibility antigens by interferon γ is associated with reduced tumorigenicity in C3H mouse fibroblasts transformed by v-Ki-ras. J Exp Med. 1991;173:193–196. doi: 10.1084/jem.173.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C, Mathis D. Regulation of major histocompatibility complex class II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- Billingham WD. Immunobiology of Trophoblast: Organization, Ultrastructure and histochemistry of the placenta: Immunological considerations. In: Edwards RG, Howe CWS, Johnson MH, editors. Cinical and Experimental Immunoreproduction. Vol. 1. Cambridge, England: Cambridge University Press; 1975. pp. 67–86. [Google Scholar]
- Boothby M, Gravallese E, Liou H-C, Glimcher LH. A DNA binding protein regulated by IL-4 and by differentiation in B cells. Science. 1988;242:1559–1562. doi: 10.1126/science.3144043. [DOI] [PubMed] [Google Scholar]
- Boothby M, Liou H-C, Glimcher LH. Differences in DNA sequence specificity among MHC class II X box binding proteins. J Immunol. 1989;142:1005–1015. [PubMed] [Google Scholar]
- Boss JM, Strominger JL. Regulation of a transfected human class II major histocompatibility complex gene in human fibroblasts. Proc Natl Acad Sci USA. 1986;83:9139–9143. doi: 10.1073/pnas.83.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-H, Fontes JD, Peterlin M, Flavell RA. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee-Hasrouni S, Lala PK. MHC antigens on mouse trophoblast cells: Paucity of Ia antigens despite the presence of H-2K and D. J Immunol. 1981;127:2070–2073. [PubMed] [Google Scholar]
- Chin K-C, Mao C, Skinner C, Riley JL, Wright KL, Moreno CS, Stark GP, Boss JM, Ting JP-Y. Molecular analysis of G1B and G3A IFNγ mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Davis MM, Cohen DI, Nielson EA, Steinmetz M, Paul WE, Hood L. Cell type specific cDNA probes and the murine I region: The localization and orientation of IAα. Proc Natl Acad Sci USA. 1984;81:2194–2198. doi: 10.1073/pnas.81.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePravel C, Lisowska-Grospierre B, Loche M, Mach B. A transacting class II regulatory gene unlinked to the MHC controls expression of HLA class II genes. Nature. 1985;218:291–293. doi: 10.1038/318291a0. [DOI] [PubMed] [Google Scholar]
- Faria TN, Soares MJ. Trophoblast cell differentiation: Establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology. 1991;129:2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- Fuji H, Iribe H. Clonal variation in tumorigenicity of L1210 lymphoma cells: Nontumorigenic variants with an enhanced expression of tumor-associated antigen and Ia antigens. Cancer Res. 1986;46:5541–5547. [PubMed] [Google Scholar]
- Glimcher LH, Kara CJ. Sequences and factors: A guide to MHC class II transcription. Ann Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- Gravellese EM, Boothby MR, Smas CM, Glimcher LH. A lipopolysaccaride-induced DNA binding protein for a class II gene in B cells is distinct from NF-kB. Mol Cell Biol. 1989;9:3184–3192. doi: 10.1128/mcb.9.8.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griscelli C, Lisowska-Grospierre B, Mach B. Immunodeficiency with defective expression in MHC class II genes. Immunodefic Rev. 1989;1:135–153. [PubMed] [Google Scholar]
- Htun H, Dahlberg JE. Single strands, triple strands, and kinks in H-DNA. Science. 1988;241:1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Hunt JS, Deb S, Faria TN, Wheaton D, Soares MJ. Isolation of phenotypically distinct trophoblast cell lines from normal rat chorioallantoic placentas. Placenta. 1989;10:161–177. doi: 10.1016/0143-4004(89)90038-6. [DOI] [PubMed] [Google Scholar]
- Hunt JS, Atherton RA, Pace JL. Differential responses of rat trophoblast cells and embryonic fibroblasts to cytokines that regulate proliferation and class I MHC antigen expression. J Immunol. 1990a;145:184–189. [PubMed] [Google Scholar]
- Hunt JS, Fishback JL, Chumbley G, Loke YW. Identification of class I MHC mRNA in human first trimester trophoblast cells by in situ hybridization. J Immunol. 1990b;144:4420–4425. [PubMed] [Google Scholar]
- Kamada S, Miwa T. A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene. 1992;119:229–236. doi: 10.1016/0378-1119(92)90276-u. [DOI] [PubMed] [Google Scholar]
- Kara CJ, Glimcher LH. In vivo footprinting of MHC class II genes: Bare promoters in the bare lymphocyte syndrome. Science. 1991;252:709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–910. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Lata JA, Tuan RS, Shepley KJ, Mulligan MM, Jackson LG, Smith JB. Localization of major histocompatibility complex class I and II mRNA in human first trimester chorionic villi by in situ hybridization. J Exp Med. 1992;175:1027–1032. doi: 10.1084/jem.175.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloberas J, Maki RA, Celada A. Repression of major histocompatibility complex I-Aβ gene expression by dbpA and dbpB (mYB-1) proteins. Mol Cell Biol. 1995;15:5092–5099. doi: 10.1128/mcb.15.9.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald GH, Itoh-Lindstrom Y, Ting JY-P. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J Biol Chem. 1995;270:3527–3533. doi: 10.1074/jbc.270.8.3527. [DOI] [PubMed] [Google Scholar]
- McNichols J, Steinmetz M, Hunkupiller T, Jones P, Hood L. DNA sequence of the gene encoding the IEα Ia polypeptide of the Balb/c mouse. Science. 1982;218:1229–1232. doi: 10.1126/science.6815800. [DOI] [PubMed] [Google Scholar]
- Medawar PB. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol. 1953;7:320–338. [Google Scholar]
- Nickoloff BJ, Turka LA. Immunological functions of non-professional antigen-presenting cells: New insights from studies of T-cell interactions with keratinocytes. Immunol Today. 1994;15:464–469. doi: 10.1016/0167-5699(94)90190-2. [DOI] [PubMed] [Google Scholar]
- O’Neill D, Bornschlegel K, Flamm M, Castle M, Bank A. A DNA-binding factor in hematopoietic cells interacts with a pyrimidine-rich domain upstream from the human δ-globin gene. Proc Natl Acad Sci USA. 1991;88:8953–8957. doi: 10.1073/pnas.88.20.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S, Thakur A, Clements V. Rejection of mouse sarcoma cells after transfection of MHC class II genes. J Immunol. 1990;144:4068–4071. [PubMed] [Google Scholar]
- Pan WT, Liu Q, Bancroft C. Identification of a growth hormone gene promoter repressor element and its cognate double- and single-stranded DNA-binding proteins. J Biol Chem. 1990;265:7022–7028. [PubMed] [Google Scholar]
- Parslow TG, Blair DL, Murphy WJ, Granner DK. Structure of the 5′ ends of immunoglobulin genes: A novel conserved sequence. Proc Natl Acad Sci USA. 1984;81:2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazmany T, Murphy SP, Gollnick SO, Brooks SP, Tomasi TB. Activation of multiple transcription factors and fos and jun gene expression in cells exposed to a single electric pulse. Exp Cell Res. 1995;221:103–110. doi: 10.1006/excr.1995.1357. [DOI] [PubMed] [Google Scholar]
- Peyman JA, Hammond GL. Localization of IFN-γ receptor in first trimester placenta to trophoblasts but lack of stimulation of HLA-DRA, -DRB or invariant chain mRNA expression by IFN-γ. J Immunol. 1992;149:2675–2680. [PubMed] [Google Scholar]
- Peyman JA, Nelson PJ, Hammond GL. HLA-DR genes are silenced in human trophoblasts and stimulation of signal transduction pathways does not circumvent interferon-γ unresponsiveness. Transplantation Proc. 1992;24:470–471. [PubMed] [Google Scholar]
- Potter H, Weir L, Leder P. Enhancer-dependent expression of human κ immunoglobulin genes introduced into mouse pre-B lymphocytes by electroporation. Proc Natl Acad Sci USA. 1984;81:7161–7165. doi: 10.1073/pnas.81.22.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith W, Satola S, Sanchez CH, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam HR, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- Reith W, Barras E, Satola S, Kobr M, Reinhart D, Sanchez CH, Mach B. Cloning of the major histocompatibility complex class II promoter binding protein affected in a hereditary defect in class II gene regulation. Proc Natl Acad Sci USA. 1989;86:4200–4204. doi: 10.1073/pnas.86.11.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JL, Westerheide SD, Price JA, Brown JA, Boss JM. Activation of class II MHC genes requires both the X box region and the class II transactivator. Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- Robinson RR, Germain RN, McKean DJ, Seidman JG. Extensive polymorphism surrounding the murine Ia Aβ chain gene. J Immunol. 1982;131:2025–2032. [PubMed] [Google Scholar]
- Salter-Cid L, Flajnik MF. Evolution and developmental regulation of the major histocompatibility complex. Critical Rev Immunol. 1995;15:31–75. doi: 10.1615/critrevimmunol.v15.i1.20. [DOI] [PubMed] [Google Scholar]
- Scholl T, Stevens MB, Mahanta S, Strominger JL. A zinc finger protein that represses transcription of the human MHC class II gene, DPA. J. Immunol. 1996:1448–1457. [PubMed] [Google Scholar]
- Silacci P, Mottet A, Steimle V, Reith W, Mach B. Developmental extinction of major histocompatibility complex class II gene expression in plasmacytes is mediated by silencing of the transactivator gene CIITA. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- Steimle V, Siegrist C-A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-γ mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- Stein B, Rahmsdorf HJ, Steffen A, Litfin M, Herrlich P. UV-induced DNA damage is an intermediate step in UV-induced expression of human immunodeficiency virus type 1, collagenase, c-fos, and metallothionein. Mol Cell Biol. 1989;9:5169–5181. doi: 10.1128/mcb.9.11.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M, Minard K, Horvath S, McNichols J, Frelinger J, Wake C, Long E, Mach B, Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982;300:35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Sun S, Stoflet ES, Cogan JG, Strauch AR, Getz MJ. Negative regulation of the vascular smooth muscle α-actin gene in fibroblasts and myoblasts: Disruption of enhancer function by sequence-specific single-stranded DNA-binding proteins. Mol Cell Biol. 1995;15:2429–2436. doi: 10.1128/mcb.15.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanuma Y, Nakabayashi H, Esumi M, Endo H. A silencer element for the lipoprotein lipase gene promoter and cognate double- and single-stranded DNA-binding proteins. Mol Cell Biol. 1995;15:517–523. doi: 10.1128/mcb.15.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraski PI, Mickey MR, Yamerake JN, Vredevoe D. Maternal fetal incompatibility. I. Incidence of HL-A antibodies and possible association with congenital abnormalities. Transplantation. 1970;9:538–545. [PubMed] [Google Scholar]
- Thanos D, Mavrothalassitis G, Papamatheakis J. Multiple regulatory regions on the 5′ side of the mouse Eα gene. Proc Natl Acad Sci USA. 1988;85:3075–3079. doi: 10.1073/pnas.85.9.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus DE. In: Promega Protocols and Applications Guide. second edition. Titus DE, editor. Madison, WI: Promega Corp.; pp. 288–289. [Google Scholar]
- Van Rood JJ, Eernisse JG, van Leeuwen A. Leukocyte antibodies in sera from pregnant women. Nature (London) 1985;181:1735–1737. doi: 10.1038/1811735a0. [DOI] [PubMed] [Google Scholar]
- Wells RD, Collier DA, Hanvey JC, Shimuzi M, Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine: Oligopyrimidine sequences. FASEB J. 1988;2:2939–2949. [PubMed] [Google Scholar]
- Zhou H, Glimcher LH. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]