Abstract
Bone morphogenetic proteins (BMPs) play important roles in cardiovascular development. However, how BMP-signaling pathways regulate cardiac gene expression is less clear. We have previously identified myocardin as a cardiac and smooth muscle–specific transcriptional cofactor for serum response factor (SRF). Myocardin potently activates target gene expression by tethering with SRF bound to SRF-responsive elements, the CArG box. Here, we show that Smad1, an effector of the BMP-signaling pathway, synergistically activates myocardin-dependent cardiac gene expression. Interestingly, the CArG box is necessary and sufficient to mediate such synergy, whereas no obvious Smad-binding element appears to be involved. Consistent with their functional interaction, we find that myocardin and Smad1 proteins interact directly. Furthermore, myocardin protein levels were dramatically increased by BMP-2 treatment in cardiomyocytes. These findings suggest myocardin participates in a BMP signaling–dependent cardiac gene transcriptional program.
Keywords: myocardin, serum response factor, bone morphogenetic protein, Smad, cardiac gene expression
Myocardin is a serum response factor (SRF) cofactor expressed in cardiac and smooth muscle (SM) cell lineages.1 Although myocardin lacks intrinsic DNA-binding ability, it forms a stable ternary complex with SRF to potently activate muscle-specific genes through the consensus sequence CC(A/T)6GG, known as a CArG box.1–3 In addition to activating cardiac gene expression and the cardiogenesis program, myocardin is also a potent transactivator for smooth muscle cell (SMC) differentiation and SM-related gene expression.1,4–13 Despite the significant role of myocardin in controlling muscle gene expression, upstream signaling pathways that regulate myocardin activity remain unknown.
Bone morphogenetic proteins (BMPs) are growth and differentiation factors of the transforming growth factor (TGF)-β superfamily.14 Signaling by this superfamily is mediated by Smad proteins. There are three classes of Smads: receptor-regulated Smads (R-Smads), co-Smad (Smad4), and inhibitory Smads. To date, 3 R-Smads (Smad1, -5, and -8) participate in BMP signaling.15 Once activated, R-Smads form a heteromeric complex with Smad4 that translocates into the nucleus to regulate expression of BMP-responsive genes.15 Smad proteins bind DNA relatively weakly, but are strongly recruited to specific target genes by interacting with other transcription factors.15
BMPs and downstream BMP signaling effectors are essential for cardiovascular development.16–21 However, it is clear BMP signaling alone is insufficient to activate the cardiac gene program because BMP-signaling pathway components are expressed in a wide range of tissues and cell types outside of cardiac muscle. Indeed, the ability of BMP signaling to commit specific mesodermal cells to a cardiac fate requires the interpretation of BMP signaling in a cell type–specific mechanism. How BMP signaling interacts with cardiac transcriptional networks is largely unknown. In light of the role of BMP signaling and myocardin in cardiovascular development, we investigated whether BMP signaling might regulate myocardin-mediated cardiac gene expression.
In this report, we show that Smad1 synergistically activates myocardin-dependent cardiac gene expression. Interestingly, the CArG box is necessary and sufficient for such synergy, whereas no obvious Smad-binding elements (SBEs) are involved. Consistent with their functional interaction, myocardin and Smad1 proteins physically interact. Myocardin transactivity was repressed by Smad7, an inhibitory Smad, and enhanced by constitutively activated activin receptor-like kinase (ALK)-3, a type I BMP receptor. Furthermore, myocardin protein expression was dramatically induced by BMP-2 treatment in cardiomyocytes. These findings suggest a role for BMP signaling in regulating myocardin expression and activity to control cardiac gene expression.
Materials and Methods
Plasmid Constructs
Myocardin and SRF-expression plasmids have been described.1,22 Myocardin C-terminal deletion mutants (M1, M2, and M3) were cloned into a pcDNA expression vector with a N-terminal Myc tag. Myocardin and Smad1 cDNAs were cloned into pGEX-KG vector to generate glutathione S-transferase (GST) fusion proteins. Smad1, Smad4, Smad7, and constitutively activated (Q233D) ALK3 (ALK3 QD) expression plasmids have been described.23–26 Smad1 cDNA was subcloned into pM1 vector to make GAL4-Smad1 fusion protein.
Cell Culture and Luciferase Reporter Assays
α-Cardiac actin (α-CA),27 myosin light chain 2V (MLC2V),28 α-myosin heavy chain (α-MHC),1 and atrial natriuretic factor (ANF)1 promoter luciferase reporters have been described. The Nkx2.5 promoter and SBE mutations have been described and were cloned into pGL3.29 Truncated ANF luciferase reporters were generated by cloning the −406, −226, and −115 to +70 regions of the ANF promoter into pGL3. The −115 ANF reporter was further truncated by deleting the −5 to +70 region. The ANF SBE mutation (−5 to −2) was generated by site-directed mutagenesis. COS7 cells were cultured as described.1 Neonatal rat cardiomyocytes prepared as described.30 Reporter assays were conducted in triplicate at least 2 times in 12-well plates. Transfections were performed with either Fugene6 (Roche) or Lipofectamine (Invitrogen) reagents. Unless otherwise indicated, 100 ng of reporter and 200 ng of activator plasmids were used. A cytomegalovirus-lacZ reporter was used as an internal control to normalize for transfection efficiencies, and total amount of DNA per well was kept constant by adding the corresponding amount of empty expression vector. Statistical analysis was performed using the Student’s t test; comparisons were considered significant where P<0.05.
GST Protein Pull-Down Assays
GST alone, GST-Smad1, and GST-myocardin proteins were expressed and purified as described and used for in vitro binding assays.12 Smad1 and myocardin proteins were in vitro translated (Promega) and [35S] methianone labeled. Pull-downs were performed by incubating radiolabeled proteins with bead-bound GST fusion proteins in buffer (20 mmol/L Tris [pH 7.3], 150 mmol/L NaCl, 0.5% Nonidet P-40, and protease inhibitors) for 2 hours at 4°C, followed by 3 washes in the same buffer. Samples were analyzed by SDS-PAGE and autoradiography.
Communoprecipitation and Western Blotting Assays
COS7 cells were transfected with Myc-myocardin and Flag-Smad1 plasmids in 10-cm plates. After 48 hours posttransfection, whole-cell extracts were prepared in 1 mL of PBS buffer containing 1 mmol/L EDTA, 0.5% Triton X-100, 1 mmol/L phenylmethylsulfonyl fluoride, and protease inhibitors. Extracts were cleared by 10 000g centrifugation for 10 minutes, incubated with anti-Flag M2 affinity gel resin (Sigma) for 2 hours at 4°C and then washed 3 times in the same buffer, and samples were subsequently analyzed by SDS-PAGE and Western blot analysis using anti-Myc (1:2500; A-14, Santa Cruz Biotechnology) or anti-Flag (1:2500; M2, Sigma) antibodies.
Electrophoretic Mobility-Shift Assay
Electrophoretic mobility-shift assays (EMSAs) were performed essentially as described using the c-fos CArG probe.31 Complementary oligonucleotides were annealed and labeled using Klenow polymerase and [α-32P]dCTP. Labeled probe was incubated with in vitro translated SRF, Myc-tagged myocardin, and/or Smad1 proteins in gel-shift buffer. Antibody supershift experiments were performed with anti-SRF (G-20, Santa Cruz Biotechnology) or anti-Myc. DNA–protein complexes were separated by gel electrophoresis on a 5% nondenaturing polyacrylamide gel and visualized by autoradiography.
BMP-Response Assay
After serum starving overnight, cardiomyocytes cultured in 10-cm plates were treated with (or without treatment in control) 20 ng/mL recombinant BMP-2 (R&D Systems) for 48 hours, then harvested in 200 μL lysis buffer composed of PBS containing 0.5% Triton X-100, 1 mmol/L EDTA, 1 mmol/L phenylmethylsulfonyl fluoride, and protease inhibitors. Twenty microliters of lysate were loaded onto SDS-PAGE for Western blot analysis. Antibodies used were anti-myocardin (1:1000; sc-21559, Santa Cruz Biotechnology); anti–myocyte enhancer factor 2 (MEF2) (1:1000; sc-313, Santa Cruz Biotechnology); anti-SRF (1:2500; sc-335, Santa Cruz Biotechnology); anti–α-actinin (1:2500; sc-17829, Santa Cruz Biotechnology); and anti–β-tubulin (1:5000; T-4026, Sigma).
Results
Synergistic Activation of Cardiac Promoters by Myocardin and Smad Proteins
Smad1 is a downstream effector of BMP signaling that mediates target gene transcription; we, therefore, tested whether myocardin and Smad1 might activate cardiac gene expression in a cooperative manner. Whereas myocardin strongly activated the ANF promoter luciferase reporter (Figure 1A),1 coexpression of myocardin and Smad1 synergistically activated this reporter in COS7 cells. In contrast, Smad1 by itself did not significantly activate the ANF reporter (Figure 1A). Myocardin and Smad1 also synergistically activated the ANF reporter in cardiomyocytes (Figure 1G). Similarly, myocardin and Smad1 synergistically activated all other cardiac promoter reporters tested (Figure 1B through 1E). Because Smad1 is known to heterodimerize with Smad4, we investigated the effects of Smad4 on myocardin/Smad1 synergy and found that Smad4 further increased myocardin/Smad1 activation (Figure 1E).
Figure 1.
Synergistic activation of cardiac promoters by myocardin and Smad1. Luciferase reporters controlled by ANF (A), α-MHC (B), MLC2V (C), α-CA (D), and Nkx2.5 (E) promoters were transfected into COS7 cells with Smad1 and/or myocardin expression plasmids. F, COS7 cells were transfected with UAS-luciferase reporter and/or GAL4-Smad1 and myocardin (400 and 800 ng as indicated) expression plasmids. G, Cardiomyocytes were transfected with ANF luciferase reporter with Smad1 and/or myocardin expression plasmids. Values are the fold increase in luciferase activity relative to activation of the reporter alone. Error bars represent SD of at least 2 experiments. Student’s t test: *P<0.05, myocardin alone vs myocardin plus Smad1.
To rule out the possibility that the myocardin/Smad1 synergy observed was an indirect effect mediated through SRF, we tested whether myocardin and GAL4-Smad1 fusion protein could activate a GAL4-dependent luciferase reporter (UAS-luciferase). GAL4-Smad1 and myocardin activated the UAS-luciferase reporter in a dose-dependent manner (Figure 1F). Similarly, Smad1 synergized with GAL4-myocardin fusion protein to activate the UAS-luciferase reporter (data not shown). These results suggest myocardin/Smad1 synergy is directly mediated by myocardin and Smad1 interaction. Together, we conclude BMP-signaling mediator Smad1 dramatically enhances myocardin transactivation of cardiac promoters.
CArG Box Is Necessary and Sufficient to Mediate Myocardin and Smad1 Synergy
Deletion analysis of the ANF promoter was performed to determine the minimal region required for myocardin/Smad1 functional interaction. Coexpression of myocardin and Smad1 increased activation of the ANF promoter reporter ≈5-fold higher than myocardin alone (Figure 2A). Truncating the ANF promoter to −406 to +70 did not significantly affect the activation by myocardin alone or the synergy between myocardin and Smad1 (Figure 2A). The −226 to +70 ANF reporter, which excluded 1 of the 2 CArG boxes present within the ANF promoter (CArG-far, −397 to −77), dramatically decreased the activation of this reporter by myocardin alone (data not shown); however, the synergy between myocardin and Smad1 was unaffected (Figure 2A). Further deletion of the ANF promoter (−115 to +70) only slightly reduced reporter activation by myocardin and Smad1 (Figure 2A). Thus, the −115 to +70 region of the ANF promoter, containing a single CArG box, is sufficient to mediate myocardin/Smad1 synergy.
Figure 2.
CArG box is necessary and sufficient to mediate myocardin and Smad1 synergy. A, COS7 cells were transfected with the indicated ANF promoter luciferase reporters and/or myocardin and Smad1 expression plasmids. Values are the fold increase of luciferase activity by myocardin and Smad1 (black bars) relative to luciferase activity by myocardin alone (gray bars), which is assigned the value of 1. B, Luciferase reporter controlled by 4 copies of a consensus CArG box was transfected into COS7 cells with myocardin and/or Smad1 expression plasmids. Values are the fold increase in luciferase activity relative to activation of the reporter alone. Error bars represent SD of at least 2 experiments. Student’s t test: *P<0.05, myocardin alone vs myocardin plus Smad1.
To test whether the CArG box is required for Smad1 and myocardin functional interaction, the 2 CArG boxes within the ANF promoter luciferase reporter were mutated.1 Whereas myocardin and Smad1 could synergistically activate the ANF luciferase reporter with a CArG-far mutation, such synergy was abolished by double CArG mutations (Figure 2A). These data indicate Smad1 and myocardin functional interaction is CArG-box dependent.
To determine whether the CArG box is sufficient to mediate myocardin/Smad1 synergy, we used a luciferase reporter controlled by 4 tandem copies of a consensus CArG box.1 Myocardin alone activated the 4×CArG reporter ≈200-fold, whereas Smad1 and myocardin increased activation to ≈625-fold (Figure 2B). Thus, we conclude the CArG box is necessary and sufficient to mediate myocardin and Smad1 synergy.
SBE Is Not Required for Myocardin and Smad1 Synergy
Most Smads weakly interact with DNA through a SBE sequence within the promoter of responsive genes.32 The ANF −115 to +70 promoter region does not contain a consensus SBE (AGAC GTCT) but does have 3 AGAC, a half-SBE previously shown to be sufficient for Smad MH1-domain binding (Figure 3A).33,34 However, Smad1 or Smad4 did not bind to the 3 AGAC and flanking ≈10 bp by EMSA (data not shown). Mutating the AGAC closest to the TATAA box (−5 to −2), the only AGAC identified in the promoter region within the −115 to +70 of the ANF regulatory region (the other 2 are in the 5′ untranslated region of this gene; Figure 3A), did not affect activation by myocardin and Smad1 (Figure 3B). Myocardin and Smad1 synergistically activated a truncated ANF reporter (−115 to −5) where all 3 AGAC sites were deleted (data not shown), suggesting the SBE site is not required for myocardin and Smad1 synergy. Furthermore, we found that Smad1 did not activate a luciferase reporter controlled by 6×SBE from SM22, nor could Smad1 synergize with myocardin on this reporter (data not shown). Finally, we tested whether Smad1 and myocardin could activate the Nkx2.5 promoter luciferase reporter containing a SBE mutation.29 As shown in Figure 3C, SBE mutation did not affect the synergy of Smad1 and myocardin. Together, these data demonstrate that Smad1 can synergistically activate cardiac target gene expression with myocardin in a SBE-independent manner.
Figure 3.
SBE is not required for myocardin and Smad1 synergy. A, DNA sequence of the −115 to +70 region of the ANF promoter. Underlined are CArG-near, TATA box, the mutated half-SBE site, and 2 other half-SBE sites (AGAC) within the 5′ untranslated region. B, Luciferase reporters controlled by the ANF promoter (gray bars) or the ANF promoter with mutated half-SBE (black bars) were transfected into COS7 cells with myocardin and/or Smad1 expression plasmids. C, Luciferase reporter controlled by the Nkx2.5 promoter containing SBE mutations was transfected into COS7 cells with myocardin and/or Smad1 expression plasmids. Values are the fold increase in luciferase activity relative to activation of the reporter alone. Error bars represent SD of at least 2 experiments. Student’s t test: *P<0.05, myocardin alone vs myocardin plus Smad1.
Myocardin and Smad1 Interact Directly
We performed coimmunoprecipitation assays to test if myocardin and Smad1 interact directly. COS7 cells were cotransfected with expression plasmids encoding Flag-tagged Smad1 and Myc-tagged myocardin (or singly transfected with each of those constructs in controls). Anti-Flag antibodies were used to immunoprecipitate Smad1. Anti-Myc antibodies were then used to detect the presence of associated myocardin. The interaction of myocardin and Smad1 was detected in lysates prepared from cells expressing both proteins (Figure 4A). Such interaction was further confirmed using a series of C-terminal deletion myocardin mutants (Figure 4B).
Figure 4.
Myocardin and Smad1 interact directly. A, Myocardin coimmunoprecipitates with Smad1. COS7 cells were transfected with plasmids encoding Flag-tagged Smad1 and/or Myc-tagged myocardin, as indicated. Smad1 was immunoprecipitated by (IP) anti-Flag antibodies, and anti-Myc antibodies were used to detect the presence of myocardin in the immunoprecipitates by Western blot (WB) analysis (top). One-fifteenth of cell extracts were directly immunoblotted to detect the presence of myocardin and Smad1 proteins (middle panels). One-fifteenth of supernatants (after immunoprecipitation) were immunoblotted to detect myocardin proteins (bottom). B, Myc-tagged myocardin aa 1 to 274 (M1), aa 1 to 351 (M2), and aa 1 to 421 (M3) were detected in Flag-Smad1 immunoprecipitates (top). Ten percent of cell extracts were directly immunoblotted to detect the presence of truncated myocardin proteins or Smad1 (middle panels). One-fifteenth of supernatants (after immunoprecipitation) were immunoblotted to detect truncated myocardin proteins (bottom). C, Myocardin specifically interacted with GST-Smad1 but not with GST alone. Coomassie-stained proteins corresponding to the amount of GST and GST-Smad1 protein used in the pull-down assay are shown below the autoradiograph, and 5% of the input protein is shown at left. D, Smad1 interacts with GST fused to myocardin aa 1 to 560 and aa 129 to 689 but not with aa 328 to 670, nor aa 669 to 935, nor with GST alone. Coomassie-stained proteins corresponding to the amounts of GST and GST-myocardin protein used in the pull-down assay are shown directly below the autoradiograph, and 1% of the input protein is shown at left. E, Myocardin and Smad1 interaction summary. Myocardin domains are abbreviated as follows: NTD indicates amino-terminal domain;++, basic domain; Q, a stretch of glutamine residues; SAP, SAF A/B, Acinus, PIAS domain; TAD, transactivation domain. F, Luciferase reporter controlled by 4 copies of a consensus CArG box and expression plasmids for myocardin, myocardin ΔSAP domain, or myocardin ΔQ domain and/or Smad1 was transfected into COS7 cells. Values are the luciferase activity by myocardin or myocardin mutants and Smad1 (black bars) relative to the activation of reporter by myocardin alone or myocardin mutants (gray bars). Error bars represent SD of at least 2 experiments. Student’s t test: *P<0.05, myocardin alone vs myocardin plus Smad1. Myc-tagged myocardin, myocardin ΔSAP, and myocardin ΔQ protein expressions are shown by Western blot.
To test whether myocardin and Smad1 interact directly in vitro, we performed GST fusion protein pull-down assays. GST-Smad1 protein was bacterially expressed and immobilized to glutathione-agarose beads and incubated with in vitro translated radiolabeled myocardin. Myocardin specifically interacted with GST-Smad1 but not with GST alone (Figure 4C).
To confirm the specificity of such interaction, as well as to determine the region of myocardin that mediates Smad1 interaction, we generated a deletion series of GST-myocardin fusion proteins and tested their interaction with Smad1 by pull-down assay. Radiolabeled Smad1 specifically interacted with amino acids (aa) 1 to 560 and 129 to 689 of myocardin, but not with GST alone or with aa 382 to 670 or aa 669 to 935 of myocardin (Figure 4D). This result suggests Smad1 directly interacts with myocardin at a region between aa 129 to 560 (Figure 4E). We have previously identified several conserved domains within this region of myocardin protein, including the basic domain (aa 243 to 260), Q domain (aa 287 to 320), and SAP domain (aa 380 to 414). 1,22 We, therefore, tested whether the functional interaction of myocardin and Smad1 is affected by mutations in those domains by luciferase reporter assay. Basic domain mutation completely abolished myocardin transactivation as well as its synergy with Smad1 (data not shown). Deletion of the SAP domain or Q domain dramatically decreased the synergy of myocardin and Smad1 (Figure 4F). The mutations did not alter the expression of those proteins (Figure 4F). Together, these data demonstrate a direct interaction between myocardin and Smad1 and suggest such physical interaction is important for their synergistic activation of cardiac promoters.
Smad Proteins Did Not Directly Affect Formation of the Myocardin/SRF/CArG Complex
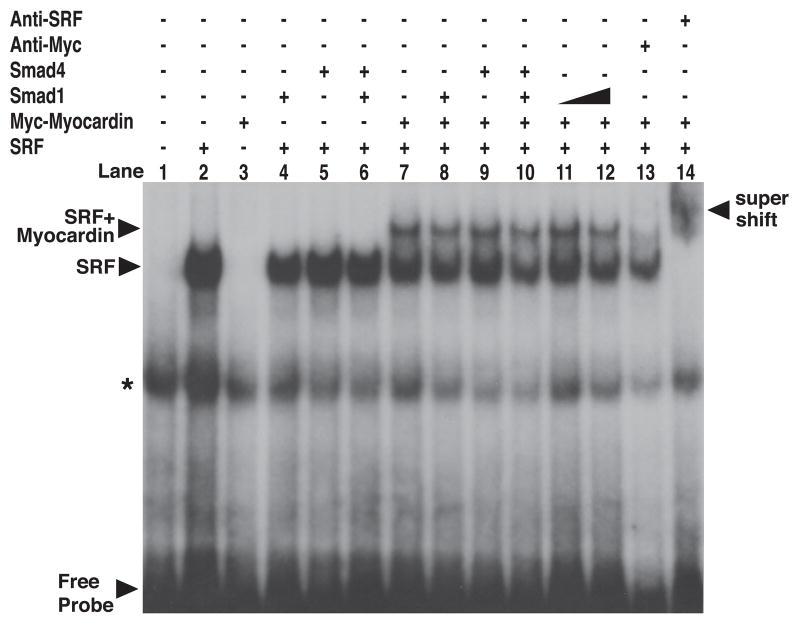
Myocardin does not bind to DNA directly, instead it is recruited to target genes by forming a stable complex with its cofactor SRF bound to DNA element, the CArG box.1 The interaction of myocardin and Smad1 led us to examine whether Smad1 directly associates with myocardin/SRF protein complexes bound to CArG box. EMSAs were performed using a radiolabeled oligonucleotide containing a consensus CArG box. Addition of in vitro translated SRF to the labeled probe resulted in a specific band (Figure 5, lane 2). Addition of both SRF and Myc-tagged myocardin resulted in an additional specific band corresponding to the myocardin/SRF/CArG complex (Figure 5, lane 7). This myocardin/SRF/CArG complex was diminished when anti-Myc antibodies were added (Figure 5, lane 13), demonstrating it contained myocardin. Similarly, both SRF and myocardin/SRF complexes were supershifted by anti-SRF antibodies, demonstrating these complexes contained SRF (Figure 5, lane 14). However, when in vitro translated Smad1 was added to the incubation mixtures, the SRF/CArG or myocardin/SRF/CArG complexes were neither supershifted nor diminished (Figure 5, lane 8). A similar result was obtained despite a several-fold increase in the amount of Smad1 relative to the fixed amounts of SRF and myocardin (Figure 5, lanes 11 and 12). We also found Smad4 alone or Smad1 plus Smad4 could not affect the myocardin/SRF/CArG complex (Figure 5, lanes 9 and 10). These results indicate, under the experimental conditions used, Smad1 is not a stable component of the myocardin/SRF/CArG complex.
Figure 5.
Smad proteins did not affect the formation of the myocardin/SRF/CArG complex. SRF, Myc-tagged myocardin, and Flag-tagged Smad1 and Smad4 proteins were translated in vitro and incubated with radiolabeled CArG probe, as described in Materials and Methods. Protein–DNA complexes were separated by nondenaturing PAGE and analyzed by autoradiography. Asterisk indicates nonspecific band.
Myocardin Activity Is Modulated by BMP Signaling
Interestingly, BMP signaling can be antagonized by inhibitory Smad6 and Smad7.14,15 Smad7 repressed myocardin transactivation of α-CA and ANF promoters in a dose-dependent fashion (Figure 6A and 6B), presumably by interfering with activation of endogenous BMP-signaling components. Constitutively activated ALKs activate BMP signaling in the absence of BMP ligands.23 To test whether upstream BMP-signaling components stimulate myocardin transactivity, we used a constitutively activated BMP receptor, ALK3 QD, in luciferase reporter assays. ALK3 QD stimulated myocardin transactivation of the α-CA and Nkx2.5 reporters (Figure 6C and 6D). Whereas stimulation of myocardin transactivity by ALK3 QD is comparable to Smads 1 and -4, cotransfection of both ALK3 QD and Smads -1 and -4 with myocardin further increased the activation of the Nkx2.5 reporter (Figure 6D). Those results suggest myocardin transactivity is stimulated by BMP signaling originating from the cell surface.
Figure 6.
Myocardin activity is modulated by BMP signaling. Myocardin and/or Smad7 (100, 150, and 200 ng) expression plasmids and α-CA (A) or ANF (B) luciferase reporter were transfected into COS7 cells. Values are the percentage of luciferase activity relative to activation of the reporter by myocardin alone. C, α-CA or Nkx2.5 luciferase reporter, Smads 1 and -4, ALK3 QD, and/or myocardin expression plasmids were transfected into COS7 cells. Values are the fold increase in luciferase activity relative to activation of the reporter alone. Error bars represent SD of at least 2 experiments. Student’s t test; P<0.05: *myocardin alone vs myocardin plus Smad7, Smad1/4, or ALK3 QD; **myocardin alone vs myocardin plus Smad1/4 and ALK3 QD.
BMP Signaling Increases Myocardin Protein Expression
BMPs induce the expression of cardiac transcription factor Nkx2.5 and other cardiac markers in treated chick embryos,18 as well as the P19CL6 cell line.19,20 We asked whether BMPs have the same effect on myocardin expression. Neonatal rat cardiomyocytes treated with BMP-2 dramatically increased myocardin and MEF2 protein expression, but not of SRF or α-actinin (Figure 7). BMP-2 did not induce global protein synthesis, as β-tubulin protein expression was unchanged in BMP-2–treated cardiomyocytes (Figure 7). This result demonstrates BMP signaling induces myocardin expression in cardiomyocytes and suggests a positive feedback mechanism for BMP signaling and myocardin to activate cardiac genes.
Figure 7.

Myocardin protein level is increased in BMP-2–treated cardiomyocytes. Rat neonatal cardiomyocytes were treated with 20 ng/mL BMP-2 (or without in negative control) for 48 hours before harvesting and lysate production for SDS-PAGE and Western Blot analysis with indicated antibodies, as described in Materials and Methods.
Discussion
In this report, we identified the molecular interaction of myocardin and the BMP-signaling pathway to synergistically activate cardiac gene expression. Along with the accompanying study demonstrating that myocardin and TGF-β–signaling pathway synergistically activate SM gene expression,35 our results clearly establish that myocardin is involved in TGF-β superfamily signaling pathways that regulate cardiac and SM-specific gene expression.
Transactivation of Cardiac Gene Expression by Myocardin and BMP Signaling
We have recently uncovered several mechanisms by which myocardin regulates cardiac and SM gene expression. GATA4 represses or activates myocardin-mediated transactivation, depending on the specificity of target genes.36 Myocardin transactivity is also positively and negatively regulated by p300 and HDAC5, suggesting an additional layer of regulation at the chromatin level.12 Interestingly, myocardin is also involved in a molecular switch controlling SRF-dependent cell differentiation versus proliferation processes, where myocardin directly competes with Elk1 for SRF association and target gene activation.13 Together, those studies indicate the transcriptional activity of myocardin is tightly controlled.
In this study, we demonstrated myocardin transactivation of cardiac gene expression is modulated by BMP signaling through a protein–protein interaction between myocardin and BMP-downstream effector Smad1, providing another novel mechanism in which myocardin is integrated into an important signaling pathway to regulate gene expression. Most importantly, we found the BMP signaling was also able to induce expression of myocardin, suggesting a potential positive-feedback mechanism. This mechanism could be used where both myocardin and BMP initiate early cardiac gene expression, whereas myocardin is later used for the maintenance of the cardiac program. Interestingly, such mechanisms exist in skeletal muscle and other biological systems.37,38
SBE Dependency or SBE Independency?
The synergy between Smad1 and myocardin in activating the ANF promoter appears to be SBE independent. Those results were distinct from the response of Smad3 and myocardin, which synergistically activate a SBE-controlled reporter.35 In contrast, myocardin and Smad1 synergistically activated a luciferase reporter driven by 4 CArG box copies, where absolutely no SBE is involved. These data suggest Smad1 can activate cardiac gene expression independently of inherent DNA binding. Similar SBE-independent mechanisms have been recently reported for a variety of target genes regulated by BMP/Smads.39–41
Then how does Smad1 activate target gene expression independent of DNA binding? Several mechanisms may apply: Smad1 could be recruited by myocardin and SRF to the CArG boxes in the ANF promoter. However, whereas we detected protein–protein interaction between myocardin and Smad1 in vitro and in vivo, we were unable to obtain evidence for the formation of a potential ternary complex among those proteins bound to DNA under our experimental conditions. Interestingly, some SRF cofactors are suggested to enhance the affinity of SRF/DNA association, despite not forming a stable ternary complex with SRF bound to DNA.3,42 Our data suggest that Smad1 enhances the activity of the myocardin/SRF transcriptional complex through alternative mechanisms. Those include the recruitment of coactivators, such as p300, or by repelling transcriptional repressors from this transcriptional complex. It will be important to investigate how the physical and functional interaction of endogenous Smad1 and myocardin proteins is influenced by BMP signaling, as we cannot rule out the possibility that Smad1 might affect myocardin/SRF/CArG–complex formation in a BMP–dependent manner.
Cardiac or Smooth Muscle?
Myocardin is a cardiac and SM-specific transcriptional cofactor for SRF and activates target gene expression in a CArG-dependent manner.1,3 It is currently unclear how myocardin discriminates between cardiac and SM target genes, although the SAP domain has been suggested to be involved.1 In this study, we showed myocardin and Smad1 interact directly to synergistically activate cardiac reporter gene expression. Such activation requires the CArG box and appears to be SBE independent. Additionally, the functional interaction between myocardin and Smad1 was completely abolished in SRF-null embryonic stem cells (T.E.C. and D.-Z.W., unpublished data, 2005), further supporting the notion of CArG box/SRF dependency. We suggest the target specificity (cardiac versus SM) for myocardin is determined, at least in part, by which upstream signals, TGF-β or BMP, and their downstream effectors, Smad3 or Smad1, are used. This hypothesis is consistent with the notion that BMPs are key regulators for cardiac gene expression, whereas TGF-β appears to play a significant role in controlling SMC gene expression.16,43,44 Interestingly, the SAP domain of myocardin appears involved in mediating the functional interaction between myocardin and Smad1 because SAP domain mutation dramatically decreased the synergy of myocardin and Smad1 on a CArG-box–dependent reporter gene (Figure 4E). Together, our data suggest the SAP domain of myocardin may serve as a nodal point to integrate BMP-signaling pathway in activating myocardin-mediated cardiac gene expression. Future investigation, in particular in vivo studies, will be needed to further clarify this issue. Nevertheless, our studies establish a direct molecular and functional interaction between myocardin and BMP signaling and suggest a molecular mechanism for the transcriptional regulation of the cardiac gene program. Given the importance of myocardin and BMPs, it is intriguing to speculate that mutations in either molecule or disruption of their functional interaction may contribute to human cardiovascular diseases.
Acknowledgments
D.-Z.W. is a Basil O’Connor Scholar of March of Dimes Birth Defects Foundation and was supported by NIH grant R01 HL75251, the Muscular Dystrophy Association, and an American Heart Association Grant-in-Aid. We thank members of the Wang laboratory for discussion and help, Dr Li Li for communicating results before publication, Dr Mark Majesky for critical reading of the manuscript and constructive advice, Dr Ching-Ling Lien for reagents, and Dr Eric Olson for continuous support.
References
- 1.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 2.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132:987–997. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- 5.Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN, Owens GK. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 9.Majesky MW. Decisions, decisions. SRF coactivators and smooth muscle myogenesis. Circ Res. 2003;92:824–826. doi: 10.1161/01.RES.0000071525.18323.4C. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 14.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 15.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 16.Monzen K, Nagai R, Komuro I. A role for bone morphogenetic protein signaling in cardiomyocyte differentiation. Trends Cardiovasc Med. 2002;12:263–269. doi: 10.1016/s1050-1738(02)00172-x. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- 18.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 19.Monzen K, Shiojima I, Hiroi Y, Kudoh S, Oka T, Takimoto E, Hayashi D, Hosoda T, Habara-Ohkubo A, Nakaoka T, Fujita T, Yazaki Y, Komuro I. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol. 1999;19:7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monzen K, Hiroi Y, Kudoh S, Akazawa H, Oka T, Takimoto E, Hayashi D, Hosoda T, Kawabata M, Miyazono K, Ishii S, Yazaki Y, Nagai R, Komuro I. Smads, TAK1, and their common target ATF-2 play a critical role in cardiomyocyte differentiation. J Cell Biol. 2001;153:687–698. doi: 10.1083/jcb.153.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh-Choudhury N, Abboud SL, Mahimainathan L, Chandrasekar B, Choudhury GG. Phosphatidylinositol 3-kinase regulates bone morpho-genetic protein-2 (BMP-2)-induced myocyte enhancer factor 2A-dependent transcription of BMP-2 gene in cardiomyocyte precursor cells. J Biol Chem. 2003;278:21998–22005. doi: 10.1074/jbc.M302277200. [DOI] [PubMed] [Google Scholar]
- 22.Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci U S A. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macias-Silva M, Hoodless PA, Tang SJ, Buchwald M, Wrana JL. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J Biol Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 24.Zhu HJ, Iaria J, Sizeland AM. Smad7 differentially regulates transforming growth factor beta-mediated signaling pathways. J Biol Chem. 1999;274:32258–32264. doi: 10.1074/jbc.274.45.32258. [DOI] [PubMed] [Google Scholar]
- 25.Pouponnot C, Jayaraman L, Massague J. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 26.Janknecht R, Wells NJ, Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CY, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky MJ, Schwartz RJ. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Garcia AV, Ross RS, Evans SM, Chien KR. A conserved 28-base-pair element (HF-1) in the rat cardiac myosin light-chain-2 gene confers cardiac-specific and alpha-adrenergic-inducible expression in cultured neonatal rat myocardial cells. Mol Cell Biol. 1991;11:2273–2281. doi: 10.1128/mcb.11.4.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2–5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 30.Nicol RL, Frey N, Pearson G, Cobb M, Richardson J, Olson EN. Activated MEK5 induces serial assembly of sarcomeres and eccentric cardiac hypertrophy. EMBO J. 2001;20:2757–2767. doi: 10.1093/emboj/20.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang PS, Li L, McAnally J, Olson EN. Muscle specificity encoded by specific serum response factor-binding sites. J Biol Chem. 2001;276:17206–17212. doi: 10.1074/jbc.M010983200. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 34.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 35.Qiu P, Ritchie R, Fu Z, Cao D, Cumming J, Miano JM, Wang D-Z, Li HJ, Li L. Myocardin enhances Smad3-mediated transforming growth factor-β1 signaling in a CArG box–independent manner. Smad-binding element is a critical cis element for SM22α transcription in vivo. Circ Res. 2005;97:983–991. doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- 36.Oh J, Wang Z, Wang DZ, Lien CL, Xing W, Olson EN. Target gene-specific modulation of myocardin activity by GATA transcription factors. Mol Cell Biol. 2004;24:8519–8528. doi: 10.1128/MCB.24.19.8519-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- 38.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 39.Brown CO, 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2–5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 40.Lee KH, Evans S, Ruan TY, Lassar AB. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development. 2004;131:4709–4723. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- 41.Blokzijl A, ten Dijke P, Ibanez CF. Physical and functional interaction between GATA-3 and Smad3 allows TGF-beta regulation of GATA target genes. Curr Biol. 2002;12:35–45. doi: 10.1016/s0960-9822(01)00623-6. [DOI] [PubMed] [Google Scholar]
- 42.Chang DF, Belaguli NS, Iyer D, Roberts WB, Wu SP, Dong XR, Marx JG, Moore MS, Beckerle MC, Majesky MW, Schwartz RJ. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 43.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 44.Qiu P, Feng XH, Li L. Interaction of Smad3 and SRF-associated complex mediates TGF-beta1 signals to regulate SM22 transcription during myo-fibroblast differentiation. J Mol Cell Cardiol. 2003;35:1407–1420. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]