Aurora B activity is inhibited when centromeric repeat sequences are absent, although kinetochores can still assemble.
Abstract
The nearly ubiquitous presence of repetitive centromere DNA sequences across eukaryotic species is in paradoxical contrast to their apparent functional dispensability. Centromeric chromatin is spatially delineated into the kinetochore-forming array of centromere protein A (CENP-A)–containing nucleosomes and the inner centromeric heterochromatin that lacks CENP-A but recruits the aurora B kinase that is necessary for correcting erroneous attachments to the mitotic spindle. We found that the self-perpetuating network of CENPs at the foundation of the kinetochore is intact at a human neocentromere lacking repetitive α-satellite DNA. However, aurora B is inappropriately silenced as a consequence of the altered geometry of the neocentromere, thereby compromising the error correction mechanism. This suggests a model wherein the neocentromere represents a primordial inheritance locus that requires subsequent generation of a robust inner centromere compartment to enhance fidelity of chromosome transmission.
Introduction
Centromeres direct chromosome segregation by coordinating physical interactions with the microtubule-based mitotic spindle (Cleveland et al., 2003). Accurate chromosome segregation requires stable connections between the mitotic kinetochore and the spindle (Cheeseman and Desai, 2008) as well as a correction mechanism to destabilize improper spindle–kinetochore connections and allow subsequent proper connections to form (Kelly and Funabiki, 2009). In the event that centromere location changes, both of these distinct centromere functions must be present at the new location to avoid compromising the fidelity of chromosome segregation. Kinetochores are built upon a chromatin domain defined by the presence of centromere protein A (CENP-A)–containing nucleosomes, whereas error correction is mediated by the aurora B–containing chromosome passenger complex that localizes to the inner centromere compartment where classical heterochromatin is present at the location of final sister chromatid cohesion. The chromosomal location of the centromere is defined epigenetically (Black and Bassett, 2008), and the most striking examples of epigenetic centromere inheritance are naturally occurring neocentromeres where CENPs, including CENP-A, occupy a new location devoid of centromeric repeat sequences (Depinet et al., 1997; du Sart et al., 1997; Warburton et al., 1997). The universal presence of repetitive DNA at normal human centromeres and their presence at the centromeres of diverse eukaryotes, along with experiments using artificial chromosomes that suggest the importance of specific types of satellite DNA in centromere acquisition (Schueler et al., 2001), imply a contribution of repetitive DNA to mitotic centromere function, but the nature of such a genetic contribution remains largely unclear.
Results and discussion
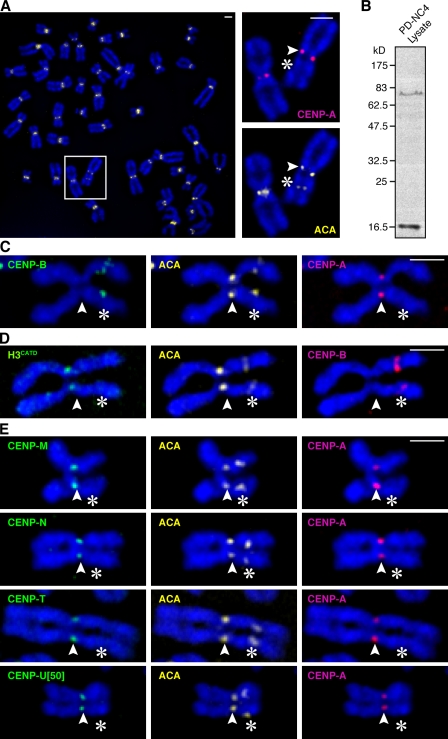
To investigate the structural features of centromeric chromatin required for its diverse functions in chromosome segregation, we examined a patient-derived cell line containing a chromosome 4 variant termed PD-NC4 (Amor et al., 2004). This chromosome variant is unaltered in DNA sequence, but its original centromere containing >1 Mbp of α-satellite DNA, the genetic hallmark of normal human centromeres (Willard, 1991), has been epigenetically silenced, and a neocentromere has formed 25 Mbp down the q arm at a site that lacks centromere repeats (Amor et al., 2004). Our PD-NC4 detection scheme exploits a preparation of anti-centromere antibodies (ACAs) that detect two centromere components, CENP-A (exclusively at the neocentromere; Amor et al., 2004) and CENP-B (exclusively at the silenced centromere; by virtue of direct binding to the 17-bp CENP-B recognition site, termed the CENP-B box, found in the αI-satellites present at the original centromere location; Fig. 1, A–C). Normal chromosomes all contain a single pair of ACA foci, whereas PD-NC4 contains two pairs of ACA foci of readily discernable identity (Fig. 1, A and C). In this way, it is possible to assess the distribution of factors that remain at the original epigenetically silenced location (such as CENP-B; Fig. 1 C, left), that have tracked with CENP-A (Fig. 1 C, right) to the neocentromere, or that exist at both sites (such as HP1α; Amor et al., 2004).
Figure 1.
Epigenetic determinants of centromere identity are intact at the neocentromere of the PD-NC4 chromosome variant. (A) ACA stains both the epigenetically silenced centromere and the neocentromere. Insets show higher magnification views of the boxed area. (B) Immunoblot of PD-NC4 lysates with ACA reveals two bands with electrophoretic mobility consistent with ∼80 kD CENP-B (Earnshaw and Rothfield, 1985) and ∼17 kD CENP-A (Earnshaw and Rothfield, 1985). (C) CENP-A and CENP-B clearly differentiate the active versus inactive ACA staining centromeres on the PD-NC4 chromosome. (D) The CATD is sufficient for targeting H3CATD to the neocentromere. (E) CENP-ANAC components, each stably expressed as LAP fusions, track with CENP-A to the neocentromere. Outer kinetochore components Mad1 (Fig. S3 A) and Ndc80 (Fig. S3 B) were recruited to the neocentromere. Arrowheads indicate the neocentromere, and asterisks indicate the silenced centromere. Bars, 2 µm.
CENP-A targeting domain (CATD) functions independently of α-satellite DNA
We previously identified the CATD, consisting of its loop 1 and α2 helix within the histone fold domain (Black et al., 2004). When substituted into conventional H3, the CATD converts H3 into a centromeric histone in terms of its subchromosomal localization (Black et al., 2004), access to the Holliday junction recognition protein–mediated centromeric chromatin assembly pathway (Foltz et al., 2009), CENP-A–like structural rigidity when incorporated into nucleosomes (Black et al., 2007a), recognition by CENP-N (Carroll et al., 2009), and ability to replace the essential role of CENP-A in mitosis (Black et al., 2007b). Together, these findings have generated a model of self-propagating centromere inheritance in which CENP-A–containing nucleosomes mark centromere location by their rigid physical properties that distinguish them from nucleosomes assembled with conventional H3 and where newly expressed CENP-A is targeted to established centromeric chromatin via the cis-acting CATD. Another possibility is that the CATD-mediated targeting of newly made CENP-A to centromeres is conferred by selective binding to α-satellite repeats. To distinguish between these models, we stably expressed an H3 chimera containing the CATD (H3CATD) in the patient-derived cells and found that it is undetectable at the silenced centromere and enriched at the neocentromere (Fig. 1 D), providing evidence for a direct role for the CATD in the epigenetic maintenance of centromere identity that is independent of a particular DNA sequence.
Neocentromeric recruitment of the CENP-A nucleosome-associated complex (CENP-ANAC)
CENP-A nucleosomes, when released from chromatin by nuclease digestion, copurify with CENP-ANAC, consisting of several proteins (including CENP-M, CENP-N, CENP-T, and CENP-U[50]) that constitute a subset of the constitutive centromere components (Foltz et al., 2006; Izuta et al., 2006; Hori et al., 2008). CENP-A nucleosomes also copurify with CENP-B, presumably as a consequence of its direct binding to the α-satellite DNA that wraps CENP-A nucleosomes at normal centromeres. It is possible that CENP-ANAC components are recruited to CENP-A nucleosomes or proximal H3-containing nucleosomes in a manner dependent on the presence of CENP-A nucleosomes and α-satellite DNA sequences or in a manner independent of α-satellite sequences but dependent on the physical properties of CENP-A nucleosomes. To distinguish how CENP-ANAC components are recruited to centromeres, we determined their localization on the PD-NC4 chromosome and found that CENP-M, CENP-N, CENP-T, and CENP-U[50] are each vacant from the silenced centromere but present at the neocentromere location (Fig. 1 E) at similar levels as those found at normal chromosomes. These findings support a model in which the neocentromeric CENP-A array that is two thirds the size of that found at the normal centromere of chromosome 4 (Amor et al., 2004) is sufficient to recruit a robust constitutive centromeric chromatin compartment independently of DNA sequence. Other newly identified constitutive centromere components more distal to CENP-A nucleosomes, such as CENP-O, CENP-P, CENP-R, and CENP-S, also track with the epigenetically defined neocentromere (Fig. S1). Thus, all centromere targeted proteins, with the exception of CENP-B, that originally copurified with CENP-A–containing nucleosomes but not bulk H3-containing nucleosomes (Foltz et al., 2006) track with CENP-A to the neocentromere. These findings provide further support for the notion that epigenetic centromere propagation and generation of a unique subchromosomal chromatin domain are linked properties conferred by the structurally rigid nucleosomes containing CENP-A (Black and Bassett, 2008).
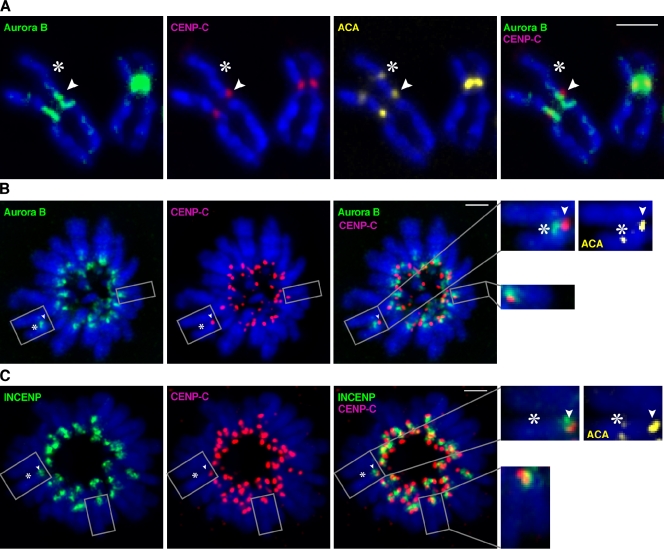
Neocentromeric recruitment of the aurora B kinase
The inner centromere localization of the aurora B kinase precisely positions it to be active in phosphorylating kinetochore targets (such as the Ndc80 complex) when improperly attached to the spindle and silent upon proper chromosome biorientation (Adams et al., 2001; Biggins and Murray, 2001; Tanaka et al., 2002; Andrews et al., 2004; Cheeseman et al., 2006; Sandall et al., 2006; Liu et al., 2009), and we reasoned that aurora B would track with the epigenetically defined centromere location because of the critical role it plays at normal centromeres in guiding faithful chromosome segregation. Aurora B indeed vacates the epigenetically silenced location on PD-NC4 and is detectable at the neocentromere (Fig. 2). The pool of aurora B stably bound to the centromere in metaphase spread preparations, however, is spatially altered on the neocentromere relative to the robust inner centromere localization on normal chromosomes (Fig. 2 A). The pool of aurora B that tracks with the neocentromere is present along ∼1 µm of each sister chromatid arm, which is internal to the kinetochore-forming chromatin but distinct from the normal single focus of inner centromere localization. The splitting into discrete aurora B signals on each sister chromatid of the PD-NC4 chromosome in metaphase spreads is likely a consequence of the altered centromeric cohesion in the PD-NC4 neocentromere that causes a 15% increase in interkinetochore distances measured in such preparations (Amor et al., 2004). Indeed, in intact mitotic cells (as opposed to chromosome spread preparations), the aurora B signal on the PD-NC4 chromosome exists as a single focus at the inner centromere as assessed either in prometaphase, metaphase, or upon metaphase arrest with the microtubule-destabilizing compound nocodazole (Fig. S2).
Figure 2.
Altered aurora B localization on a neodicentric chromosome. (A) In mitotic spread preparations, aurora B is present on normal centromeres at the inner centromere location, whereas on the PD-NC4 chromosome, the kinase has vacated the silenced centromere and now tracks to an ∼1-µm region of chromosome 4 adjacent to the neocentromere that runs along the length of each metaphase chromatid arm. Image capturing to robustly detect aurora B on the neodicentric chromosome on metaphase spread preparations overexposes aurora B staining at normal centromeres. (B and C) In intact, monastrol-arrested cells with monopolar spindles, the total pool sizes of neocentromeric aurora B (B) and INCENP (C) are within the range observed at normal centromeres in the same cells (aurora B: normal chromosome range, 5,623–11,249 arbitrary fluorescence units; PD-NC4, 7,019; INCENP: normal chromosome range, 2,871–7,173; PD-NC4, 3,650). In monastrol-treated cells, many of the chromosomes are rotated and have small interkinetochore distances, so for consistency, the enlargements (B and C) are examples where the sister centromeres overlap in x and y planes. Arrowheads indicate the neocentromere, and asterisks indicate the silenced centromere. Insets show higher magnification views of the boxed areas. Bars, 2 µm.
We also used the reversible Eg5 inhibitor monastrol to arrest cells with monopolar spindle-attached chromosomes and found that both aurora B (Fig. 2 B) and its binding partner/activator, inner CENP (INCENP; Fig. 2 C), are each present as a single focus on the PD-NC4 neocentromere. Despite the spatial rearrangement of aurora B on the DNA proximal to the centromere, the neocentromere nonetheless recruits a mitotic pool of aurora B that is comparable in size with that found on normal centromeres. Although the exact mechanism of centromere targeting of the aurora B–containing chromosome passenger complex remains unknown even for normal centromeres, our results indicate a requirement for centromere activity, not underlying DNA sequences. Indeed, the PD-NC4 or other neocentromeres represent potential contexts in which to define the precise means by which aurora B localizes to inner centromeres.
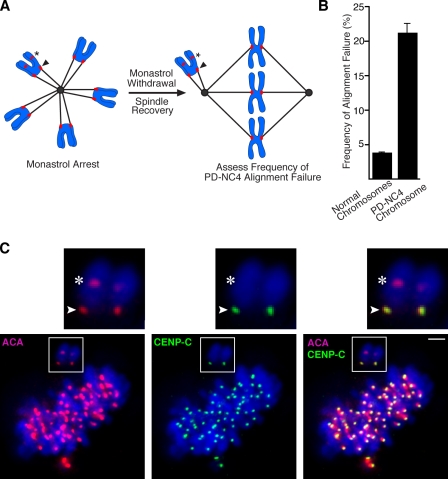
Defective neocentromeric error correction in mitosis
To investigate whether the altered spatial positioning of aurora B on the chromatin adjacent to the neocentromere disrupts centromere function or whether the recruitment of normal levels of aurora B observed in intact cells (Fig. 2 and Fig. S2) is sufficient for its error correction function, we used an assay that first generates a high frequency of attachment errors by monastrol treatment that, after monastrol withdrawal, must be corrected before proper biorientation on the metaphase plate is achieved (Lampson et al., 2004). After monastrol withdrawal, we measured the frequency at which the PD-NC4 chromosome failed to correct misattachments and align at the metaphase plate (Fig. 3 A). The PD-NC4 chromosome is greater than fivefold more likely than normal chromosomes to fail to rapidly correct attachment errors (Fig. 3, B and C), indicating that the aurora B–dependent error correction mechanism is severely compromised at the neocentromere. The PD-NC4 chromosome is found in all of the 100 cells analyzed from patient-derived lymphoblast lines even after prolonged cell culture (Amor et al., 2004), indicating its mitotic stability, selection against chromosome 4 monosomy in culture, or both. Moreover, the PD-NC4 neodicentric and its normal chromosome 4 counterpart yielded pooled anaphase lag rates similar to other chromosomes (Amor et al., 2004). Nonetheless, the finding that the PD-NC4 chromosome inefficiently corrects monastrol-induced spindle connection errors indicates a deficiency in its aurora B–dependent mitotic quality control.
Figure 3.
Aurora B–dependent error correction is crippled on the neodicentric chromosome. (A) Experimental scheme to detect the rate of failure of the PD-NC4 to biorient on the metaphase plate as cells recover from monastrol treatment. (B) The PD-NC4 chromosome has an elevated rate of alignment failure after spindle recovery after initial monastrol treatment. The measured values from three independently performed experiments are plotted showing SEM. (C) An example of the PD-NC4 chromosome failing to align when the normal chromosomes have corrected syntelic attachments after monastrol withdrawal and are nearly aligned on the metaphase plate. Insets show higher magnification views of the boxed areas. Arrowheads indicate the neocentromere, and asterisks indicate the silenced centromere. Bar, 2 µm.
Aurora B is unable to properly sense neocentromeric misattachments to the mitotic spindle
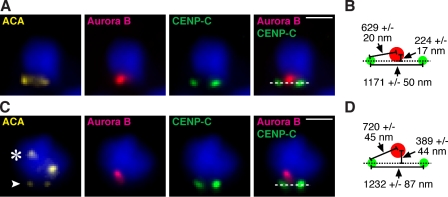
The geometry of the centromere is central to current models of the aurora B–dependent error correction mechanism wherein improper attachments are destabilized by aurora B phosphorylation of kinetochore substrates, whereas proper attachments are stabilized by silencing aurora B kinase activity at the kinetochore. The mechanism for silencing aurora B kinase activity at properly bioriented kinetochores may involve removal of microtubules from the inner centromere that activate the kinase on maloriented chromosomes (Sandall et al., 2006; Rosasco-Nitcher et al., 2008), conformational changes to inner centromeric chromatin that silence the kinase upon the centromere stretching that accompanies chromosome biorientation on the spindle (Tanaka et al., 2002), or spatial separation of the kinase from its kinetochore substrates upon biorientation (Tanaka et al., 2002; Andrews et al., 2004; Liu et al., 2009). The latter mechanism is somewhat distinct from conventional views of kinase silencing but represents an intriguing proposal because of the magnitude of the centromere stretching that occurs only upon proper chromosome biorientation and extends the distance from the inner centromeric chromatin where aurora B accumulates to its substrates. Inner centromere localization is key because artificial repositioning of aurora B throughout the αI-satellite DNA that extends from the inner centromere outward to the CENP-A–containing nucleosomes at the inner kinetochore (as opposed to the natural discrete position of aurora B at the inner centromere) is sufficient to destabilize normal spindle microtubule–kinetochore attachments (Liu et al., 2009). We investigated aurora B localization at centromeres in need of error correction to determine its position relative to kinetochores. We first found that among the small number of normal chromosomes that fail to align after initial erroneous connection, aurora B is concentrated at a location that is nearly centered between the sister kinetochores (Fig. 4, A and B). For unaligned PD-NC4 chromosomes, aurora B is concentrated at a site that is 165 nm further removed from the central axis of the centromere (P < 0.005), a conformation that positions the kinase >90 nm further than normal from the kinetochore (P < 0.05; Fig. 4, C and D). Together with the dilution of stable aurora B–binding sites along pericentromeric chromatin revealed in mitotic chromosome spread preparations (Fig. 2 A) and the compromised error correction of the PD-NC4 chromosome (Fig. 3), these findings indicate that the atypical positioning of aurora B at the PD-NC4 neocentromere places the kinase at a location that is disadvantageous for sensing attachment errors.
Figure 4.
Measuring the mispositioning of aurora B at a location adjacent to the neocentromere of the PD-NC4 chromosome. (A–D) Aurora B location on misoriented normal chromosomes (A and B) or misoriented PD-NC4 chromosomes (C and D) was determined by indirect immunofluorescence in intact cells that were fixed after monastrol treatment and recovery as in Fig. 3. The indicated distance measurements (mean ± SD) for misoriented normal chromosomes (B) and misoriented PD-NC4 chromosomes (D) reveal mispositioning in the PD-NC4 chromosome of aurora B away from its kinetochore targets. The arrowhead indicates the neocentromere, and the asterisk indicates the silenced centromere. Bar, 2 µm.
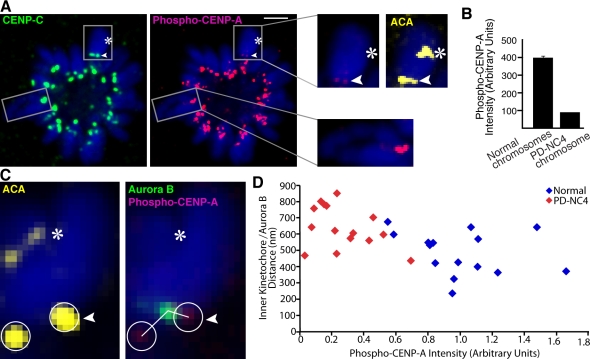
To directly test whether or not aurora B kinase activity at the neocentromere is inappropriately silenced, we measured the phosphorylation of a specific kinetochore substrate site (Ser7 of CENP-A; Zeitlin et al., 2001a,b). Phosphorylation of this site depends on chromosome–spindle attachment state with about fivefold higher levels of phospho–CENP-A signal present on misattached chromosomes than on properly bioriented chromosomes (Liu et al., 2009). In monopolar cells, where spindle biorientation is impossible, normal chromosomes exhibit high levels of aurora B kinase activity at their kinetochores, but the neocentromere of PD-NC4 is silenced (Fig. 5 A) with a more than fourfold reduction in phospho–CENP-A (Fig. 5 B). This substantial decrease is caused by aurora B silencing and is not a result of lower levels of the CENP-A substrate because CENP-A levels at the PD-NC4 neocentromere are comparable with those at normal centromeres (Fig. 1 A). Simultaneous measurement in monopolar cells of phospho–CENP-A levels and the distance from aurora B to the inner kinetochore (Fig. 5, C and D) reveal that the increased distances from aurora B to its substrates correspond to a marked decrease in CENP-A phosphorylation on the PD-NC4 centromere. Thus, the inner centromere architecture of the neocentromere is insufficient to support the robust mechanisms to sense chromosome–spindle misconnections present at centromeres with normal inner centromere geometry.
Figure 5.
Mispositioned aurora B renders the neocentromere insensitive to monastrol-induced attachment errors. (A) In monopolar cells after monastrol treatment, aurora B activity at the PD-NC4 kinetochore, measured by phospho–CENP-A detection, is inappropriately silenced. Insets show higher magnification views of the boxed areas. Bar, 2 µm. (B) The phospho–CENP-A intensity values in cells treated as in A are plotted as SEM. (C and D) When measured simultaneously (C), the longer kinetochore–aurora B distances and reduced phospho–CENP-A intensity on the PD-NC4 chromosome segregate away from the shorter kinetochore–aurora B distances and higher phospho–CENP-A intensity of normal chromosomes (D). The circles indicate the area of measurement of phospho–CENP-A intensities at each centromere. Arrowheads indicate the neocentromere, and asterisks indicate the silenced centromere.
Conclusions
The chromatin generated by an array of nucleosomes containing CENP-A is thought to specify the location of the centromere independent of DNA sequence. Although the mechanism remains unclear of the initial seeding of a CENP-A–containing nucleosome array at a chromosome arm site, the rigidity CENP-A imposes to the neocentromeric chromatin, along with the CATD pathway for targeting newly expressed CENP-A to the centromere, maintains the mark in perpetuity (Black and Bassett, 2008). Our effort in this study provides evidence that CENP-A marking, the CATD, and the proximal constitutive CENPs that require its presence are all intact at an epigenetically defined neocentromere but vacant from a silenced centromere on the same chromosome. Our data further suggest that this neocentromere is a primordial centromere; it has sufficient functional capacity to confer substantial chromosome stability but lacks the robust inner centromere of its mature counterparts that efficiently corrects spindle attachment errors. Although the genes along the chromosome arms are highly syntenic, genome-wide analyses of diverse mammals have shown that centromeres move their location more rapidly than nearly any other part of the chromosome (Murphy et al., 2005), and centromere repositioning has been proposed as a mechanism contributing to speciation events (Amor et al., 2004; Ventura et al., 2007). Neocentromeres, either in the context of chromosomal rearrangements that separate a chromosome arm fragment from its natural centromere or by repositioning the centromere, lack satellite sequences. The recent finding that one normal horse chromosome (chromosome 11) contains an evolutionarily new centromere that lacks any centromeric satellite DNA (Wade et al., 2009) strongly suggests that satellite-independent modes of generating a robust inner centromere compartment exist. However, it is likely that inner centromere formation depends on the local chromatin environment present before neocentromere formation, suggesting that multiple modes of centromere maturation may exist. Eventual acquisition of repetitive DNA sequences and concomitant heterochromatin formation would be a simple mechanism to enhance inner centromere function of an epigenetically defined centromere. In this way, maturation of a centromere would increase the probability of its own propagation, generating a chromosomal inheritance locus that is fortified over the timescales of speciation events.
Materials and methods
Cell lines
Patient-derived fibroblasts harboring the PD-NC4 chromosome variant (PD-NC4 fibroblasts; Amor et al., 2004) were generously provided by A. Choo (Murdoch Childrens Research Institute, Parkville, Victoria, Australia) and cultured in DME supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 100 µg/ml G418. Derivative cell lines expressing YFP-H3CATD, localization and purification (LAP; Cheeseman and Desai, 2005)–tagged CENP-M, -N, -O, -P, -R, -S, -T, and -U[50] were generated by stable integration via Moloney murine leukemia retroviral delivery. pBABE-BLAST (Shah et al., 2004)–derived retroviral plasmids expressing YFP-H3CATD, LAP–CENP-O, LAP–CENP-P, LAP–CENP-R, LAP–CENP-S, LAP–CENP-T, and pBABE-PURO (Morgenstern and Land, 1990)–derived retroviral plasmids expressing LAP–CENP-M, LAP–CENP-N, and LAP–CENP-U[50] have been described previously (Foltz et al., 2006; Black et al., 2007b). Each retroviral plasmid was cotransfected using Effectene (QIAGEN) with the vesicular stomatitis virus glycoprotein pseudotyping plasmid into 293GP cells (which express the retroviral gag and pol genes) to generate amphotropic retrovirus (Morgenstern and Land, 1990). Virus-containing supernatant was harvested 2 d after transfection, passed through a 0.45-µm filter, mixed with hexadimethrine bromide (Polybrene; 8 µg/ml), and incubated with PD-NC4 fibroblasts overnight. 2.5 µg/ml blasticidin S (EMD) or 250 ng/ml puromycin (EMD) selection for pBABE-BLAST or pBABE-PURO derivatives, respectively, was introduced 2–3 d after infection and continued for 10–18 d.
Chromosome spreads
Cells arrested in mitosis with 400 ng/ml nocodazole (Sigma-Aldrich) were harvested by agitation, hypotonically swollen in 75 mM KCl, spun onto glass slides using a centrifuge (Cytospin; Thermo Fisher Scientific) at 2,000 rpm for 4 min, fixed in 4% formaldehyde for 10 min, and processed for indirect immunofluorescence. Anti–CENP-A (provided by K. Yoda, Nagoya University, Nagoya, Japan) tissue culture supernatant was used at a dilution of 1:100, anti–CENP-B (provided by D. Cleveland, University of California, San Diego, La Jolla, CA; Earnshaw et al., 1987) ascites was used at 1:200, affinity-purified anti–CENP-C rabbit polyclonal antibody was used at 0.5 µg/ml, anti–aurora B (anti-AIM1; BD) purified mAb was used at 0.25 µg/ml, affinity-purified anti-GFP rabbit polyclonal antibody was used at 0.5 µg/ml, anti-Mad1 tissue culture supernatant (provided by A. Musacchio, Istituto Europeo di Oncologia, Milan, Italy) was used at 1:20, and human ACAs (Antibodies, Inc.) were used at 4 µg/ml. FITC-, Cy3-, and Cy5-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. Samples were stained with DAPI before mounting with Vectashield medium (Vector Laboratories). Digital images were captured at 23°C using software (LAF; Leica) by a charge-coupled device camera (ORCA AG; Hamamatsu Photonics) mounted on an inverted microscope (DMI6000B; Leica) with a 100× 1.4 NA objective. For each sample, images were collected at 0.2-µm z sections that were subsequently deconvolved using identical parameters. The z stacks were projected as single two-dimensional images and assembled using PhotoShop (version 9; Adobe) and Illustrator (version 12; Adobe).
Immunoblotting
Whole cell extracts from 5 × 104 PD-NC4 fibroblasts were separated by SDS-PAGE and transferred to nitrocellulose. ACA was used at 2 µg/ml. Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc.
Monastrol treatments
Cells were incubated with 100 µM monastrol (Tocris) for 2 h to arrest them in mitosis with monopolar spindles (Kapoor et al., 2000). The spindle bipolarization assay allowed us to examine the frequency of chromosome alignment failure. Cells were incubated with monastrol as above. Monastrol was withdrawn, and cells were incubated with the proteasome inhibitor, 10 µM MG132 (Tocris), for an additional 30 min to allow spindle bipolarization and chromosome alignment on the metaphase plate. After these treatments, cells were fixed and processed for indirect immunofluorescence microscopy using the aforementioned antibodies and imaging parameters. Anti-INCENP (Active Motif)–purified mAb was used at 0.1 µg/ml, and anti-Ndc80 (Abcam)–purified mAb was used at 0.2 µg/ml. To determine the frequency of alignment failure, cells containing one to five misaligned chromosomes were randomly selected, imaged, and the identity of each chromosome was determined (PD-NC4 or normal chromosome). A total of 140 misaligned chromosomes from 72 cells in three independent experiments were inspected in this manner. The frequency of misalignment was calculated by dividing the number of misaligned chromosomes by the number of each chromosome type (PD-NC4 or normal) present in the cell. To measure the distances between kinetochore pairs (using the inner kinetochore protein CENP-C as a marker), between aurora B and the kinetochore, and between the center of aurora B accumulation and the centromere axis, images of several misaligned normal or PD-NC4 chromosomes from multiple experiments were acquired as described above and deconvolved and analyzed in MMAF software (Leica). CENP-C and aurora B isosurfaces were rendered so that the three-dimensional coordinates of the centroid of each corresponding immunofluorescence signal could be identified. Interkinetochore distances and aurora B–kinetochore distances were measured directly using the multidimensional tools within the MMAF software. The distance between the center of aurora B accumulation and the centromere axis was determined by first calculating the angle between the centromere axis and a line connecting aurora B and the kinetochore, and then multiplying the sine of this angle by the distance of aurora B from the kinetochore (i.e., trigonometric function for right angles).
Phospho–CENP-A detection of aurora B kinase activity at kinetochores
Phospho–CENP-A staining to detect aurora B kinase activity at misattached kinetochores has been described previously (Liu et al., 2009). In brief, cells were first incubated with 100 µM monastrol for 1.5 h to arrest cells with monopolar spindles before addition of 3.3 µM reversible aurora B kinase inhibitor ZM447439 (Tocris; Ditchfield et al., 2003) and 10 µM MG132 for an additional 1.5 h. ZM447439 was withdrawn to reactivate aurora B kinase activity, and cells were incubated in 100 µM monastrol and 10 µM MG132 for an additional 45 min before being fixed and processed for indirect immunofluorescence microscopy using the aforementioned imaging parameters. Antiphospho–CENP-A (Ser7) rabbit polyclonal antibody (Millipore) was used at 2 µg/ml. To quantify phospho–CENP-A levels on PD-NC4 and normal chromosomes, the mean intensity per centromere was measured using MMAF software with a total of 30 PD-NC4 chromosomes and 591 normal chromosomes (randomly selected in the ACA channel).
Online supplemental material
Fig. S1 shows that CENP-ACAD components track with CENP-A to the neocentromere of the PD-NC4 neocentromere. Fig. S2 shows aurora B at the PD-NC4 neocentromere in prometaphase, metaphase, and nocodazole-arrested cells. Fig. S3 shows kinetochore proteins at the neocentromere of the PD-NC4 chromosome. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001035/DC1.
Acknowledgments
We thank A. Choo, K. Yoda, D. Cleveland, and A. Musacchio for the kind gifts of reagents. We also thank D. Liu and M. Lampson (University of Pennsylvania, Philadelphia, PA) for many helpful discussions throughout these experiments and A. Stout (University of Pennsylvania) for advice on image analysis.
This work was supported by a grant from the National Institutes of Health (GM82989), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and a Rita Allen Foundation Scholar Award to B.E. Black. E.A. Bassett is supported by the University of Pennsylvania Structural Biology Training grant (National Institutes of Health grant GM08275) and a predoctoral fellowship from the American Heart Association.
Footnotes
Abbreviations used in this paper:
- ACA
- anti-centromere antibody
- CATD
- CENP-A targeting domain
- CENP
- centromere protein
- CENP-ANAC
- CENP-A nucleosome-associated complex
- INCENP
- inner CENP
References
- Adams R.R., Eckley D.M., Vagnarelli P., Wheatley S.P., Gerloff D.L., Mackay A.M., Svingen P.A., Kaufmann S.H., Earnshaw W.C. 2001. Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma. 110:65–74 10.1007/s004120100130 [DOI] [PubMed] [Google Scholar]
- Amor D.J., Bentley K., Ryan J., Perry J., Wong L., Slater H., Choo K.H. 2004. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA. 101:6542–6547 10.1073/pnas.0308637101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A.W. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118–3129 10.1101/gad.934801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Bassett E.A. 2008. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 20:91–100 10.1016/j.ceb.2007.11.007 [DOI] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Jr., Cleveland D.W. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- Black B.E., Brock M.A., Bédard S., Woods V.L., Jr., Cleveland D.W. 2007a. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 104:5008–5013 10.1073/pnas.0700390104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Jansen L.E.T., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., Cleveland D.W. 2007b. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 25:309–322 10.1016/j.molcel.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C.C., Godek K.M., Jansen L.E.T., Straight A.F. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., Sullivan K.F. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Depinet T.W., Zackowski J.L., Earnshaw W.C., Kaffe S., Sekhon G.S., Stallard R., Sullivan B.A., Vance G.H., Van Dyke D.L., Willard H.F., et al. 1997. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum. Mol. Genet. 6:1195–1204 10.1093/hmg/6.8.1195 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D., Cancilla M.R., Earle E., Mao J.I., Saffery R., Tainton K.M., Kalitsis P., Martyn J., Barry A.E., Choo K.H. 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 16:144–153 10.1038/ng0697-144 [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 91:313–321 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Sullivan K.F., Machlin P.S., Cooke C.A., Kaiser D.A., Pollard T.D., Rothfield N.F., Cleveland D.W. 1987. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104:817–829 10.1083/jcb.104.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E.T., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E.T., Bailey A.O., Yates J.R., III, Bassett E.A., Wood S., Black B.E., Cleveland D.W. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. 2006. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684 10.1111/j.1365-2443.2006.00969.x [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., Mitchison T.J. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. 2009. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 21:51–58 10.1016/j.ceb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern J.P., Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 10.1093/nar/18.12.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., et al. 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 309:613–617 10.1126/science.1111387 [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., Stukenberg P.T. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I.X., Yates J.R., III, Oegema K., Hyman A., Desai A. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 127:1179–1191 10.1016/j.cell.2006.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler M.G., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F. 2001. Genomic and genetic definition of a functional human centromere. Science. 294:109–115 10.1126/science.1065042 [DOI] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D.W. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952 [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Ventura M., Antonacci F., Cardone M.F., Stanyon R., D’Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M. 2007. Evolutionary formation of new centromeres in macaque. Science. 316:243–246 10.1126/science.1140615 [DOI] [PubMed] [Google Scholar]
- Wade C.M., Giulotto E., Sigurdsson S., Zoli M., Gnerre S., Imsland F., Lear T.L., Adelson D.L., Bailey E., Bellone R.R., et al. ; Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 326:865–867 10.1126/science.1178158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton P.E., Cooke C.A., Bourassa S., Vafa O., Sullivan B.A., Stetten G., Gimelli G., Warburton D., Tyler-Smith C., Sullivan K.F., et al. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901–904 10.1016/S0960-9822(06)00382-4 [DOI] [PubMed] [Google Scholar]
- Willard H.F. 1991. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1:509–514 10.1016/S0959-437X(05)80200-X [DOI] [PubMed] [Google Scholar]
- Zeitlin S.G., Barber C.M., Allis C.D., Sullivan K.F., Sullivan K. 2001a. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 114:653–661 [DOI] [PubMed] [Google Scholar]
- Zeitlin S.G., Shelby R.D., Sullivan K.F. 2001b. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155:1147–1157 10.1083/jcb.200108125 [DOI] [PMC free article] [PubMed] [Google Scholar]