Correlative light and electron microscopy provides support for the linear amalgamation of yeast prion proteins.
Abstract
Yeast prion [PSI+] is caused by aggregated structures of the Sup35 protein. Although Sup35 forms typical amyloid fibrils in vitro, there is no direct evidence for the fibrillar structures of Sup35 in vivo. We analyzed [PSI+] cells in which Sup35 fused with green fluorescent protein (GFP) formed aggregates visible by fluorescence microscopy using thin-section electron microscopy (EM). Rapid-freeze EM combined with an immunogold-labeling technique as well as correlative light EM, which allows high-resolution imaging by EM of the same structure observed by light (fluorescence) microscopy, shows that the aggregates contain bundled fibrillar structures of Sup35-GFP. Additional biochemical and fluorescent correlation spectroscopy results suggest that the Sup35 oligomers diffused in the [PSI+] lysates adopt fibril-like shapes. Our findings demonstrate that [PSI+] cells contain Sup35 fibrillar structures closely related to those formed in vitro and provide insight into the molecular mechanism by which Sup35 aggregates are assembled and remodeled in [PSI+] cells.
Introduction
Amyloids are highly ordered fibrillar protein aggregates with a cross β-sheet structure (Chiti and Dobson, 2006), which binds Congo red and thioflavin dyes (Klunk et al., 1989; Naiki et al., 1989). Amyloids were originally defined as extracellular deposits in several human diseases, but the nomenclature now also includes numerous protein aggregates with nonpathological origins, raising the possibility that amyloid formation is an inherent property of many different proteins with diverse biological functions. (Chiti and Dobson, 2006). Representative examples of such nonpathological amyloid formation are budding yeast Saccharomyces cerevisiae prion proteins such as Sup35 and Ure2 (Cox, 1965).
Sup35 normally functions as a translation termination factor required for cell survival, but aggregated forms of Sup35 are the determinant of a prion phenotype [PSI+]. [PSI+] propagation is strictly dependent on the amount of Hsp104 (Chernoff et al., 1995), a chaperone which disassembles protein aggregates in an ATP-dependent manner with the aid of the Hsp70/40 system (Doyle and Wickner, 2009). Sup35 fused with GFP in [PSI+] cells gives rise to the formation of visible spherical aggregates, called foci, in the cytosol (e.g., Patino et al., 1996; Satpute-Krishnan and Serio, 2005; Song et al., 2005; Kawai-Noma et al., 2006), which are one of the indicators of the [PSI+] phenotype (Tuite and Cox, 2003). The foci are not dead-end complexes as our previous time-lapse imaging revealed that the spherical foci, which are formed by the expression of the N-terminal (N) and middle (M) domains of Sup35 fused with GFP (Sup35NM-GFP) in [PSI+] cells, can be dispersed throughout the cytoplasm as diffuse oligomers (Kawai-Noma et al., 2006; Taguchi and Kawai-Noma, 2010). Besides the spherical foci, rod- or ring-shaped aggregates may also be formed when Sup35NM-GFP is overexpressed in [psi−] cells (Zhou et al., 2001; Ganusova et al., 2006) and in [PSI+] cells treated with 3–5 mM guanidine hydrochloride (GuHCl; Tuite et al., 1981), which is known to cure [PSI+] by perturbing Hsp104 (Zhou et al., 2001; Kawai-Noma et al., 2009).
In vitro, recombinant Sup35 proteins bearing the prion-determining N-terminal domain form fibrils with a diameter of ∼10–20 nm, depending on the Sup35 constructs used in the studies (Glover et al., 1997; Kishimoto et al., 2004; Krzewska and Melki, 2006). The fibrils meet many of the criteria for typical amyloids such as binding of Congo red and thioflavin dyes, seed-dependent self-propagation, and cross β-sheet structures, as revealed by x-ray fiber diffraction (Glover et al., 1997; Kishimoto et al., 2004; Krzewska and Melki, 2006).
The ability of recombinant Sup35 proteins to form amyloid fibrils with cross β-sheet structures in vitro led to an assumption about the amyloid structures of Sup35 in vivo. Actually, several previous studies suggested the existence of amyloid fibrils in vivo. First, Sup35 aggregates in [PSI+] cells are stained by an amyloid-staining dye, thioflavin S, suggesting the presence of cross β-sheet structures in [PSI+] cells (Kimura et al., 2003). Second, an EM analysis of Sup35 oligomers isolated from [PSI+] lysates revealed ∼20-nm-wide barrels and larger structures (bundles; Bagriantsev et al., 2008). Finally, the induction of the [PSI+] phenotype by the expression of recombinant Sup35 implied that [PSI+] cells contained amyloid fibrils originating from Sup35 (Tanaka et al., 2004).
However, so far there is no direct evidence for the presence of amyloid fibrils of Sup35 in [PSI+] cells, and the detailed structure of the aggregates such as foci in [PSI+] cells is not known. Elucidating the structures of Sup35 aggregates in [PSI+] cells would provide new clues for understanding the molecular details of prion transmission in cells, which are usually not accessible by conventional genetics and cell biology. In particular, detailed information about the in vivo structures of Sup35 aggregates would directly address the important question of how Sup35 aggregates are assembled, remodeled, and transmitted in cells. In this study, we provide direct evidence for the existence of fibrillar structures of Sup35 prions in cells by using correlative light (fluorescence) EM (CLEM), an immunogold-labeling EM, and biochemical analysis combined with fluorescence correlation spectroscopy (FCS).
Results
Dotlike aggregates in [PSI+] cells contain bundles of Sup35 fibrils
Sup35 aggregates have been visualized in living [PSI+] cells by conventional fluorescence microscopy using Sup35-GFP fusion proteins (Patino et al., 1996; Zhou et al., 2001; Ganusova et al., 2006; Kawai-Noma et al., 2006). Despite the advantage of molecular selectivity, fluorescence microscopy typically provides only low-resolution images, which are insufficient to elucidate structural details. To improve the spatial resolution of images of Sup35 aggregates, we used several imaging techniques such as rapid-freeze EM, immuno-EM, and CLEM. By comparing light and electron microscopic images, CLEM provides high-resolution imaging of fluorescently labeled subcellular structures (Haraguchi et al., 2008) and is therefore well-suited for detecting the ordered structure of the aggregated form of prion proteins within a cell.
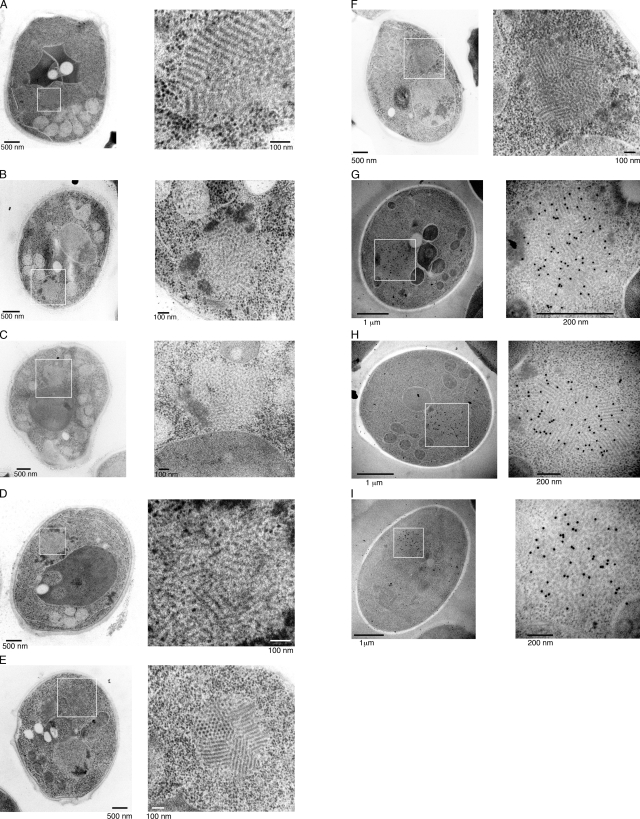
We first performed rapid-freeze EM analysis to obtain images of the Sup35NM-GFP aggregates in [PSI+] cells as well as in G74-D694 [PSI+] cells, in which GFP was inserted between the N and M domains of the endogenous Sup35 protein (Kawai-Noma et al., 2009). We found many spherical particles with ordered fibrillar structures (Fig. 1, A–F). Several types of structures were observed in [PSI+] cells, including fibrils aligned in parallel (Fig. 1 A), amorphous bundles (Fig. 1, B and C), and a mixture of both (Fig. 1 D). The fibrillar structures were specifically observed in [PSI+] cells: a total of ∼10%, i.e., 18 of 200 sections, contained those fibrillar structures, whereas no obvious fibrillar structures were found in 200 sections of [psi−] cells (Fig. S1, A and B). We also searched for such ordered structures in G74-D694 [PSI+] cells. G74-D694 [PSI+] cells expressing Sup35NM-GFP contained distinct, ordered particles in which perfectly aligned fibrils were randomly assembled (Fig. 1, E and F). The diameter of each fibril in these cells was around 20 nm, which is similar to the diameter of the full-length Sup35 fibrils formed in vitro (Glover et al., 1997; Krzewska and Melki, 2006). In some particles, we observed honeycomb structures, which likely correspond to sectioned Sup35 fibrils generated by thin slicing (Fig. 1 E and see Fig. 4). To determine whether the fibrillar structures observed in the rapid-freeze EM images contained Sup35NM-GFP, we conducted immunogold labeling of the thin sections using an anti-GFP antibody. As shown in the gallery (Fig. 1, G–I), the antibody detected Sup35NM-GFP within the fibrillar structures. Gold labeling occurred but was not clustered in control experiments using [psi−] cells (Fig. S1, C and D), indicating that Sup35-GFP proteins are present but not assembled in [psi−] cells. These results indicated that the fibrillar structures found using rapid-freeze EM contain Sup35NM-GFP in [PSI+] cells.
Figure 1.
Rapid-freeze EM images of spherical aggregates in [PSI+] and G74-D694 [PSI+] cells. (A–F) Rapid-freezing EM images of spherical aggregates in [PSI+] (A–D) and G74-D694 [PSI+] (E and F) cells. The cells contain laterally aligned fibrils (A), less-ordered, amorphously bundled fibrils (B and C), a mixture of the less-ordered and laterally aligned fibrils (D), and assemblies of well-ordered, laterally aligned fibrils (E and F). (G–I) Immunogold labeling of the thin-section EM images of aggregates in G74-D694 [PSI+] cells. An anti-GFP antibody was used to attach the gold. (A–I) The boxed areas in the left images are magnified on the right.
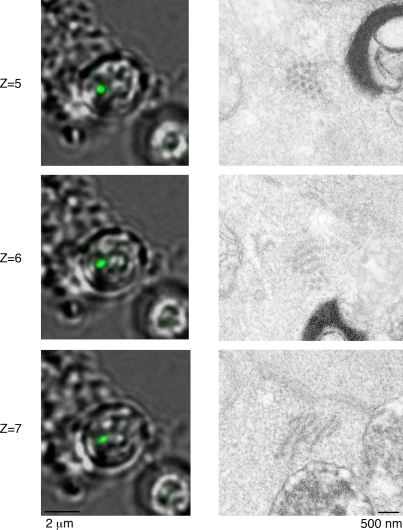
Figure 4.
Three-dimensional CLEM images of a rodlike aggregate in [PSI+] cells treated with GuHCl. Serial z-section fluorescence images (1 µm in depth) that span the rodlike aggregates in [PSI+] cells treated with GuHCl (left). The corresponding EM image for each fluorescence image is shown on the right.
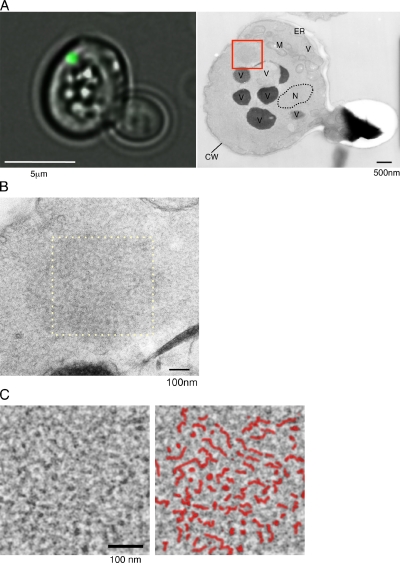
Next, we performed CLEM for aggregates of Sup35NM-GFP in [PSI+] cells, as described in Materials and methods. Fluorescence imaging of cells expressing Sup35NM-GFP showed that Sup35NM-GFP formed spherical dotlike aggregates in [PSI+] cells (Fig. 2 A, left; and Fig. S2, left). Thin-section EM images for the same cell revealed the structure of the same aggregate identified by fluorescence microscopy (Fig. 2 A, right, boxed region). We observed some dense structures in the magnified image (Fig. 2 B and Fig. S2, right), which are similar to structures observed using rapid-freeze EM techniques (Fig. 1, B and C). We processed the structure by Fourier transformation and found that it appeared to contain some fibril-like structures (Fig. 2 C; compare this with Fig. 1, C and D). We note that we did not find such fibrillar structures in serial sections of [psi−] cells using CLEM (Fig. S1 E). Collectively, CLEM analysis directly demonstrated that fluorescent foci of Sup35-GFP in [PSI+] cells contain fibrillar structures.
Figure 2.
CLEM images of spherical aggregates in a [PSI+] cell. (A) Fluorescence (left) and EM (right) images of a Sup35NM-GFP aggregate in a [PSI+] cell. Cells expressing Sup35NM-GFP were fixed after fluorescence imaging (left), and the thin section containing the same spherical aggregate identified by the fluorescence microscopy was subjected to EM analysis (right). CW, cell wall; M, mitochondrion; N, nucleus; V, vacuole. (B) The area within the red box in A is magnified. (C) Fourier-filtered image (left) of the area within the yellow dashed box in B. The spherical aggregate of Sup35NM-GFP in B was processed, using the fast Fourier transform plug-in in ImageJ software. In the right panel, drawings are superimposed on the Fourier transform image indicating fibril-like structures (red lines).
Rodlike aggregates in GuHCl-treated [PSI+] cells contain bundles of long Sup35 fibrils
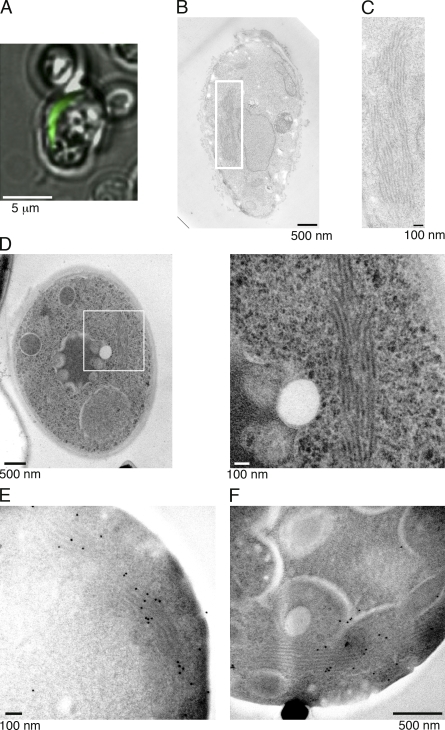
Because the shape of the aggregate can be switched from a spherical to rodlike form by a treatment with GuHCl, an inhibitor of Hsp104 (Zhou et al., 2001; Kawai-Noma et al., 2009), we also analyzed the structure of rodlike aggregates by CLEM. We treated [PSI+] cells with GuHCl, and about half of the cells contained the rod-shaped aggregates upon Sup35NM-GFP expression (Kawai-Noma et al., 2009), as shown in Fig. 3 A. CLEM analysis revealed that the rod-shaped fluorescent aggregates were composed of bundled fibrils that were longer than those observed in [PSI+] and G74-D694 [PSI+] cells without GuHCl treatment (compare Fig. 3 [B and C] with Figs. 1 and 2). The fibrils were aligned laterally, and the lengths of the bundled structures ranged from ∼0.5 to ∼2 µm. We also observed similar laterally bundled fibrils by rapid-freeze EM (Fig. 3 D). Immunogold labeling in the rapid-freeze EM revealed preferential gold labeling on the fibrillar structures, confirming that the laterally bundled fibrils contained Sup35NM-GFP (Fig. 3, E and F).
Figure 3.
CLEM and rapid-freeze images of rodlike aggregates in [PSI+] cells treated with GuHCl. (A–C) Fluorescence (A) and EM (B and C) images of a rodlike Sup35NM-GFP aggregate in a [PSI+] cell treated with GuHCl for 15 h. Cells expressing Sup35NM-GFP were fixed after fluorescence imaging (A), and the thin section containing the same rodlike aggregate identified by the fluorescence microscopy was subjected to EM analysis (B). (C) Magnified image of the boxed area in B. (D) Rapid-freeze EM images of a rodlike aggregate in a [PSI+] cell treated with GuHCl. The boxed areas in the left image are magnified on the right. (E and F) Immunogold labeling of the thin-section EM images of aggregates in [PSI+] cells treated with GuHCl. An anti-GFP antibody was used to attach the gold.
To obtain additional structural information about fibrillar bundles, we examined other rod-shaped aggregates that were perpendicularly localized to the sectioning plane in GuHCl-treated [PSI+] cells by CLEM. As shown in Fig. 4, deconvolved three-dimensional fluorescence images, covering 1 µm in depth (2 sections × 2 z sections; see Z = 5 and Z = 6), revealed that a dotlike aggregate continued toward the z axis to form a rod-shaped aggregate. Correlative EM images of the fluorescent structure showed an ordered honeycomb structure (Fig. 4, Z = 5 and Z = 6, right). Each dot of the honeycomb structure was fibrous, rather than spherical, because a continuous structure from each dot was observed in EM images of serial sections (Fig. 4, Z = 7, right), suggesting that a rod-shaped aggregate was formed with parallel fibrils aligned laterally to create a honeycomb structure, as also found in the spherical aggregates (Fig. 1, E and F).
Sup35NM-GFP oligomers in the [PSI+] lysate have fibril-like shapes
The EM analyses clearly showed that the visible spherical or rodlike aggregates in [PSI+] cells are laterally associated Sup35 fibrillar bundles. We previously demonstrated that the spherical aggregates are dynamic, not static, structures (Kawai-Noma et al., 2006). Time-lapse imaging of single living cells revealed that the visible spherical foci exist in equilibrium with oligomers dispersed throughout the cytoplasm (Kawai-Noma et al., 2006). We also showed that the cytoplasm in GuHCl-treated cells containing rod-shaped aggregates also contained diffuse oligomers (Kawai-Noma et al., 2009). In addition, time-lapse imaging of single living cells revealed that the rod-shaped aggregates changed into spherical aggregates, which have a dynamic nature, when the GuHCl was removed to reactivate Hsp104 activity (Fig. S3). This result raised the question of which structure is adopted by diffuse oligomers after the dispersion from spherical or rod-shaped aggregates.
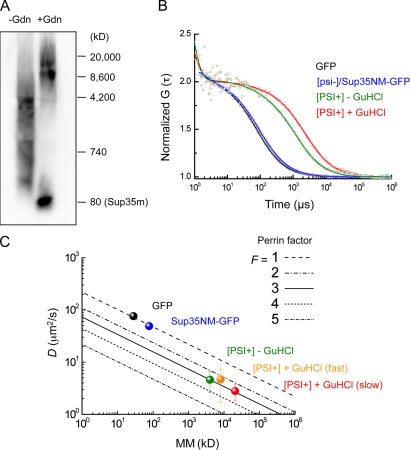
To clarify the shape of diffuse oligomers, we conducted a simulation based on the results of semidenaturing detergent agarose gel electrophoresis (SDD-AGE; Kryndushkin et al., 2003) and FCS (Lippincott-Schwartz et al., 2001; Pack et al., 2006). SDD-AGE analysis of [PSI+] lysate treated with 1% SDS, but without GuHCl treatment, revealed that the major fraction of Sup35NM-GFP oligomers ranged from ∼700 to 4,000 kD (Fig. 5 A, left lane). Assuming that the oligomers are globular in shape, diffusion constants ranging from 25 to 14 µm2/s were calculated from the molecular masses. However, an FCS analysis of the diffuse oligomers in the lysate (Fig. 5 B, green curve) revealed that the actual diffusion constants of the oligomers were much lower, ∼4.6 µm2/s on average. We also found a discrepancy in the diffusion constants in the [PSI+] lysates treated with GuHCl (Fig. 5, A [right lane] and B [red curve]), in which the size of the diffuse oligomers was larger than that of the [PSI+] lysate without the GuHCl treatment, as previously reported (Kryndushkin et al., 2003). Note that the diffusion constants of monomeric GFP and Sup35NM-GFP in the SDS-treated lysates of [psi−] cells measured by FCS, in addition to the diffusion constants of synthesized and dye-labeled proteins in solution, were consistent with those expected from their molecular masses with the assumption that the molecules are globular (Fig. 5, B [black and blue curves] and C; Pack et al., 2006).
Figure 5.
Simulation of shapes of Sup35NM-GFP oligomers in [PSI+] lysates. (A) SDD-AGE of yeast lysates obtained from [PSI+] cells treated with (right) or without (left) GuHCl. An anti-Sup35 antibody was used for Western blotting detection. Sup35m represents the monomeric form of Sup35NM-GFP. (B) Representative normalized FAFs of GFP prepared from lysates of [PSI+] cells (black), Sup35NM-GFP from [psi−] cells (blue), Sup35NM-GFP from [PSI+] cells (green) without GuHCl treatment, and Sup35NM-GFP from [PSI+] cells with GuHCl treatment (red) are shown. Solid lines show the FAF fitting curves. (C) Relation between diffusion constants and molecular masses (mm). Oblique lines indicate simulated plots for Perrin’s factors (F = 1–5). Mean diffusion constants were calculated from three independent FCS experiments. The molecular masses of GFP (black) and Sup35NM-GFP (blue) were calculated from their amino acid sequences. A molecular mass of 4,200 kD was used as the mean for the Sup35NM-GFP oligomers (green), which had a mean diffusion constant of 4.6 ± 2 µm2/s, calculated from the corresponding FCS data. The diffusion constant of Sup35NM-GFP in the [PSI+] lysate with GuHCl treatment was categorized into two groups because the FAFs fit well to a two-component model. Molecular masses of 8,000 kD (red) and 20,000 kD (orange) were used as representatives for the two different diffusion constants of 4.7 ± 3 µm2/s and 2.8 ± 1 µm2/s, respectively.
The discrepancy between the results of the SDD-AGE and FCS analyses of the [PSI+] lysates can be reconciled by the assumption that the oligomers are not globular, but fibrillar in shape. The Perrin equation is frequently used to estimate whether molecules with equivalent molecular masses are globular or prolate in shape (Björling et al., 1998; Pack et al., 2006). The Perrin factor, F, which correlates the axial ratios of the molecules (Fig. S4 A), was calculated as ∼3 for both types of oligomers from [PSI+] cells with or without GuHCl treatment (Fig. 5 C, green, orange, and red symbols). A Perrin factor of ∼3, which corresponds to an axial ratio (length/diameter) of ∼65 (Fig. S4 A), indicates that the Sup35NM-GFP oligomers in [PSI+] lysates are fibrillar in shape, suggesting that the oligomers are fragmented forms of the fibrils that were observed in the EM images. We also note that almost the same Perrin factors were obtained for different molecular masses of the diffuse oligomers from [PSI+] cells with or without GuHCl treatment (Fig. 5 C), implying that the aggregates containing Sup35NM-GFP oligomers in GuHCl-treated cell lysates are both longer and thicker than those in GuHCl-untreated cells because the axial ratio should be the same in both oligomers (Fig. S4 B).
Discussion
Evidence for the fibrillar structures in yeast [PSI+] cells
Sup35, one of the yeast prion proteins, forms amyloid fibrils in vitro. However, there is no direct evidence for fibrillar structures in [PSI+] cells. Our study aimed to find the missing link between the in vitro and in vivo results by generating several lines of evidence for the existence of fibrillar structures of Sup35 prions in cells. The CLEM analysis demonstrated that the visible fluorescent aggregates, which are often regarded as a hallmark of prions, were composed of bundled fibrils. The appearance of the bundled structures varied from less-ordered, amorphously bundled structures to well-ordered, laterally associated fibrils, with some assembled in a honeycomb structure. Fibrillar structures were observed irrespective of the different fixation methods used in this study, confirming that the fibrillar structures were not caused by an artifact of fixation.
A previous thin-section EM analysis of another yeast prion, [URE3], revealed that [URE3] prion cells overexpressing Ure2 contain irregularly associated filaments composed of Ure2 (Speransky et al., 2001). Such filamentous aggregates of Ure2 seem to be similar to the less-ordered, amorphous bundled fibrils in [PSI+] cells (Fig. 1, B and C; and Fig. 2), suggesting that this less-ordered, amorphous bundled fibril is a common structure among prions, although we also observed other types of aggregates, including highly ordered fibrils, in [PSI+] cells (Fig. 1, A, E, and F).
In addition to the fibrillar structures visualized by EM, the FCS analysis combined with SDD-AGE revealed that the diffuse Sup35 oligomers, which cannot be visualized by EM or conventional fluorescent microscopy, are prolate in shape, suggesting that the diffuse Sup35 oligomers are also rodlike (fibrillar) structures in cells. Using EM analysis, Bagriantsev et al. (2008) observed ∼20-nm-wide bundles of Sup35 oligomers (also called polymers), which were prolate in shape, isolated from [PSI+] lysates, supporting the fibrillar structures of the diffuse Sup35 oligomers in [PSI+] cells. The dimensions of the 20-nm-wide structures of the bundles observed by Bagriantsev et al. (2008) are the same as those of the fibrils observed in our study, suggesting that the fragmented fibrils are diffuse in the cytoplasm of [PSI+] cells.
Our EM analysis revealed that the structural assembly of the fibrils in the spherical aggregates in [PSI+] or G74-D694 [PSI+] cells ranged from less ordered to highly ordered. Although the mechanism for generating these distinct assemblies is unclear, we suggest the possibility that the composition of the aggregates might differ in these distinct aggregates. Because the fibrils with a diameter of ∼20 nm are tightly packed in the highly ordered aggregates, other components, such as chaperones, would be excluded from the aggregates. However, the less-ordered aggregates might contain associated proteins.
Connection of fibrils in [PSI+] cells with amyloid fibrils
We could not immediately connect the fibrillar structures in [PSI+] cells with amyloid structures that contain cross β-sheet structures, as our EM analyses relied on the morphologies of the aggregates. However, the following points prompted us to propose that the fibrillar structures are amyloid fibrils. First, Kimura et al. (2003) reported that spherical Sup35-GFP aggregates were stained by an amyloid-staining dye, thioflavin S, suggesting that the spherical aggregates we analyzed were composed of amyloid-containing structures. Second, the diameters of the fibrils in the images were around 20 nm, which resembles the dimensions of the full-length Sup35 fibrils formed in vitro (Glover et al., 1997; Krzewska and Melki, 2006), also suggesting that the fibril structures we observed are amyloid fibrils. Finally, the resistance of the fibrillar-shaped oligomers detected by FCS and SDD-AGE to 2% SDS implies that the oligomers are amyloid because the SDS resistance is one of the necessary conditions for amyloid structures.
The rod-shaped aggregates we observed in GuHCl-treated cells were composed of bundled longer fibrils, reaching up to ∼2 µm in length. It is important to emphasize that, irrespective of whether the aggregates appear to be either spherical or rod-shaped by fluorescent microscopy, the aggregates are composed of similar ∼20-nm-thick fibrils, as determined by EM analyses. Because the ∼20-nm-thick fibrils, which were found in spherical aggregates, were amyloids, as discussed in this section, the longer fibrils in the rod-shaped aggregates should also be amyloids.
Implications for Hsp104 remodeling activity
Because GuHCl inhibits Hsp104, which can fragment Sup35 fibrils in vitro (Inoue et al., 2004; Shorter and Lindquist, 2004), the loss of fibril fragmentation caused by impaired Hsp104 function would predictably generate longer fibrils observed in the rod-shaped aggregates. We showed that the rod-shaped aggregates were dynamically converted to spherical aggregates when we removed GuHCl from the medium, suggesting that the recovery of Hsp104 function would fragment the longer fibrils into shorter ones. These shorter fibrils might then be assembled into the spherical aggregates.
Based on a simulation in which the diffuse oligomers in GuHCl-treated [PSI+] cells were both longer and thicker than those in [PSI+] cells without the GuHCl treatment (Fig. 5), we propose that Hsp104 promotes the dissociation of laterally associated bundled fibrils, in addition to their fragmentation, for the following reason. If Hsp104 only fragmented the fibrils, impaired Hsp104 function would result in longer fibrils, leading to a larger axial ratio, as the diameter of the fibrils is the same. However, this is not the case because the axial ratios calculated from the Perrin factors were the same, irrespective of the presence of GuHCl. To maintain the same axial ratios with or without the Hsp104 function, we have to assume that the lateral association of fibrils is enhanced in the Hsp104-impaired cells, resulting in the bundled, and thus thicker, fibrils (Fig. S4 B). Actually, EM analyses revealed that the fibrils are assembled in laterally associated forms in [PSI+] cells (Figs. 1–4), supporting our hypothesis that Hsp104 has the ability to dissociate the laterally associated fibrils, besides the fibril-cleaving activity.
Conclusion
In conclusion, using several EM imaging methods and fluorescence microscopy, we found that the spherical and rod-shaped Sup35 aggregates in [PSI+] cells are composed of fibrillar structures that correspond to those formed in vitro. In addition, Sup35 oligomers that are more diffuse in the cytoplasm are also fibrillar, suggesting that the diffuse oligomers are fragmented forms of the fibrils that were observed in EM images. This in vivo evidence for the fibrillar structures of Sup35 prions in yeast cells highlights the importance of amyloid fibrils in vivo and provides further opportunities to incorporate in vitro studies on amyloid fibrils into in vivo prion biology.
Materials and methods
Yeast strains and media
The strains used in this study were 74-D694 (MATa ade1-14 leu2-3112 his3-Δ200 trp1-289 ura3-52 [PSI+] and [psi−]) strains. GFP was inserted into the chromosomal SUP35 gene of G74-D694, in between the N and M domains. G74-D694 was constructed by transforming 74-D694 with the integration plasmid YIp211-NGMC, which is a URA3-based plasmid carrying SUP35NGMC (Kawai-Noma et al., 2009). SRaf-Leu medium, a synthetic complete medium (using Difco yeast nitrogen base) lacking Leu (SC-Leu), was used (Sherman, 2002). SRaf-Leu medium contained 2% raffinose instead of glucose. To induce the expression of the GAL1 promoter, galactose was added to a final concentration of 2% (wt/vol). For the GuHCl treatment, GuHCl was added to the medium at 5 mM. Yeast strains were grown at 30°C.
Plasmids
The yeast plasmid YCp-GAL1p-SUP35(NM)–GFP [LEU2], expressing the Sup35NM domain conjugated to GFP by the galactose-inducible GAL1 promoter, was previously described (Kawai-Noma et al., 2006, 2009).
Rapid-freeze EM
Cells were mounted on a copper grid (HF51; Pyser-SGI) to form a thin layer and immersed in liquid propane (−187°C) cooled with liquid nitrogen in an EM cryoworkstation (CPC; Leica). The frozen cells were transferred to anhydrous acetone containing 2% OsO4 (Merck) at −80°C in an EM automatic freeze-substitution apparatus (AFS; Leica), maintained at −80°C for 78 h, warmed gradually to 0°C over 11.4 h, maintained at 0°C for 1.5 h, warmed gradually to 23°C over 3.9 h, and incubated at 23°C for 2 h. After washing three times with anhydrous acetone, the samples were infiltrated with increasing concentrations of Spurr’s resin in anhydrous acetone and finally with 100% Spurr’s resin. After polymerization (5 h at 50°C plus 60°C for 2–3 d) in capsules, ultrathin sections were obtained with a microtome (Ultracut UCT; Leica) and stained with uranyl acetate (Merck) and Reynolds lead citrate (Reynolds, 1963). Immunostaining with anti-GFP polyclonal antibody (1:200 or 1:400 dilution; a gift from T. Kuno, Kobe University, Chuo-ku, Kobe, Japan) was performed on ultrathin sections and picked up on nickel grids (grid type G200HH; Gilder Grids) and an immunogold conjugated (15-nm gold) EM goat anti–rabbit IgG (1: 80 dilution; BBInternational) and then were stained with 3% uranyl acetate for 2 h. The sections were then viewed on an electron microscope (H-7600; Hitachi) at 100 kV. Negatives were scanned and converted to tif images with an Advantage HR (Advanced Microscopy Techniques, Corp.). Photoshop CS4 (version11.0.1; Adobe) was used for image processing (Fig. 1; Fig. 3, D–F; and Fig. S1, A–D).
CLEM
Cells were fixed with glutaraldehyde (Polysciences, Inc.) at a final concentration of 2% in 0.1 M of phosphate buffer, pH 7.2, for 2 h at 4°C. After four washes with the buffer, the cells were digested with zymolyase 100T (Seikagaku Co.) at a final concentration of 0.1 mg/ml in the buffer for 30–90 min at 30°C. The cells were then placed in a 35-mm culture dish with a grid (grid size 500 µm; ibidi GmbH). Before use, the dishes were coated with an aqueous solution of 0.2% concanavalin A (Sigma-Aldrich) for 15 min and air-dried after the excess solution was removed. Three-dimensional images (10 focal planes at 0.5-µm intervals) of a cell mounted in 0.1 M of phosphate buffer, pH 7.2, were obtained at 26°C using a microscope system (DeltaVision RT; Applied Precision) equipped with a charge-coupled device (CoolSNAP HQ; Photometrics) and with a Plan-Apochromat 60× NA 1.4 oil immersion objective lens (Olympus) and were computationally processed by three-dimensional deconvolution (Agard et al., 1989) using SoftWoRx software (Applied Precision) on the DeltaVision system (Figs. 2 A, 3 A, and 4). Low-melting agarose (final concentration of 1%) was placed on the cells to prevent the loss of cells from the dish during subsequent manipulations, but this step can be eliminated. EM observation of the same cells was performed as follows. The cells were postfixed with an aqueous solution of 2% OsO4 (Nisshin EM Inc.) for 2 h, washed briefly with distilled water, and stained with 1% uranyl acetate (Merck) in water for 1 h. After washes with distilled water, the cells were sequentially dehydrated with increasing concentrations of ethanol from 50 to 100%, and for embedding, the cells were sequentially incubated in acetone for 10 min, 50% (vol/vol) Epon812 (TAAB) in acetone for 1 h, 66.7% Epon812 in acetone for 1 h, and 100% Epon812 overnight. After a further incubation in fresh 100% Epon812 for 3–4 h, the resin was polymerized at 60°C. The epoxy block containing the same cells observed by fluorescence microscopy was trimmed according to the address on the coverslip. Serial sections with an 80-nm thickness were obtained, stained with 4% uranyl acetate and a commercial ready to use solution of lead citrate (Sigma-Aldrich), and analyzed by a transmission electron microscope (JEM-1200EXS; JEOL) with an acceleration voltage of 80 kV. Images were taken on negative films. After development, the negative films were subjected to the image scanning to convert the analogue data to digital data using a scanner (GT-X978 and Epson File Manager for X software; Epson), and the image data obtained by scanning were saved as a tif file. Fast Fourier transformation was performed using the ImageJ software (National Institutes of Health). A Fourier bandpass filter with a low-frequency cutoff at 0.016 nm−1 and a high-frequency cutoff at 0.24 nm−1 was applied for Fig. 2 C (corresponding to 60 pixels and 4 pixels, respectively, for cutoff parameters in the ImageJ software). Photoshop 11.0.1 was used to adjust the brightness and contrast for figure production (Fig. 2, A–C; Fig. 3, B and C; and Fig. 4). Negatives were scanned and converted to tif images with an Advantage HR. Photoshop CS4 (version 11.0.1) was used for image processing (Fig. 1; Fig. 3, D–F; and Fig. S1, A–D).
Cell lysis
Yeast strains containing YCp-GAL1p-SUP35(NM)–GFP or YCp-GAL1p–GFP were grown to mid–log phase in SRaf-Leu. After 2% galactose (Wako) was added, the yeast were incubated for 4 h at 30°C. Cells were collected by centrifugation, broken with glass beads (Sigma-Aldrich) by vortexing for 1 min at 4°C in lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, pH 8.0, 1 mM DTT, and Complete protease inhibitor cocktail EDTA-free [Roche]), and incubated for 1 min on ice. This procedure was repeated for 4–6 cycles. The crude lysates were clarified by centrifugation.
SDD-AGE
The lysates were incubated in sample buffer (0.5% Tris-acetate-EDTA, 2% SDS, 5% glycerol, and 0.05% bromophenol blue) for 5 min at 37°C (Kryndushkin et al., 2003) and were fractionated by electrophoresis using horizontal 1.8% agarose gels in Tris-acetate-EDTA buffer containing 0.1% SDS. The proteins were transferred to a polyvinylidene fluoride membrane (Immobilon; Millipore) and were analyzed by Western blotting. The bound antibody (anti-Sup35 antibody; Patino et al., 1996) was detected by chemiluminescence (Immobilon Western; Millipore).
FCS measurements and simulations of molecular shapes
All FCS measurements were performed at 25°C on a confocal microscope (LSM510; Carl Zeiss, Inc.) combined with a ConfoCor 2 (Carl Zeiss, Inc.). Details of the analysis of fluorescence auto-correlation functions (FAFs) and the simulation of the molecular shapes of various proteins were described in our previous studies (Pack et al., 1999, 2006; Kawai-Noma et al., 2006, 2009). FAFs in aqueous solutions were fitted to a one- or two-component model with or without a triplet term. In practice, the FAFs of GFP in solution samples prepared from [psi−] and [PSI+] cells and the FAFs of Sup35NM-GFP in solution from [psi−] cells have only one diffusional component. Moreover, the FAFs of Sup35NM-GFP from [PSI+] were best fitted by a one-component model, but occasionally by a two-component model, indicating that it is monodisperse in solution. In contrast, the FAFs of Sup35NM-GFP from [PSI+] cells with GuHCl treatment were best fitted by a two- or three-component model and exhibited a large range of diffusion times, from monomer to much larger oligomers.
On-chip cultivation system
The microcultivation chamber array chip consisted of microchambers positioned on 0.2-mm-thick glass slides (24 × 36 mm; Matsunami). By enclosing the cells in the microchambers, we were able to observe them in liquid medium for a long time without them leaving the field of vision of the objective lens (Inoue et al., 2001; Umehara et al., 2003; Ayano et al., 2004; Kawai-Noma et al., 2006). The microchamber array, which was made of negative photoresist SU-8 10 (Microlithography Chemical Co.), was photolithographically microfabricated on a glass slide (Ayano et al., 2004). Each microchamber in the array was designed with a surface area of 1,500 µm2 and a 10-µm-high wall. The cultivation system was used with a brightfield optical microscopy system (IX-71 inverted microscopy with a UPlan-Apochromat 100× NA 1.35 oil immersion objective lens; Olympus) with an EM charge-coupled device camera (iXon; Andor Technology) to obtain differential interference contrast and fluorescent images. During on-chip cultivation, fresh medium was continuously supplied to the chamber system at a constant rate of 1 ml/min by a peristaltic pump. Yeast cells were grown in SRaf-Leu medium. The temperature of the system was maintained at 30°C throughout the observations by a heated chamber surrounding the microscope. Photoshop CS4 (version 11.0.1) was used for image processing (Fig. S3).
Online supplemental material
Fig. S1 shows EM images of thin sections in [psi−] cells. Fig. S2 shows CLEM images of spherical aggregates in a [PSI+] cell. Fig. S3 shows the dynamics of a rod-shaped aggregate in a living cell after the removal of GuHCl, monitored by an on-chip cultivation system. Fig. S4 shows a schematic representation of the relationship between Perrin’s factor, F, and the axial ratio, as well as an example of two different sizes of cylinder-shaped molecules with the same axial ratio. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201002149/DC1.
Acknowledgments
We thank Dr. Yoshikazu Ohya for his continuous support, Drs. Mamiko Sato and Shou Waga for sharing their electron microscope facilities, Dr. Shouhei Kobayashi for helpful discussions, and Dr. Takayoshi Kuno for anti-GFP polyclonal antibody.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology to H. Taguchi (19058002), to Y. Hiraoka (20114002), and to T. Haraguchi (21370094).
Note added in proof. During the review process of this manuscript, Tyedmers et al. (2010. Proc. Natl. Acad. Sci. USA. doi:10.1073/pnas.1003895107) published a paper describing fibrillar structures of Sup35 in [PSI+] cells.
Footnotes
Abbreviations used in this paper:
- CLEM
- correlative light EM
- FAF
- fluorescence auto-correlation function
- FCS
- fluorescence correlation spectroscopy
- GuHCl
- guanidine hydrochloride
- SDD-AGE
- semidenaturing detergent agarose gel electrophoresis
References
- Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30:353–377 10.1016/S0091-679X(08)60986-3 [DOI] [PubMed] [Google Scholar]
- Ayano S., Noma S., Yoshida M., Taguchi H., Yasuda K. 2004. On-chip single-cell observation assay for propagation dynamics of yeast Sup35 prionlike proteins. Jpn. J. Appl. Phys. 43:L1429–L1432 10.1143/JJAP.43.L1429 [DOI] [Google Scholar]
- Bagriantsev S.N., Gracheva E.O., Richmond J.E., Liebman S.W. 2008. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol. Biol. Cell. 19:2433–2443 10.1091/mbc.E08-01-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björling S., Kinjo M., Földes-Papp Z., Hagman E., Thyberg P., Rigler R. 1998. Fluorescence correlation spectroscopy of enzymatic DNA polymerization. Biochemistry. 37:12971–12978 10.1021/bi980694c [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist S.L., Ono B., Inge-Vechtomov S.G., Liebman S.W. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 268:880–884 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75:333–366 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- Cox B.S. 1965. [PSI], a cytoplasmic suppressor of supersuppression in yeast. Heredity. 20:505–521 10.1038/hdy.1965.65 [DOI] [Google Scholar]
- Doyle S.M., Wickner S. 2009. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34:40–48 10.1016/j.tibs.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Ganusova E.E., Ozolins L.N., Bhagat S., Newnam G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O. 2006. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 26:617–629 10.1128/MCB.26.2.617-629.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Kowal A.S., Schirmer E.C., Patino M.M., Liu J.J., Lindquist S. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 89:811–819 10.1016/S0092-8674(00)80264-0 [DOI] [PubMed] [Google Scholar]
- Haraguchi T., Kojidani T., Koujin T., Shimi T., Osakada H., Mori C., Yamamoto A., Hiraoka Y. 2008. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 121:2540–2554 10.1242/jcs.033597 [DOI] [PubMed] [Google Scholar]
- Inoue I., Wakamoto Y., Moriguchi H., Okano K., Yasuda K. 2001. On-chip culture system for observation of isolated individual cells. Lab Chip. 1:50–55 10.1039/b103931h [DOI] [PubMed] [Google Scholar]
- Inoue Y., Taguchi H., Kishimoto A., Yoshida M. 2004. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 279:52319–52323 10.1074/jbc.M408159200 [DOI] [PubMed] [Google Scholar]
- Kawai-Noma S., Ayano S., Pack C.G., Kinjo M., Yoshida M., Yasuda K., Taguchi H. 2006. Dynamics of yeast prion aggregates in single living cells. Genes Cells. 11:1085–1096 10.1111/j.1365-2443.2006.01004.x [DOI] [PubMed] [Google Scholar]
- Kawai-Noma S., Pack C.G., Tsuji T., Kinjo M., Taguchi H. 2009. Single mother-daughter pair analysis to clarify the diffusion properties of yeast prion Sup35 in guanidine-HCl-treated [PSI] cells. Genes Cells. 14:1045–1054 10.1111/j.1365-2443.2009.01333.x [DOI] [PubMed] [Google Scholar]
- Kimura Y., Koitabashi S., Fujita T. 2003. Analysis of yeast prion aggregates with amyloid-staining compound in vivo. Cell Struct. Funct. 28:187–193 10.1247/csf.28.187 [DOI] [PubMed] [Google Scholar]
- Kishimoto A., Hasegawa K., Suzuki H., Taguchi H., Namba K., Yoshida M. 2004. beta-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem. Biophys. Res. Commun. 315:739–745 10.1016/j.bbrc.2004.01.117 [DOI] [PubMed] [Google Scholar]
- Klunk W.E., Pettegrew J.W., Abraham D.J. 1989. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. 37:1273–1281 [DOI] [PubMed] [Google Scholar]
- Kryndushkin D.S., Alexandrov I.M., Ter-Avanesyan M.D., Kushnirov V.V. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278:49636–49643 10.1074/jbc.M307996200 [DOI] [PubMed] [Google Scholar]
- Krzewska J., Melki R. 2006. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 25:822–833 10.1038/sj.emboj.7600985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Snapp E., Kenworthy A. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444–456 10.1038/35073068 [DOI] [PubMed] [Google Scholar]
- Naiki H., Higuchi K., Hosokawa M., Takeda T. 1989. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177:244–249 10.1016/0003-2697(89)90046-8 [DOI] [PubMed] [Google Scholar]
- Pack C.G., Nishimura G., Tamura M., Aoki K., Taguchi H., Yoshida M., Kinjo M. 1999. Analysis of interaction between chaperonin GroEL and its substrate using fluorescence correlation spectroscopy. Cytometry. 36:247–253 [DOI] [PubMed] [Google Scholar]
- Pack C., Saito K., Tamura M., Kinjo M. 2006. Microenvironment and effect of energy depletion in the nucleus analyzed by mobility of multiple oligomeric EGFPs. Biophys. J. 91:3921–3936 10.1529/biophysj.105.079467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino M.M., Liu J.J., Glover J.R., Lindquist S. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 273:622–626 10.1126/science.273.5275.622 [DOI] [PubMed] [Google Scholar]
- Reynolds E.S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Serio T.R. 2005. Prion protein remodelling confers an immediate phenotypic switch. Nature. 437:262–265 10.1038/nature03981 [DOI] [PubMed] [Google Scholar]
- Sherman F. 2002. Getting started with yeast. Methods Enzymol. 350:3–41 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 304:1793–1797 10.1126/science.1098007 [DOI] [PubMed] [Google Scholar]
- Song Y., Wu Y.X., Jung G., Tutar Y., Eisenberg E., Greene L.E., Masison D.C. 2005. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell. 4:289–297 10.1128/EC.4.2.289-297.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speransky V.V., Taylor K.L., Edskes H.K., Wickner R.B., Steven A.C. 2001. Prion filament networks in [Ure3] cells of Saccharomyces cerevisiae. J. Cell Biol. 153:1327–1336 10.1083/jcb.153.6.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi H., Kawai-Noma S. 2010. Amyloid oligomers: diffuse oligomer-based transmission of yeast prions. FEBS J. 277:1359–1368 10.1111/j.1742-4658.2010.07569.x [DOI] [PubMed] [Google Scholar]
- Tanaka M., Chien P., Naber N., Cooke R., Weissman J.S. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature. 428:323–328 10.1038/nature02392 [DOI] [PubMed] [Google Scholar]
- Tuite M.F., Cox B.S. 2003. Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 4:878–890 10.1038/nrm1247 [DOI] [PubMed] [Google Scholar]
- Tuite M.F., Mundy C.R., Cox B.S. 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics. 98:691–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara S., Wakamoto Y., Inoue I., Yasuda K. 2003. On-chip single-cell microcultivation assay for monitoring environmental effects on isolated cells. Biochem. Biophys. Res. Commun. 305:534–540 10.1016/S0006-291X(03)00794-0 [DOI] [PubMed] [Google Scholar]
- Zhou P., Derkatch I.L., Liebman S.W. 2001. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol. Microbiol. 39:37–46 10.1046/j.1365-2958.2001.02224.x [DOI] [PubMed] [Google Scholar]