This virus takes a less-travelled cytoskeletal road both to reach its replication site in the nucleus and to get back to the plasma membrane to escape the host cell.
Abstract
Most viruses move intracellularly to and from their sites of replication using microtubule-based mechanisms. In this study, we show that nucleocapsids of the baculovirus Autographa californica multiple nucleopolyhedrovirus undergo intracellular motility driven by actin polymerization. Motility requires the viral P78/83 capsid protein and the host Arp2/3 complex. Surprisingly, the virus directs two sequential and coordinated phases of actin-based motility. Immediately after cell entry, motility enables exploration of the cytoplasm and collision with the nuclear periphery, speeding nuclear entry and the initiation of viral gene expression. Nuclear entry itself requires transit through nuclear pore complexes. Later, after the onset of early gene expression, motility is required for accumulation of a subpopulation of nucleocapsids in the tips of actin-rich surface spikes. Temporal coordination of actin-based nuclear and surface translocation likely enables rapid transmission to neighboring cells during infection in insects and represents a distinctive evolutionary strategy for overcoming host defenses.
Introduction
Intracellular pathogens have evolved mechanisms for rapidly navigating the cell interior to replicate and spread in a timely fashion while evading host defenses. Viruses generally use microtubule-based mechanisms to mediate transport to and from sites of replication (Greber and Way, 2006), whereas bacteria use actin-based motility for transit and cell to cell spread (Gouin et al., 2005). Even vaccinia virus, which undergoes actin-based motility after replication and release in its extracellular form, uses microtubules for intracellular movement and egress (Hollinshead et al., 2001; Rietdorf et al., 2001; Ward and Moss, 2001).
We propose that baculoviruses are an exception to this convention. These nuclear replicating DNA viruses release their nucleocapsids into the cytoplasm after endocytosis and must immediately transit from the periphery into the nucleus (Rohrmann, 2008). Moreover, during baculovirus infection of epithelial cells within the insect midgut, a subpopulation of nucleocapsids has been proposed to bypass the nucleus and transit directly to the basolateral surface without replicating to enable rapid spread (Granados and Lawler, 1981; Washburn et al., 1999). Unlike many viruses, baculoviruses do not require microtubules for the production of progeny budded virus (Volkman and Zaal, 1990), although microtubules may play a role in this process (Fang et al., 2009). In contrast, the actin cytoskeleton is essential for budded virus production (Ohkawa and Volkman, 1999). Nucleocapsids from the baculovirus type species Autographa californica multiple nucleopolyhedrovirus (AcMNPV) were seen to associate with cable-like actin structures immediately after release into the cytoplasm (Charlton and Volkman, 1993), hinting at a possible actin-based means of transport. Moreover, the AcMNPV capsid protein P78/83 was shown to be a nucleation-promoting factor that activates actin polymerization by the host Arp2/3 complex (Goley et al., 2006), and P78/83 and the Arp2/3 complex were required for actin assembly within the nucleus late in infection. However, it has remained unclear whether, how, or why baculoviruses might use actin for intracellular transport early in infection.
In this study, we demonstrate that AcMNPV nucleocapsids undergo motility driven by actin polymerization during the early stage of infection. Movement requires the ability of P78/83 to stimulate actin nucleation with the host Arp2/3 complex. In the first phase of infection, nucleocapsid actin-based motility is critical for efficient transit to the nucleus, enabling translocation through nuclear pore complexes. In the next phase of infection, after the onset of viral gene expression, motility is required for accumulation of a distinct set of nucleocapsids at the cell periphery. Coordination of these two phases of motility may provide a mechanistic basis for accelerated passage through infected insect tissues, which is vital to evasion of host defenses.
Results and discussion
AcMNPV undergoes actin-based motility
To visualize AcMNPV movements in cells, we generated a virus encoding an inserted copy of the major viral capsid protein VP39, fused at its C terminus to three copies of the monomeric red fluorescent protein mCherry (Fig. S1). This virus (named 3mC) produced budded progeny at rates similar to wild type (WT; Fig. S2) and had sufficient incorporation of VP39–triple mCherry for nucleocapsids to be visualized by fluorescence microscopy. The 3mC virus was used to infect High Five cells that were transfected with a plasmid expressing EGFP-actin so that the virus and actin could be observed simultaneously.
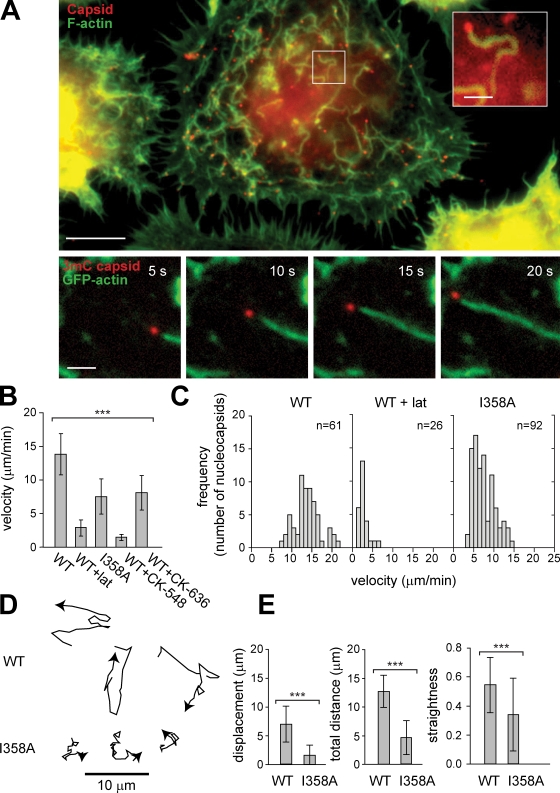
Remarkably, in infected cells at very early times (5–30 min) after infection, nucleocapsids moved rapidly within the cytoplasm and were trailed by actin comet tails (Fig. 1 A, bottom; and Video 1). The mean velocity of movement was 14 ± 3 µm/min (mean ± SD; n = 61) with a range of 7–22 µm/min and was not affected by expression of EGFP-actin (13 ± 3 µm/min in untransfected cells; n = 38). Movement depended on actin assembly, as it was inhibited by the actin-depolymerizing agent latrunculin A (latA; Fig. 1 B). Baculovirus actin-based motility was similar to that of the bacterial pathogens Listeria monocytogenes and Shigella flexneri and vaccinia virus with respect to its velocity (10–26 µm/min depending on pathogen and cell type; Gouin et al., 1999; Rietdorf et al., 2001) and the association of moving viruses with actin tails. However, in contrast to vaccinia virus, which undergoes actin-based motility after replication and release in its extracellular form (Hollinshead et al., 2001; Rietdorf et al., 2001; Ward and Moss, 2001), baculovirus motility occurred immediately after entry and release into the cytoplasm and before replication.
Figure 1.
AcMNPV undergoes actin-based motility within the cell cytoplasm. (A, top) High Five cell 30 min after infection with AcMNPV stained for viral capsids (red; anti-VP39 immunofluorescence) and actin (green; FITC-phalloidin). Bar, 10 µm. (inset) Magnified nucleocapsid (of boxed region) with associated actin tail. Bar, 2 µm. (bottom) Time series in 5-s intervals of AcMNPV 3mC virus (red) and EGFP-actin (green) in a High Five cell. Bar, 2 µm. (B) Mean velocity ± SD for AcMNPV 3mC (WT; n = 61), 3mC in the presence of latA (WT + lat; n = 26), 3mC I358A (I358A; n = 92), and 3mC in the presence of Arp2/3 inhibitor CK-548 (WT + CK-548; n = 29) or CK-636 (WT + CK-636; n = 37). ***, P < 0.0001 (by one-way analysis of variance). (C) Histograms of nucleocapsid velocities for 3mC (WT), 3mC in the presence of latA (WT + lat), and 3mC I358A (I358A). (D) Representative tracks of 3mC (WT; top) and 3mC I358A (I358A; bottom) over 135 s. (E) Displacement (straight line distance from initial location to destination), total distance traveled (along path length), and straightness (displacement/total distance traveled) over 50 s for 3mC (WT; n = 58) and 3mC I358A (I358A; n = 58). ***, P < 0.0001 (by unpaired t test). Error bars indicate mean ± SD.
Actin-based motility of L. monocytogenes is mediated by the bacterial surface–associated nucleation-promoting factor ActA, which activates the host Arp2/3 complex to promote actin polymerization (Gouin et al., 2005). Because baculovirus nucleocapsids contain the nucleation-promoting factor P78/83, we sought to determine whether P78/83 and Arp2/3 complexes play a role in viral actin-based motility. We first generated a mutant 3mC virus bearing a substitution of a conserved residue in the predicted Arp2/3-binding region of P78/83 (Ile358 → Ala358 [I358A]) that results in a partial defect in actin polymerization by the purified P78/83 protein in vitro (Goley et al., 2006). Interestingly, the 3mC I358A mutant moved at a reduced velocity (8 ± 3 vs. 14 ± 3 µm/min for WT; P < 0.0001; Fig. 1 B and Video 2) with a distribution of rates skewed toward that observed in the presence of latA (Fig. 1 C). Moreover, the actin comet tails formed by 3mC I358A were shorter and more difficult to visualize (Video 3), and the movement paths were more erratic with more frequent changes in direction (Fig. 1 D and Video 4). To quantitatively measure the quality of motion, we calculated the straightness of the path by dividing the net displacement by the total distance traveled. By this measure, the mean straightness of I358A paths was only 63% of WT (P < 0.0001). We further confirmed involvement of the Arp2/3 complex by treating infected cells with the Arp2/3 chemical inhibitors CK-548 and CK-636 (Nolen et al., 2009), which decreased the velocity of viral movement to the values observed for I358A or in latA-treated cells (Fig. 1 B). Thus, the ability of P78/83 to activate the Arp2/3 complex drives actin assembly and AcMNPV motility, and reducing P78/83 activity impairs the ability of the virus to move rapidly and progressively in a single direction.
Early AcMNPV actin-based motility is critical for translocation to the nucleus
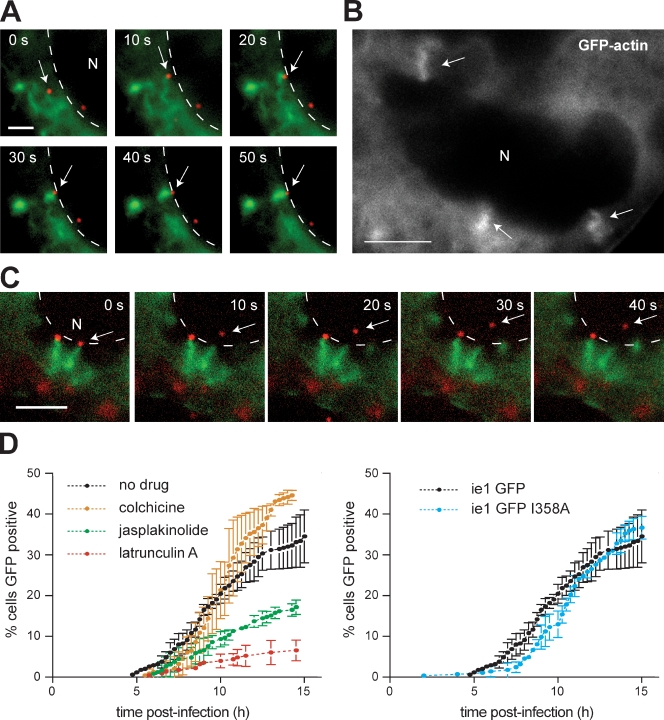
As nuclear-replicating viruses, baculoviruses rapidly travel to the nucleus within 1 h after inoculation (Granados and Lawler, 1981). Because the immediate onset of actin-based motility corresponded with the time of nuclear translocation, we wondered whether motility might promote nucleocapsid interactions with the nucleus. While visualizing movements in the vicinity of the nucleus, we noticed that nucleocapsids frequently collided with and stuck to the nuclear periphery. Strikingly, these viruses continued to polymerize actin, which radiated from the nucleus and underwent corkscrew-like motions (Fig. 2 A and Videos 5 and 6). In many instances, the nuclear membrane indented at the point of impact, suggesting that significant force was exerted (Fig. 2 B and Video 7). Nucleocapsids remained docked at the nuclear periphery for 3 ± 1 min (n = 5) after collisions with the nuclear envelope and then separated from their actin corkscrews and entered the nucleus (Fig. 2 C and Video 8). This resulted in a termination of actin assembly and depolymerization of actin on the nuclear periphery. After separation, virus particles moved a short distance within the nucleus, presumably to sites of capsid uncoating and gene expression. These results suggested that actin-based motility plays a role in transit of baculoviruses to the nucleus.
Figure 2.
Actin-based motility facilitates viral nuclear entry. (A) Time series of actin (green; EGFP-actin) and a nucleocapsid (red; 3mC virus; arrows) encountering the nuclear envelope (N; dashed lines). Bar, 2 µm. (B) Actin (EGFP-actin) corkscrew structures (arrows) associated with the periphery of the nucleus (N) are shown. Bar, 5 µm. (C) Time series of a nucleocapsid (red; 3mC; arrows) separating from its actin corkscrew (green; EGFP-actin) and entering the nucleus (N; nuclear periphery marked by dashed lines). Bar, 5 µm. (D) Graph of the percentage of High Five cells infected with AcMNPV ie-1 GFP that express visible EGFP at different times after infection. (left) ie-1 GFP in the absence or presence of cytoskeleton-disrupting drugs. (right) ie-1 GFP compared with ie-1 GFP I358A. Error bars indicate mean ± SD (n = 3 replicates).
To test the importance of actin-based motility for translocation to nuclei, we constructed a virus that expresses EGFP behind the immediate-early baculovirus promoter ie-1 and evaluated the kinetics of EGFP expression. When cells were infected with this ie-1 GFP virus and observed by time-lapse microscopy, GFP expression was initially observed at 5 h postinfection (hpi), with the percentage of cells initiating GFP expression increasing steadily over the first 15 hpi (Fig. 2 D). Treating cells with the microtubule depolymerizing drug colchicine did not affect the kinetics of GFP expression (Fig. 2 D), confirming that microtubules are not important for nuclear translocation of baculovirus. In contrast, disrupting actin dynamics by treating cells with either latA or the actin-stabilizing drug jasplakinolide slowed the kinetics dramatically (Fig. 2 D), indicating that actin-based motility is crucial for efficient viral transit to the nucleus. We obtained similar results after allowing endocytosis to occur for 5 min before drug addition, ruling out the possibility that latA treatment interfered with endocytosis rather than actin-based motility. To test the role of rapid and directionally persistent motility in transit to the nucleus, we also introduced the P78/83 I358A mutation into the ie-1 GFP virus to generate an ie-1 GFP I358A variant. This mutant virus exhibited delayed GFP expression in the early stage of infection (6–10 hpi) but ultimately reached WT levels by 12 h (Fig. 2 D). In keeping with an ability of the P78/83 mutant to reach the nucleus, 3mC I358A made actin corkscrews when associated with the nuclear periphery (Video 9). These results demonstrate that actin-based motility is crucial for cytoplasmic translocation to the nucleus but that even the more erratic movement of the I358A mutant is sufficient for the virus to reach the nucleus.
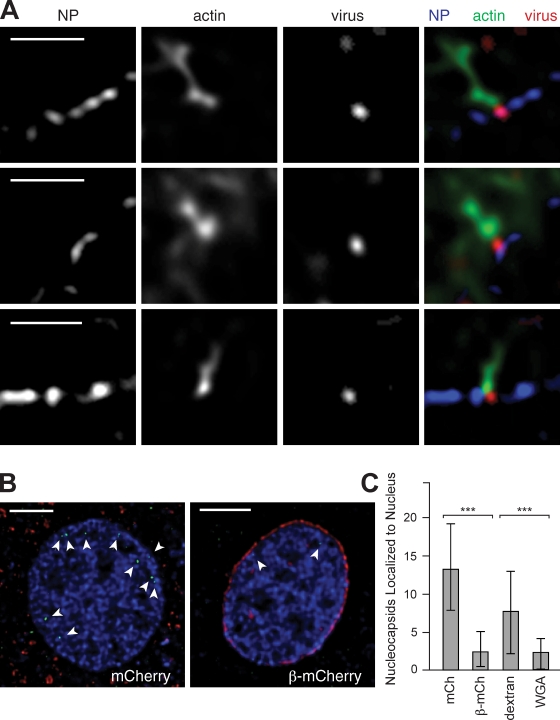
Although our results implicated actin-based motility in virus transit to the nuclear periphery, the mechanism of translocation across the nuclear envelope remained unclear. We sought to distinguish whether the virus crossed the envelope through nuclear pores, as was hypothesized based on electron micrographs of the virus docked on pores (Granados and Lawler, 1981), or whether it entered the nucleus by a different means. To evaluate the role of nuclear pores, we first determined whether nucleocapsids at the nuclear periphery colocalized with pores. Deconvolution microscopy revealed that 3mC nucleocapsids were either colocalized with (Fig. 3 A, top; and Fig. S3 A), adjacent to (Fig. 3 A, middle; and Fig. S3 B), or between pores (Fig. 3 A, bottom; and Fig. S3 C). The variable associations of nucleocapsids with pores may represent temporal stages in the translocation pathway, with an initial collision occurring between pores followed by pore engagement and translocation. This notion is consistent with the observed 3 ± 1 min delay between collision and entry into the nucleus as well as the tight fit for the 30–60-nm diameter nucleocapsid (Cunningham, 1968; Hughes, 1977) to squeeze through the 50-nm diameter pore (Elad et al., 2009).
Figure 3.
AcMNPV enters the nucleus through nuclear pores. (A) Deconvolved images showing virus (red; 3mC), actin (green; FITC-phalloidin), and nuclear pores (NP; blue; anti–nuclear pore immunofluorescence). (top) Viral colocalization with a pore. (middle) Virus adjacent to a pore. (bottom) Virus on the nuclear membrane in the space between pores. Bars, 2 µm. (B) Deconvolved sections of nuclei from cells expressing mCherry (left) or importin-β71–876–mCherry (right) infected with AcMNPV at 2 h after infection. Colors show virus (green; anti-VP39 immunofluorescence), nuclear DNA (blue; DAPI), and mCherry or importin-β71–876–mCherry (red). Arrowheads indicate capsids within the nucleus. Bars, 5 µm. (C) Numbers of capsids per nucleus (mean ± SD) counted from z stacks of nuclei in infected cells expressing either mCherry (mCh; n = 30) or importin-β71–876–mCherry (β-mCh; n = 30) or microinjected with dextran tetramethylrhodamine (n = 49) or WGA (n = 47). ***, P < 0.0001 (by unpaired t test).
To further determine whether nuclear pores function as the main route of entry, we examined the effect of expressing a dominant-negative human importin-β truncation derivative, importin-β71–876 (amino acid residues 71–876), that blocks transport through nuclear pores (Chi et al., 1997; Kutay et al., 1997). Cells were transfected with a plasmid that expresses importin-β71–876–mCherry or mCherry alone as a control and infected with AcMNPV. The number of capsids per nucleus was quantified at 2 h after infection (Fig. 3 B). Cells expressing importin-β71–876–mCherry showed a 78% decrease in the number of detectable capsids per nucleus compared with cells expressing mCherry (Fig. 3 C). To confirm this result, we microinjected cells with WGA, a well-established inhibitor of nuclear transport (Finlay et al., 1987), or with dextran as a control. Microinjection of WGA caused a 70% decrease in the number of capsids per nucleus compared with controls (Fig. 3 C). These results demonstrate that the main route of nuclear entry of nucleopolyhedroviruses is through nuclear pores.
Later actin-based motility results in prereplication accumulation of nucleocapsids at the cell surface
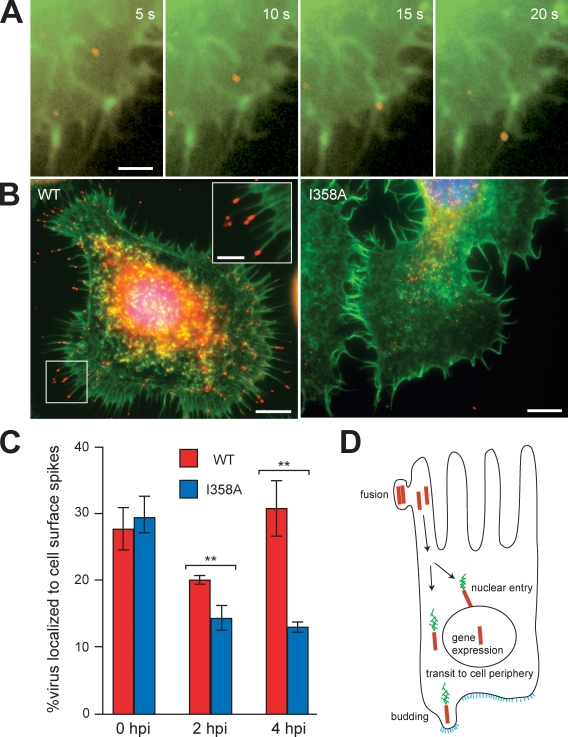
The use of actin-based motility by AcMNPV for transit to the nucleus is unprecedented, as other pathogens use cytoplasmic actin-based motility to propel themselves to the cell surface to facilitate cell to cell spread (Gouin et al., 2005). Therefore, we wondered whether actin-based motility might also play a role in viral transit to the cell surface. In High Five cells infected with 3mC at 0–2 hpi, we observed nucleocapsids moving to the cell periphery and entering into preexisting actin-rich cell surface spikes (Fig. 4 A and Video 10). However, these associations with the surface were transient, and in fixed cells, nucleocapsids were observed primarily in the cytoplasm associated with comet tails (Fig. 1 A). In contrast, after the onset of early viral gene expression (2–4 hpi), nucleocapsids accumulated at the tips of cell surface spikes (Fig. 4 B, left). These nucleocapsids were from the inoculum and were not newly synthesized progeny, as the timing was too early for virus production, and nucleocapsids accumulated at the cell surface in the presence of aphidicolin, which blocks DNA synthesis (Fig. S3 D). To determine whether nucleocapsid accumulation in surface spikes required actin-based motility, we examined the localization of the 3mC I358A virus in fixed cells at 4 hpi. Although WT virus was localized to both the cell body and the tips of surface spikes, 3mC I358A primarily localized in the cell body (Fig. 4 B, right) and exhibited a 58% reduction in the percentage localized to cell surface spikes compared with WT (Fig. 4 C). This dramatic impairment in cell surface localization of 3mC I358A indicates that accumulation in surface spikes requires rapid movement in straight paths in contrast to nuclear translocation, which occurred even with the mutant’s slower and more erratic movements.
Figure 4.
AcMNPV actin-based motility drives virus localization to cell surface spikes. (A) Time series in 5-s intervals showing actin (green; EGFP-actin) and a 3mC virus (red) entering a cell surface spike. Bar, 2 µm. (B) Virus (red; anti-capsid immunofluorescence), actin (green; FITC-phalloidin), and DNA (blue; DAPI) in High Five cells infected with AcMNPV (left) or I358A AcMNPV (right) at 2 hpi, showing virus accumulation in cell surface spikes. Bars, 5 µm. Inset shows a magnified view of the boxed region. Bar, 2 µm. (C) Quantification of the percentage of 3mC (red) or 3mC I358A (blue) nucleocapsids localized to cell surface spikes at 0, 2, and 4 hpi in High Five cells. Error bars indicate mean ± SD. **, P < 0.01 (by unpaired t test). (D) Model of the roles of actin-based motility in baculovirus transport during the early phase of infection of a midgut epithelial cell. Colors represent nucleocapsids (red), actin (green), and viral glycoprotein GP64 (blue).
Conclusions and models
Our results demonstrate that AcMNPV utilizes actin-based motility in two key transport processes for which other viruses require microtubules. In the initial phase of infection, motility enables the virus to efficiently probe the cytoplasm, promoting frequent collision with and translocation into the nucleus. After early gene expression, motility enables nucleocapsids to accumulate at the tips of actin-rich cell surface spikes. We propose a model in which these two motility processes are coordinated to enable rapid transmission to neighboring cells during infection in insects (Fig. 4 D). The multiple nucleopolyhedroviruses, including AcMNPV, are defined by the presence of multiple nucleocapsids packaged within a single envelope in the occlusion-derived virus form. During oral infection of caterpillars by occlusion-derived virus, midgut epithelial cells receive a dose of several nucleocapsids, which is similar to the high MOI conditions of our experiments. It was previously hypothesized (Granados and Lawler, 1981; Washburn et al., 1999) that some nucleocapsids transit to the nucleus and initiate early gene expression, whereas others move to the basal cell surface. Upon early gene expression by the nuclear population, the budding protein GP64 is expressed, and the surface-associated nucleocapsids bud, enabling them to pass through epithelial cells and infect secondary targets. Our discovery that actin-based motility enables successive transport to the nucleus and the cell surface, coordinated by patterns of early gene expression, supports this hypothesis and provides a mechanistic explanation for how these transport events are performed and coordinated. The selective pressure driving the evolution of this pass-through behavior is likely to be the need to overcome the caterpillar’s defense response of frequent apoptosis and sloughing of midgut epithelial cells by rapidly spreading to secondary targets to establish a productive systemic infection (Engelhard and Volkman, 1995; Washburn et al., 1998, 1999, 2003). Interestingly, baculoviruses exploit a common molecular mechanism of actin assembly to enable cytoplasmic actin-based motility (this study) and nuclear actin assembly (Goley et al., 2006), both involving the viral P78/83 protein and the host Arp2/3 complex, indicating that this pathway can be adapted to enable diverse behaviors during infection. Thus, AcMNPV is a fascinating pathogen that has appropriated the actin cytoskeleton for multiple functional roles and represents a model for understanding the diverse functions of the cytoskeleton in pathogenesis.
Materials and methods
Cells and viruses
Sf9 cells were maintained in Grace’s insect media (Gemini Bio-Products) with 10% FBS (and 1% Pluronic F-68 [Invitrogen] for shaker cultures) or ESF 921 media (Expression Systems) at 28°C. High Five cells were maintained in Hink’s insect media (SAFC Biosciences) with 10% FBS (and 1% Pluronic F-68 for shaker cultures) at 28°C. AcMNPV WOBpos (Goley et al., 2006) was the parental bacmid strain used for this study.
Plasmid DNA constructions
To generate the vp39 gene fusion to three copies of the gene encoding mCherry (3mC; Fig. S1 A), the mCherry gene was PCR amplified from pIZ-mCherry (Goley et al., 2006) and the vp39 gene from the AcMNPV E2 HinDIII C fragment. The native vp39 gene promoter was derived from 1.34 kb of the HinDIII C fragment covering the vp39 promoter and part of the vp39 ORF, which was ligated to vp39-mCherry. The native promoter vp39–triple mCherry was cloned into pWOBCAT and pWOBGent6 to generate 3mC transfer vectors for selection with either chloramphenicol or gentamycin (Fig. S1, A and B). These plasmids target insertion of gene constructs just upstream of the polyhedrin gene in the viral genome.
The ie-1 GFP transfer vector was constructed by removal of the actin gene from ie-1 GFP-actin/pWOBGent3 (Fig. S1 C). pWOBGent3 targets insertion of gene constructs upstream of both the kanamycin resistance cassette and the polyhedrin gene (Fig. S1 D).
For the importin-β71–876–mCherry construct, the importin-β71–876 gene (Chi et al., 1997; Kutay et al., 1997) was PCR amplified out of pET 71–876 (provided by K. Weis, University of California, Berkeley, Berkeley, CA) and ligated to mCherry. This importin-β71–876–mCherry fusion gene was subcloned into pACT (Ohkawa et al., 2002). For the negative control that expresses mCherry alone, mCherry was subcloned into pACT. All sequences were confirmed by DNA sequencing.
Recombinant bacmid generation and virus verification
To generate the 3mC and ie-1 GFP viruses, linearized 3mC/pWOBCAT or ie-1 GFP/pWOBGent3 DNA was coelectroporated with WOBpos viral DNA into BW25113pKD46 Escherichia coli as described previously (Goley et al., 2006). Selection was performed on plates containing kanamycin and chloramphenicol (3mC) or kanamycin and gentamycin (ie-1 GFP). To generate the 3mC I358A and ie-1 GFP I358A viruses, WOBpos containing the p78/83 I358A mutation (Goley et al., 2006) was used in place of WOBpos. Insertion of 3mC and ie-1 GFP into the correct location in the viral genome was verified by restriction digestion and/or PCR, and the presence of the I358A mutation was verified by DNA sequencing. Bacmid DNAs were transfected into Sf9 cells using Cellfectin (Invitrogen), and the resulting virus was amplified and titered (Volkman and Goldsmith, 1982). One-step growth curves comparing WOBpos (WT) with 3mC and ie-1 GFP viruses were performed in triplicate as described previously (Ohkawa and Volkman, 1999).
Imaging of fixed cells
High Five cells were seeded onto coverslips 1 d before infection with AcMNPV WOBpos at an MOI of ∼200. Cells were chilled for 15 min, and chilled virus was added and allowed to adsorb at 4°C for 1 h. After the inoculum was removed, the cells were washed, and fresh media at 28°C were added (this point was defined as time 0). In the experiment with aphidicolin (Sigma-Aldrich), drug was included at a concentration of 5 µg/ml. Coverslips were processed at 30 min or 2 hpi by either (a) fixing in 4% formaldehyde in PHEM (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9), permeabilizing in 0.15% Triton X-100/PHEM, and staining with 0.1 µg/ml DAPI and 100 nM TRITC-phalloidin (Invitrogen), or (b) preextracting in 0.15% Triton X-100/PHEM containing 0.3 µM TRITC-phalloidin and 0.1 µg/ml DAPI. Cells were blocked with 3.3% normal goat serum in PHEM and stained with mouse monoclonal anti-VP39 antibody P10C6 (1:250; Whitt and Manning, 1988) and 4 µg/ml FITC anti–mouse secondary antibody (Invitrogen). To visualize nuclear pores, cells were stained with 100 nM FITC-phalloidin, 2 µg/ml primary antibody Mab414 (Covance; Aris and Blobel, 1989), and 4 µg/ml Alexa Fluor 350 anti–mouse secondary antibody (Invitrogen).
For deconvolution microscopy, cells were seeded onto coverslips 1 d in advance of transfection (Cellfectin) with importin-β71–876/pACT or mCherry/pACT DNA, or in the case of nuclear pore imaging, were seeded 1 d in advance of infection with 3mC at an MOI of 60 for 30 min before processing. Transfected cells were infected 1 or 2 d after transfection with AcMNPV WOBpos at an MOI of 60, and after 2 h, they were fixed, stained, and mounted using Prolong Gold Antifade (Invitrogen).
Images were captured at 25°C using a microscope (IX71; Olympus) with a 60× 1.40 NA or 100× 1.35 NA Plan Apo oil objective, a camera (CoolSNAP HQ; Photometrics), and MetaMorph software (MDS Analytical Technologies). Image files were in 8-bit tagged image file format. Deconvolution microscopy was performed at 25°C on a DeltaVision 4 system (Applied Precision) with a 100× 1.35 NA Plan Apo oil objective and SoftWoRx software (version 3.3.6; Applied Precision). Deconvolution was performed using Huygens Professional software (version 3.1.0p0; Scientific Volume Imaging), and images were exported as 32-bit tagged image file format files. Images files were converted to 8-bit and processed with ImageJ (National Institutes of Health) and Photoshop (Adobe), with brightness/contrast levels adjusted without altering γ settings.
Time-lapse imaging
High Five cells were seeded onto coverslip dishes (MatTek), and 1 d later, were transfected using Cellfectin with a plasmid expressing EGFP–B mori A4 actin (GA/pACT; Ohkawa et al., 2002). 2 d after transfection, dishes were chilled for 15 min at 4°C, and freshly harvested, chilled 3mC or 3mC I358A virus was added. After 1 h of adsorption at 4°C, the inoculum was removed, cells were washed with Grace’s media, and fresh 28°C media with antibiotics (penicillin/streptomycin and fungizone) were added (this point was defined as time 0). For drug treatments, 0.5 ml media containing each drug (latA; Enzo Life Sciences, Inc.), CK-548 and CK-636 (Cytokinetics) or DMSO control (Sigma-Aldrich), was added to cells in 0.5 ml drug-free media to bring the final drug concentrations to 8 µM latA, 100 µM CK-548 or CK-636, or 1% DMSO. Images were captured at 5-s intervals using a microscope (IX71; Olympus) as described in the previous paragraph. Velocities of virus particles were calculated after measuring the distance traveled over at least six 5-s intervals. Velocities were measured seconds after latA addition or 30 min after addition of Arp2/3 inhibitors. Displacement was defined as the distance between the initial and final location in a 50-s interval. Total distance traveled was the total path length in the same 50-s interval. Straightness was defined as displacement/total distance traveled. Speed/distance measurements were made using the manual tracking plug-in of ImageJ.
To analyze the relative efficiency of viral nuclear entry, High Five cells were inoculated with ie-1 GFP or ie-1 GFP I358A virus at 4°C as described in the previous paragraph. 28°C Hink’s media with 10% FBS and antibiotics with or without 8 µM latA, 1 µM jasplakinolide (EMD), or 10 µM colchicine (Sigma-Aldrich) was added (defined as time 0). Time-lapse imaging was performed at 25°C using a microscope (IX71; Olympus) equipped with a 20× objective over 15 h with images taken at 15-min intervals as described for the imaging of fixed cells. Fields containing 65–122 cells were analyzed for the time of onset of detectable GFP expression in individual cells.
Microinjections
High Five cells were seeded 1 d in advance of injection. Cells were microinjected using Femtotips with a FemtoJet microinjector controlled with an InjectMan micromanipulator (Eppendorf) using a pressure of 80 hPa, a time of 0.4 s, and a compensation pressure of 30 hPa. WGA Alexa Fluor 488 (Invitrogen) or dextran tetramethylrhodamine (70,000 molecular mass; lysine fixable; Invitrogen) was diluted to 0.5 mg/ml in PBS, centrifuged for 15 min, and filtered through a 0.2-µm filter (Millipore). After injection, cells were inoculated with AcMNPV WOBpos at an MOI of 60 as described for time-lapse imaging. Cells were stained with DAPI and P10C6 and processed for deconvolution microscopy as described for imaging of fixed cells.
Statistical analyses
All statistical analyses were conducted using Prism (GraphPad Software, Inc.). Nucleocapsid velocities (Fig. 1 B) were analyzed by a one-way analysis of variance test. Displacement, total distance traveled, and straightness data (Fig. 1 E) were analyzed by unpaired t tests. Mean nucleocapsids/nucleus data from deconvolution stacks (Fig. 3 C) were analyzed using an unpaired t test. The percentage of nucleocapsids localized to cell surface spikes at each time point (Fig. 4 C) was analyzed by an unpaired t test.
Online supplemental material
Fig. S1 shows the fluorescent protein gene constructs used for generation of the recombinant viruses used in this study. Fig. S2 shows viral growth curves comparing the WT virus with 3mC and ie-1 GFP. Fig. S3 shows lower magnification versions of the images shown in Fig. 3 (deconvolved images of virus associations with nuclear pore complexes in infected High Five cells) and the accumulation of nucleocapsids in surface spikes in the presence of aphidicolin. Video 1 shows AcMNPV actin-based motility in High Five cells, Video 2 shows a comparison of the motility of AcMNPV and the I358A mutant, Video 3 shows the motility of the I358A mutant, Video 4 shows a comparison of the movement tracks of AcMNPV and the I358A mutant, and Videos 5, 6, and 7 show AcMNPV collision with the nuclear periphery. Video 8 shows viral entry into the nucleus, Video 9 shows I358A virus association with the nuclear periphery, and Video 10 shows AcMNPV moving into cell surface spikes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001162/DC1.
Acknowledgments
We thank S.G. Kamita for Sf9 and High Five cells, R. Tsien for the mCherry gene, J. Hartman and Cytokinetics for the Arp2/3 inhibitors, R. Kern and P. Kalab for advice on microinjection, K. Weis for the importin-β71–876 construct and helpful discussion, K. Campellone and K. Weis for comments on the manuscript, and the Welch laboratory for discussion.
This study was supported by the United States Department of Agriculture Cooperative State Research, Education, and Extension Service (grant 2004-35607-14906) and the National Institutes of Health/National Institute of General Medical Sciences (grant R01 GM059609 to M.D. Welch).
Footnotes
Abbreviations used in this paper:
- AcMNPV
- Autographa californica multiple nucleopolyhedrovirus
- hpi
- h post-infection
- latA
- latrunculin A
- WT
- wild type
References
- Aris J.P., Blobel G. 1989. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J. Cell Biol. 108:2059–2067 10.1083/jcb.108.6.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton C.A., Volkman L.E. 1993. Penetration of Autographa californica nuclear polyhedrosis virus nucleocapsids into IPLB Sf 21 cells induces actin cable formation. Virology. 197:245–254 10.1006/viro.1993.1585 [DOI] [PubMed] [Google Scholar]
- Chi N.C., Adam E.J.H., Adam S.A. 1997. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J. Biol. Chem. 272:6818–6822 10.1074/jbc.272.10.6818 [DOI] [PubMed] [Google Scholar]
- Cunningham J.C. 1968. Serological and morphological identification of some nuclear-polyhedrosis and granulosis viruses. J. Invertebr. Pathol. 11:132–141 10.1016/0022-2011(68)90063-3 [DOI] [Google Scholar]
- Elad N., Maimon T., Frenkiel-Krispin D., Lim R.Y.H., Medalia O. 2009. Structural analysis of the nuclear pore complex by integrated approaches. Curr. Opin. Struct. Biol. 19:226–232 10.1016/j.sbi.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Engelhard E.K., Volkman L.E. 1995. Developmental resistance in fourth instar Trichoplusia ni orally inoculated with Autographa californica M nuclear polyhedrosis virus. Virology. 209:384–389 10.1006/viro.1995.1270 [DOI] [PubMed] [Google Scholar]
- Fang M., Nie Y., Theilmann D.A. 2009. AcMNPV EXON0 (AC141) which is required for the efficient egress of budded virus nucleocapsids interacts with β-tubulin. Virology. 385:496–504 10.1016/j.virol.2008.12.023 [DOI] [PubMed] [Google Scholar]
- Finlay D.R., Newmeyer D.D., Price T.M., Forbes D.J. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189–200 10.1083/jcb.104.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E.D., Ohkawa T., Mancuso J., Woodruff J.B., D’Alessio J.A., Cande W.Z., Volkman L.E., Welch M.D. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science. 314:464–467 10.1126/science.1133348 [DOI] [PubMed] [Google Scholar]
- Gouin E., Gantelet H., Egile C., Lasa I., Ohayon H., Villiers V., Gounon P., Sansonetti P.J., Cossart P. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112:1697–1708 [DOI] [PubMed] [Google Scholar]
- Gouin E., Welch M.D., Cossart P. 2005. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8:35–45 10.1016/j.mib.2004.12.013 [DOI] [PubMed] [Google Scholar]
- Granados R.R., Lawler K.A. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology. 108:297–308 10.1016/0042-6822(81)90438-4 [DOI] [PubMed] [Google Scholar]
- Greber U.F., Way M. 2006. A superhighway to virus infection. Cell. 124:741–754 10.1016/j.cell.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Hollinshead M., Rodger G., Van Eijl H., Law M., Hollinshead R., Vaux D.J.T., Smith G.L. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389–402 10.1083/jcb.200104124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K.M. 1977. Observations on the morphology of nuclear polyhedrosis viruses from 6 forest insects. Can. Entomol. 109:759–762 10.4039/Ent109759-5 [DOI] [Google Scholar]
- Kutay U., Izaurralde E., Bischoff F.R., Mattaj I.W., Görlich D. 1997. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153–1163 10.1093/emboj/16.6.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., Pollard T.D. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa T., Volkman L.E. 1999. Nuclear F-actin is required for AcMNPV nucleocapsid morphogenesis. Virology. 264:1–4 10.1006/viro.1999.0008 [DOI] [PubMed] [Google Scholar]
- Ohkawa T., Rowe A.R., Volkman L.E. 2002. Identification of six Autographa californica multicapsid nucleopolyhedrovirus early genes that mediate nuclear localization of G-actin. J. Virol. 76:12281–12289 10.1128/JVI.76.23.12281-12289.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietdorf J., Ploubidou A., Reckmann I., Holmström A., Frischknecht F., Zettl M., Zimmermann T., Way M. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992–1000 10.1038/ncb1101-992 [DOI] [PubMed] [Google Scholar]
- Rohrmann G.F. 2008. Baculovirus Molecular Biology. National Library of Medicine (US), National Center for Biotechnology Information; Bethesda, MD: 154 pp. [Google Scholar]
- Volkman L.E., Goldsmith P.A. 1982. Generalized immunoassay for Autographa californica nuclear polyhedrosis virus infectivity in vitro. Appl. Environ. Microbiol. 44:227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman L.E., Zaal K.J.M. 1990. Autographa californica M nuclear polyhedrosis virus: microtubules and replication. Virology. 175:292–302 10.1016/0042-6822(90)90211-9 [DOI] [PubMed] [Google Scholar]
- Ward B.M., Moss B. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651–11663 10.1128/JVI.75.23.11651-11663.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn J.O., Kirkpatrick B.A., Haas-Stapleton E., Volkman L.E. 1998. Evidence that the stilbene-derived optical brightener M2R enhances Autographa californica M nucleopolyhedrovirus infection of Trichoplusia ni and Heliothis virescens by preventing sloughing of infected midgut epithelial cells. Biol. Control. 11:58–69 10.1006/bcon.1997.0572 [DOI] [Google Scholar]
- Washburn J.O., Lyons E.H., Haas-Stapleton E.J., Volkman L.E. 1999. Multiple nucleocapsid packaging of Autographa californica nucleopolyhedrovirus accelerates the onset of systemic infection in Trichoplusia ni. J. Virol. 73:411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn J.O., Chan E.Y., Volkman L.E., Aumiller J.J., Jarvis D.L. 2003. Early synthesis of budded virus envelope fusion protein GP64 enhances Autographa californica multicapsid nucleopolyhedrovirus virulence in orally infected Heliothis virescens. J. Virol. 77:280–290 10.1128/JVI.77.1.280-290.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt M.A., Manning J.S. 1988. A phosphorylated 34-kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology. 163:33–42 10.1016/0042-6822(88)90231-0 [DOI] [PubMed] [Google Scholar]