PIPKI-α does a job other PIPKI isoforms cannot; it recruits Rac1 to the plasma membrane upon integrin activation, spatially regulating the actin-organizing GTPase during migration.
Abstract
Phosphatidylinositol-4,5-bisphosphate (PI4,5P2) is a critical regulator of cell migration, but the roles of the type I phosphatidylinositol-4-phosphate 5-kinases (PIPKIs), which synthesize PI4,5P2, have yet to be fully defined in this process. In this study, we report that one kinase, PIPKI-α, is a novel upstream regulator of Rac1 that links activated integrins to the regulation of cell migration. We show that PIPKI-α controls integrin-induced translocation of Rac1 to the plasma membrane and thereby regulates Rac1 activation. Strikingly, this function is not shared with other PIPKI isoforms, is independent of catalytic activity, and requires physical interaction of PIPKI-α with the Rac1 polybasic domain. Consistent with its role in Rac1 activation, depletion of PIPKI-α causes pronounced defects in membrane ruffling, actin organization, and focal adhesion formation, and ultimately affects the directional persistence of migration. Thus, our study defines the role of PIPKI-α in cell migration and describes a new mechanism for the spatial regulation of Rac1 activity that is critical for cell migration.
Introduction
Directed cell migration is fundamental to many biological processes, including embryogenesis, wound healing, the immune response, and cancer metastasis (Ridley et al., 2003). This process is initiated in response to extracellular or internal cues and is driven by the localized polymerization of F-actin, leading to cell polarization, the extension of a leading edge lamellipodium, and migration in the direction of the leading edge (Small et al., 2002; Pollard and Borisy, 2003).
F-actin assembly is induced in response to the localized activation of Rac1 at the leading edge (Nobes and Hall, 1999; Kraynov et al., 2000; Wheeler et al., 2006). In turn, Rac1 initiates and maintains polarized protrusive activity by stimulating Arp2/3-dependent de novo actin nucleation and by generating free barbed actin filament ends (Takenawa and Miki, 2001; Small et al., 2002; Pollard and Borisy, 2003). Rac1 also promotes the formation of nascent focal adhesion complexes, which stabilize membrane protrusions and generate the traction force necessary for migration (Nobes and Hall, 1999; Pankov et al., 2005; Guo et al., 2006; Vidali et al., 2006). Therefore, the localization of Rac1 activity to specific membrane domains is critical for directional migration, but the underlying mechanisms are poorly understood.
Rac1, like other small GTPases, undergoes cycles of activation and deactivation that are catalyzed by GTP exchange factors (GEFs) and GTPase-activating proteins in response to growth factor and integrin receptor signaling pathways (Etienne-Manneville and Hall, 2002; Rossman et al., 2005). Rac1 signaling is also controlled through the regulated translocation of Rac1 from a cytosolic pool to the plasma membrane (del Pozo et al., 2002). This event is induced in response to integrin-mediated adhesion of cells and is essential for Rac1 coupling to downstream effectors (del Pozo et al., 2000, 2002). Because activated integrins are concentrated at the cell front (Moissoglu and Schwartz, 2006), the integrin-induced subcellular targeting of Rac1 is likely to play a key role in cellular processes that depend on the polarized activation of Rac1, such as directed cell migration. However, the mechanisms that control the targeting of Rac1 to the plasma membrane and the significance of this process for cell migration are unclear.
Recently, Rac1 was reported to form a complex with two members of the type I phosphatidylinositol-4-phosphate 5-kinase (PIPKI) family, designated PIPKI-α and PIPKI-β (Tolias et al., 1998, 2000; van Hennik et al., 2003). PIPKIs synthesize the signaling molecule phosphatidylinositol-4,5-bisphosphate (PI4,5P2), which is a central regulator of actin and adhesion dynamics during cell migration (Yin and Janmey, 2003; Ling et al., 2006). Interestingly, Rac1 binds the two kinases independent of its GTP-loading status through its C-terminal polybasic domain (Tolias et al., 1998, 2000). This hypervariable region is distal to the effector-binding domains of Rac1 and is positioned just upstream of the CAAX box that mediates the attachment of a lipid anchor. PIPKI–Rac1 complex formation via this domain is necessary for stimulation of PI4,5P2 synthesis and actin assembly (Tolias et al., 1998, 2000). Thus, PIPKI-α and PIPKI-β are widely thought to be effectors of Rac1. Consistent with such an idea, PI4,5P2 synthesis and actin filament uncapping, which are induced in response to expression of constitutively active Rac1V12, could be blocked by simultaneous expression of a dominant-negative PIPKI-β mutant (Tolias et al., 1998, 2000). Surprisingly, expression of a corresponding kinase-dead mutant of PIPKI-α had no inhibitory effect on Rac1V12 signaling to the actin cytoskeleton (Tolias et al., 1998, 2000; van Hennik et al., 2003). This finding raised the possibility that the roles of PIPKI-α and PIPKI-β in the Rac1 pathway are not equivalent and that the physiological significance of the PIPKI-α–Rac1 interaction remains yet to be established.
Intriguingly, recent evidence suggests that the PIPKI-α interaction domain in Rac1 controls the subcellular targeting and activation of Rac1 in response to integrin-mediated adhesion (del Pozo et al., 2002; van Hennik et al., 2003). Accordingly, Rac1 mutants in the polybasic domain fail to translocate to the plasma membrane and also fail to activate downstream signaling responses (del Pozo et al., 2002). Given that the same mutations in the Rac1 polybasic domain that prevent Rac1 membrane targeting also abrogate the Rac1–PIPKI-α interaction (Tolias et al., 2000), the question arises as to whether the PIPKI-α–Rac1 interaction provides a means to regulate the localization of Rac1 activity.
In this study, we present evidence in support of such a hypothesis. We show that PIPKI-α plays a pivotal, isoform-specific role in regulating the translocation of Rac1 to the plasma membrane. Furthermore, we show that PIPKI-α thereby regulates Rac1 activation and downstream signaling processes necessary for F-actin assembly, focal adhesion formation, and directed cell migration.
Results
PIPKI-α is necessary for adhesion-induced Rac1 membrane translocation
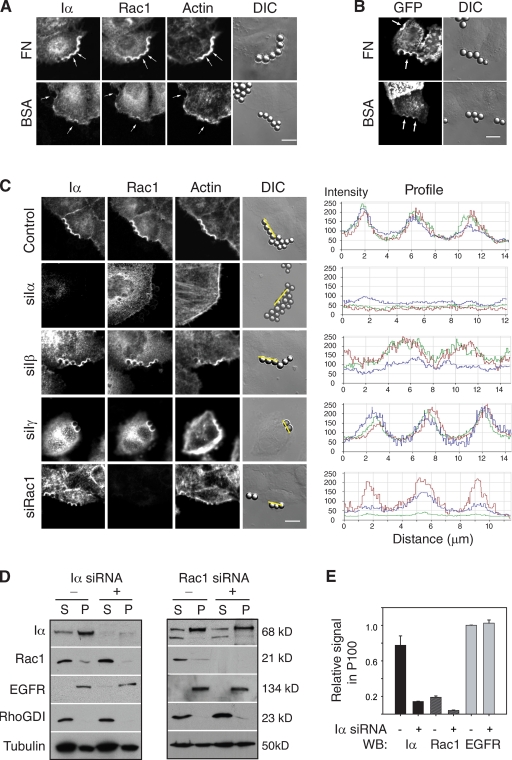
To examine whether PIPKI-α modulates the localization of Rac1 to the plasma membrane in response to integrin activation, we incubated HeLa cells with 5 µm polystyrene beads coated with fibronectin (FN), which induce the local activation and clustering of integrins at the cell surface (del Pozo et al., 2002). As previously reported (del Pozo et al., 2002), FN-coated but not BSA-coated control beads resulted in the accumulation of endogenous Rac1 and F-actin at bead–cell contact sites (Fig. 1 A). FN-coated beads also led to the clustering of endogenous PIPKI-α and the up-regulation of PI4,5P2 synthesis at bead–cell contact sites, as revealed by the localization of a GFP–PLC-δ–pleckstrin homology (PH) reporter protein (Fig. 1 B). Significantly, Rac1 clustering and F-actin assembly at bead–cell contact sites were markedly decreased by RNAi-mediated knockdown of PIPKI-α (Fig. 1 C), suggesting that integrin-induced Rac1 translocation to the membrane needs PIPKI-α. This was revealed by measuring the ratio of mean GFP pixel density at bead–cell contact sites relative to beadless areas on the membrane. This analysis showed that 87 ± 5% of beads on control siRNA–treated cells induced increased Rac1 membrane signal (n = 45), whereas only 11 ± 3% of beads (n = 55) induced Rac1 signal in PIPKI-α–depleted cells, and even around the positive beads, the signal intensity was significantly decreased (Fig. 1 C). The loss of Rac1 membrane recruitment in PIPKI-α–depleted cells was confirmed by biochemical fractionation experiments, which revealed a decrease in Rac1 protein levels in the particulate, membrane-containing fraction (Fig. 1, D and E). Conversely, depletion of Rac1 did not markedly affect the membrane association of PIPKI-α, as judged by biochemical (Fig. 1, D and E) and immunofluorescence experiments (Fig. 1 C).
Figure 1.
PIPKI-α is required for adhesion-induced Rac1 plasma membrane targeting. (A) HeLa cells were incubated for 20 min with 5 µm FN- or BSA-coated beads shown in the phase-contrast image (differential interference contrast [DIC]) and marked by arrows. Cells were fixed and stained with Alexa Fluor 647 phalloidin to visualize F-actin or with appropriate primary and secondary antibodies to detect endogenous PIPKI-α and Rac1. (B) HeLa cells, transiently transfected with GFP–PLC-δ–PH as a marker for PI4,5P2, were incubated with FN- or BSA-coated beads as described above, and GFP fluorescence and DIC images were recorded. (C) Cells transfected with control siRNAs or siRNAs targeting PIPKI-α, -β, -γ, or Rac1 were incubated with FN-coated beads, fixed, and stained as described in A. (right) Fluorescence intensity profiles showing the extent of marker colocalization were determined along the lines. Colors indicate PIPKI-α (red), Rac1 (green), and F-actin (blue) fluorescence. Images in A–C are representative examples of three separate experiments. (D) Subcellular fractionation of HeLa cell lysates into particulate (P) and soluble (S) fractions. HeLa cells transfected with the indicated siRNA pools were plated on FN-coated substrate and serum starved to eliminate contribution from growth factor signals. Cell lysates were generated, subfractionated into 100,000 g pellet (P100; membrane) and supernatant (S100; cytosol) fractions, and analyzed by Western blotting with antibodies against PIPKI-α and Rac1. EGFR and RhoGDI were detected as markers for membrane and cytosol fractions, respectively. A representative experiment is shown. Note that the amount of protein analyzed per lane represents 5% of the total soluble fraction but 20–50% of the particulate fraction. (E) Quantification of Rac1 membrane localization by quantitative Western blot (WB) analysis from D is shown. Data are normalized according to the total levels of each marker and the calculated percentage in the membrane fraction. Values are means from two independent experiments. Error bars indicate SEM. Bars, 10 µm.
Notably, the role of PIPKI-α in Rac1 membrane targeting was isoform specific. Neither PIPKI-β nor PIPKI-γ expression was necessary for Rac1 accumulation at bead–cell contact sites, as 80 ± 6% of beads in PIPKI-β–depleted cells (n = 38) and 82 ± 7% of beads in PIPKI-γ–depleted cells (n = 34) had increased Rac1 signal when compared with beadless areas (Fig. 1 C). Collectively, these data suggest that integrin-dependent membrane translocation of Rac1 is dependent on PIPKI-α. The finding that endogenous Rac1 localized with PIPKI-α at actin-rich membrane ruffles in control cells but became delocalized from the plasma membrane upon depletion of PIPKI-α (Fig. S2) further supports the idea that Rac1 membrane localization requires PIPKI-α.
Integrin-dependent Rac1 membrane targeting involves the polybasic domain of Rac1
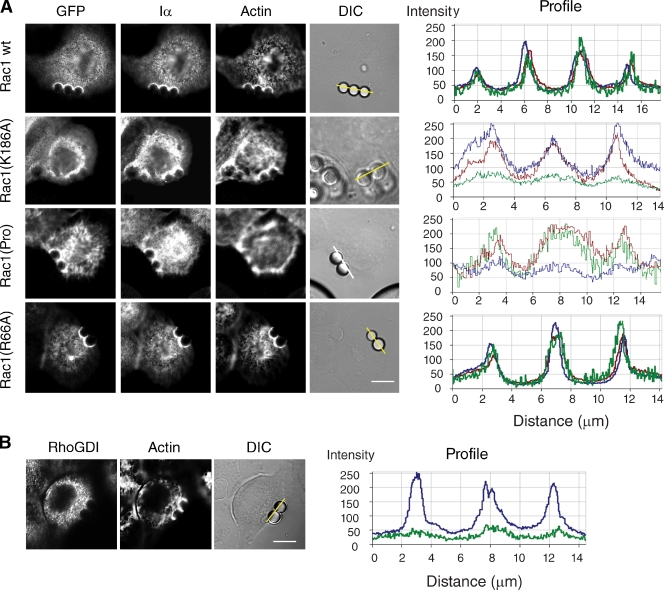
It was previously reported that the polybasic domain of Rac1 mediates the integrin-dependent translocation of Rac1 to the membrane (del Pozo et al., 2002; van Hennik et al., 2003). Consistent with this, a Rac1 variant containing a single amino acid mutation (K186A) within the polybasic domain failed to efficiently accumulate at FN–bead–cell contact sites in HeLa cells (Fig. 2 A). However, other regions in the hypervariable C-terminal domain of Rac1 have also been reported to modulate Rac1 targeting and activation. In particular, a proline-rich motif adjacent to the polybasic domain is required for Rac1 association with the Rac1 GEF β-PIX (van Hennik et al., 2003; ten Klooster et al., 2006), and this interaction has been implicated in targeting Rac1 to focal adhesions and in mediating Rac1 activation (ten Klooster et al., 2006). Therefore, we examined the localization of a Rac1 mutant, designated Rac1(Pro), which is defective for β-PIX binding (ten Klooster et al., 2006). As shown in Fig. 2 A, this mutant localized as efficiently as wild-type Rac1 to FN–bead–cell contact sites but appeared to attenuate actin assembly around beads. These results suggest that Rac1 membrane targeting can occur independently of β-PIX, but subsequent activation of Rac1 at the membrane may require interaction with β-PIX.
Figure 2.
Integrin-mediated translocation of Rac1 to the membrane involves the polybasic domain of Rac1. (A) HeLa cells expressing the indicated GFP-tagged Rac1 wild-type (wt) or mutant variants were incubated with FN-coated beads for 20 min (shown in the phase-contrast image; DIC). Cells were fixed and stained with appropriate antibodies to visualize endogenous PIPKI-α (Iα) and with Alexa Fluor 647–conjugated phalloidin to visualize actin. GFP fluorescence was recorded directly. Fluorescence intensity profiles showing the extent of GFP (green), PIPKI-α (red), and F-actin (blue) colocalization were determined along the lines drawn and are shown to the right. (B) HeLa cells were incubated with FN-coated beads for 20 min. Cells were fixed and immunostained for endogenous RhoGDI. Actin was visualized with Alexa Fluor 647–conjugated phalloidin. (right) Line scans of fluorescence intensity of RhoGDI (green) and F-actin (blue) signals are shown. Images are representative examples of three separate experiments. Bars, 10 µm.
Rac1 membrane association is also regulated by Rho guanine nucleotide dissociation inhibitor (RhoGDI), which extracts Rac1 from the plasma membrane (del Pozo et al., 2002; Dransart et al., 2005; Moissoglu and Schwartz, 2006). However, a Rac1 mutant (Rac1Q66A) that is unable to bind RhoGDI accumulated in a manner similar to wild-type Rac1 around FN–bead–cell contact sites (Fig. 2 A). Thus, RhoGDI does not contribute to the effects of PIPKI-α on Rac1 membrane targeting. This conclusion was further supported by biochemical and immunofluorescence experiments that showed that endogenous RhoGDI is predominantly cytosolic (Fig. 1, D and E) and not detected at bead–cell contact sites (Fig. 2 B).
PIPKI-α controls Rac1 membrane targeting through a kinase-independent mechanism
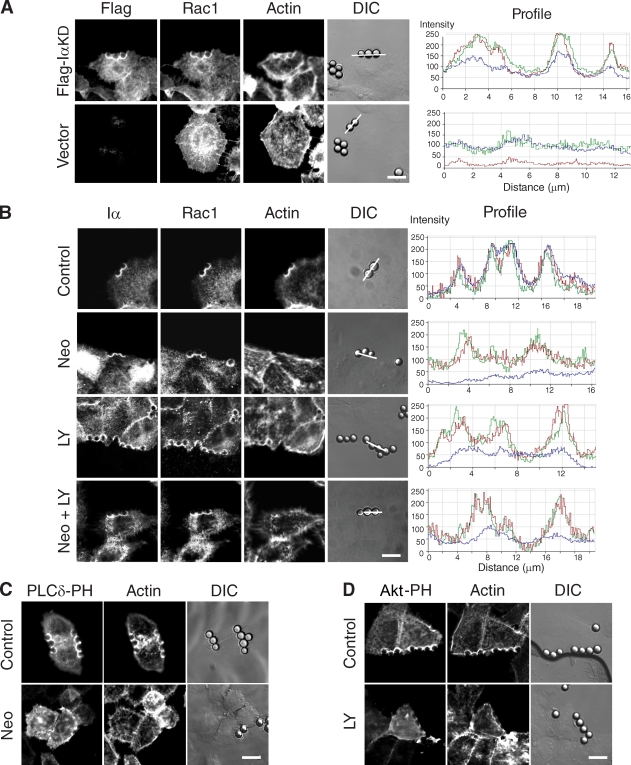
We next examined whether the polybasic domain of Rac1 mediates membrane targeting through direct physical interaction with PIPKI-α or through binding to PI4,5P2 or PI3,4,5P3 as shown for other small GTPases (Ueyama et al., 2005; Heo et al., 2006). To distinguish between these two possibilities, we tested whether integrin-induced Rac1 membrane recruitment could be restored to PIPKI-α–depleted cells by reexpression of a PIPKI-α mutant (IαKD) lacking catalytic activity. Consistent with a kinase-independent role of PIPKI-α in Rac1 plasma membrane targeting, we found that expression of IαKD restored the accumulation of endogenous Rac1 signal at FN–bead–cell contact sites in PIPKI-α–depleted cells (Fig. 3 A). IαKD expression also partially restored F-actin polymerization at these sites (Fig. 3 A), suggesting that PIPKI-α stimulates adhesion-dependent actin polymerization in part through its effect on Rac1. No Rac1 or F-actin signal was observed at FN–beads in cells transfected with empty vector (Fig. 3 A). Thus, PIPKI-α appears to control adhesion-induced Rac1 membrane targeting independently of its lipid kinase activity.
Figure 3.
Adhesion-induced Rac1 plasma membrane translocation requires PIPKI-α protein but not polyphosphoinositide synthesis. (A) PIPKI-α–depleted HeLa cells transfected with Flag-IαKD or empty vector (top) were incubated with FN-coated beads for 20 min, fixed, and stained with appropriate primary and secondary antibodies to visualize the Flag epitope and endogenous Rac1, respectively. F-actin was stained with Alexa Fluor 647 phalloidin. Fluorescence and DIC images are shown. Fluorescence intensity profiles showing the extent of Flag-IαKD (red), Rac1 (green), and F-actin (blue) colocalization were determined along the line drawn and are shown to the right. (B) HeLa cells were pretreated with 10 mM neomycin (Neo), 50 µM LY294002 (LY), or the combination of neomycin and LY294002 (Neo + LY). Cells were incubated with FN-coated beads for 20 min, fixed, and stained with Alexa Fluor 647 phalloidin or the appropriate primary and secondary antibodies to detect endogenous PIPKI-α and Rac1, respectively. (right) Fluorescence intensity profiles showing the extent of Flag–PIPKI-α (red), Rac1 (green), and F-actin (blue) colocalization were determined along the line drawn. (C and D) HeLa cells transfected with EGFP–PLC-δ–PH (C) or EGFP-AKT-PH (D) were treated with vehicle (DMSO; top) or 10 mM neomycin (C, bottom) for 60 min or with 50 µM LY294002 for 20 min (D, bottom). Cells were incubated with FN-coated beads as described above and stained with Alexa Fluor 594 phalloidin after fixation. Fluorescence and DIC images were recorded directly. Bars, 10 µm.
Consistent with this, the pharmacological inhibition of PI4,5P2 or PI3,4,5P3 production by treatment of cells with the PI4,5P2-sequestering agent neomycin or the phosphatidylinositol 3-kinase inhibitor LY294002, either alone or in combination, had no effect on Rac1 clustering around FN-coated beads (Fig. 3 B). These treatments were effective because they led to a significant reduction in F-actin assembly (Fig. 3 B) and caused the delocalization from the plasma membrane of the PH domain fusions GFP–PLC-δ–PH and GFP-Akt-PH (Fig. 3, C and D), which are biosensors for PI4,5P2 and PI3,4,5P3, respectively (Balla Várnai, 2002). Thus, synthesis of PI4,5P2 and PI3,4,5P3 is necessary for adhesion-dependent F-actin polymerization but dispensable for Rac1 plasma membrane targeting.
PIPKI-α modulates Rac1 subcellular targeting through interaction with the Rac1 polybasic domain
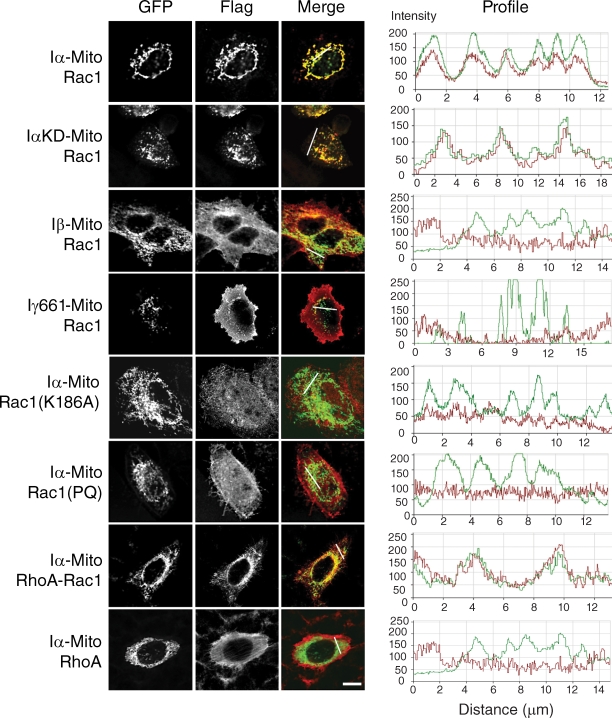
To test whether PIPKI-α directly controls the subcellular localization of Rac1, we targeted PIPKI-α to the surface of mitochondria away from the site of normal PIPKI-α and Rac1 localization by fusing PIPKI-α to the transmembrane domain of the outer mitochondrial membrane protein OMP25 (Nemoto and De Camilli, 1999). When Iα-Mito was coexpressed with Flag-tagged Rac1 in HeLa cells, Rac1 protein was sequestered to mitochondria (Fig. 4 A). Rac1 was also recruited to mitochondria in cells that expressed a PIPKI-α kinase-dead mutant (IαKD-Mito; Fig. 4 A), indicating that Rac1 sequestration to mitochondria required physical interaction with PIPKI-α but not its catalytic activity. Indeed, there appeared to be a direct correlation between the ability to bind Rac1 with high affinity in vitro and the ability to modulate Rac1 subcellular targeting in the mitochondrial sequestration assay. Accordingly, PIPKI-β, which bound Rac1 with lower apparent affinity relative to PIPKI-α (Fig. S3 B), was much less efficient in sequestering Rac1 to mitochondria (Fig. 4 A), and PIPKI-γ, which failed to bind Rac1 in vitro (Fig. S3 B), failed to recruit Rac1 to mitochondria (Fig. 4 A).
Figure 4.
PIPKI-α modulates Rac1 subcellular localization through the Rac1 polybasic domain. HeLa cells were cotransfected with mitochondrially targeted EGFP-tagged PIPK variants (Iα-Mito, Iβ-Mito, or Iγ661-Mito) and plasmids encoding Flag-tagged wild-type or mutant Rac1 variants or RhoA. Cells were fixed and stained with mouse anti-Flag antibody followed by Cy3-conjugated goat anti–mouse antibodies. GFP signal was recorded directly. (right) Fluorescence intensity profiles showing the extent of Flag (red) and EGFP (green) colocalization were determined along the line drawn. Representative examples of cells are shown (n = 3). Bar, 10 µm.
Importantly, the Rac1 polybasic domain was not only necessary but also sufficient to induce the mitochondrial sequestration of Rac1. Thus, the Rac1(PQ) or Rac1(K186A) mutations in the Rac1 polybasic domain that abrogated binding to PIPKI-α in vitro (Fig. S3 C) also prevented the recruitment of Rac1 to mitochondria (Fig. 4). Conversely, a RhoA-Rac1 chimera, which contained the polybasic region of Rac1, efficiently bound PIPKI-α in vitro (Fig. S3 D) and colocalized with Iα-Mito to mitochondria, whereas wild-type RhoA failed to do so (Fig. 4). Collectively, these results suggest that the mitochondrial sequestration of Rac1 is dependent on its direct interaction with PIPKI-α, although we cannot exclude the possibility that an unknown docking factor mediates the interaction between PIPKI-α and Rac1.
Notably, expression of Iα-Mito also caused the sequestration of endogenous Rac1 to mitochondria and the concomitant inhibition of chemotaxis compared with cells transfected with empty vector (Fig. S4, A and B). Because the chemotaxis defect induced by overexpression of Iα-Mito could be suppressed by simultaneous overexpression of Rac1, these results suggest that the PIPKI-α–directed mislocalization of Rac1 attenuated Rac1 signaling (Fig. S4 C). These data show that PIPKI-α can modulate Rac1 function by regulating its subcellular localization.
PIPKI-α regulates Rac1 activation by modulating Rac1 membrane targeting
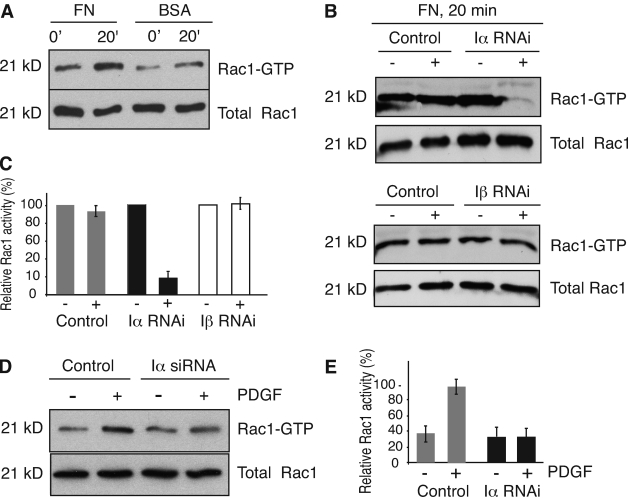
Integrins independently regulate Rac1 plasma membrane targeting and GTP loading (del Pozo et al., 2002). To determine whether PIPKI-α depletion only affects adhesion-induced Rac1 membrane translocation or also its activation, we measured Rac1-GTP loading by pull-down assay with the p21-binding domain of PAK1 kinase (Benard and Bokoch, 2002). As expected (del Pozo et al., 2000), attachment of HeLa cells to FN substrate stimulated a significant increase in Rac1-GTP levels in control siRNA–treated cells within a 10- to 20-min period relative to cells plated on BSA (Fig. 5, A and B). In contrast, little stimulation of Rac1-GTP loading was observed in PIPKI-α knockdown cells plated on FN (Fig. 5, B and C). Importantly, PIPKI-β–depleted cells exhibited an increase in Rac1-GTP levels in response to FN, which is similar to control cells (Fig. 5, B and C). Thus, PIPKI-α plays a unique role in the integrin-stimulated activation of Rac1.
Figure 5.
PIPKI-α is necessary for Rac1 activation. (A) Rac1 activation in response to attachment and spreading of HeLa cells on FN substrate or BSA. Cells were plated on FN or BSA for the indicated times, lysed, and Rac1-GTP present in the cell lysates was isolated by pull-down assay using PBD-PAK1-GST. Total and GTP-bound Rac1 were detected by Western blotting. (B) Rac1 activity in HeLa cells transfected with control siRNAs or siRNAs targeting either PIPKI-α or PIPKI-β are shown. Mock-treated (−) or siRNA-treated (+) cells were allowed to attach to FN for 20 min. Rac1-GTP was isolated from cell lysates and detected along with total Rac1 protein as described above. (C) Quantification of Rac1-GTP levels from B is shown. (D) Rac1 activation in response to PDGF treatment. Quiescent control or PIPKI-α–depleted HT1080 cells grown on plastic dishes were stimulated with 10 ng/ml PDGF (+) or left untreated (−). Rac1-GTP was isolated from cell lysates and detected along with total Rac1 protein by Western blotting as described in A. (E) A quantification of the ratios of GTP-bound to total Rac1 protein from D is shown. Values are normalized to control, and error bars indicate SEM (n = 3).
To determine whether PIPKI-α also regulates Rac1 activation in response to growth factor activation of cells, we measured Rac1-GTP levels in response to PDGF. We switched to HT1080 cells for these experiments because these cells respond to PDGF and the role of PIPKI-α in adhesion-induced Rac1 membrane targeting, and activation is conserved (unpublished data). We found that PDGF treatment of quiescent control but not of PIPKI-α–depleted cells stimulated Rac1-GTP loading (Fig. 5, D and E). Therefore, PIPKI-α appears to be necessary for both growth factor– and adhesion-induced Rac1 activation.
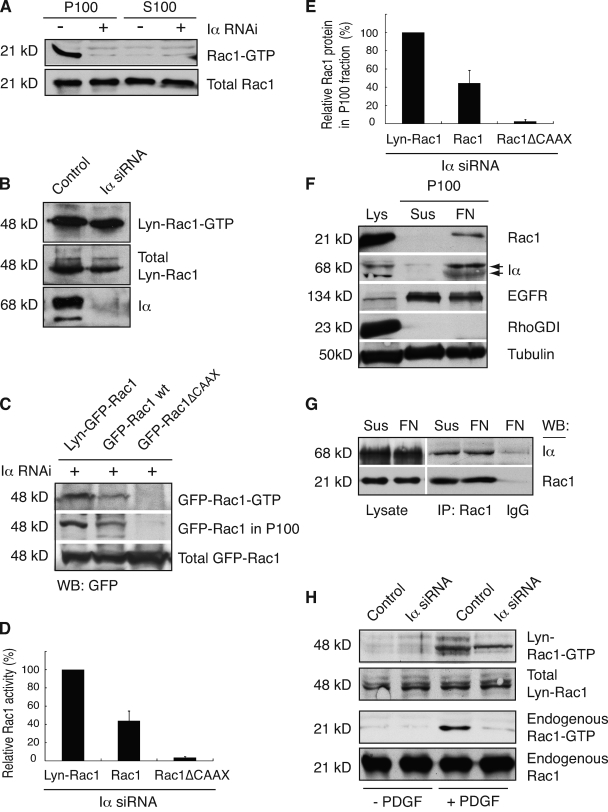
Finally, we aimed at elucidating how PIPKI-α affects Rac1 activity. PI4,5P2 is the precursor of PI3,4,5P3, which activates several of the known Rac1 GEFs, including Dock180, Vav, and Tiam (Han et al., 1998; Baumeister et al., 2003; Côté et al., 2005). Thus, one way by which PIPKI-α depletion may block Rac1 activation is by decreasing GEF activity. Alternatively, the PIPKI-α–dependent targeting of Rac1 to the plasma membrane may be a prerequisite for its interaction with a membrane-localized GEF (Moissoglu et al., 2006). Consistent with the latter concept, we found that only the membrane, but not the cytosol fraction from adherent HT1080 (Fig. 6 A) or HeLa cells (unpublished data), contained Rac1-GTP. To further assess whether PIPKI-α regulates Rac1-GTP loading through its effects on Rac1 membrane targeting, we examined whether we could bypass the requirement for PIPKI-α and restore Rac1 activation by artificially increasing Rac1 plasma membrane levels. To do so, we engineered a variant of Rac1 that contained a second plasma membrane–anchoring sequence from Lyn kinase (Kovárová et al., 2001) at its N terminus. When Lyn-Rac1 was expressed in PIPKI-α–depleted cells that were grown on FN and serum starved to exclude signals by growth factors, GTP loading occurred even in the absence of PIPKI-α (Fig. 6 B). Subcellular fractionation of cell lysates showed that Lyn-Rac1 was present in the membrane fraction in PIPKI-α–depleted cells (Fig. 6 C). This likely reflects the PIPKI-α–independent targeting of Lyn-EGFP-Rac1 to the plasma membrane, as indicated by immunofluorescence analysis of Lyn-EGFP-Rac1–expressing PIPKI-α–deficient cells (Fig. S5). Indeed, there appeared to be a close relationship between the extent of plasma membrane association and the extent of GTP loading. For example, Rac1 membrane association and GTP loading could also be restored in part by high level overexpression of wild-type Rac1 in PIPKI-α knockdown cells (Fig. 6, C–E), presumably because Rac1 overexpression drives the localization of Rac1 to the membrane by mass action (Fig. S5) because of the presence of the C-terminal CAAX sequence. Indeed, a Rac1 variant containing a mutation in the C-terminal CAAX box that abrogates lipid modification failed to undergo GTP loading, and this mutant was also absent from the membrane fraction and localized mainly to the nucleus (Fig. 6, C–E; and Fig. S5). Collectively, these findings are consistent with the idea that PIPKI-α effects on GTP loading are secondary to its effects on Rac1 membrane localization.
Figure 6.
PIPKI-α modulates Rac1 activation by regulating Rac1 membrane translocation. (A) Rac1-GTP is contained in the membrane fraction. HT1080 cells were plated on FN and serum starved to eliminate contribution from growth factor signals. Cell lysates were generated and fractionated into particulate (P) and soluble (S) fractions, and Rac1-GTP levels were determined. A representative example (n = 3) of Rac1-GTP and total Rac1 levels as detected by Western blot analysis using anti-Rac1 antibody is shown. (B) Enhanced membrane targeting restores integrin-dependent Rac1-GTP loading to PIPKI-α–deficient cells. Quiescent control or PIPKI-α–depleted HT1080 cells expressing Lyn-EGFP-Rac1 (Lyn-Rac1) were plated on FN, and GTP-loaded Lyn-EGFP-Rac1 was isolated from cell lysates by pull-down assay. Lyn-Rac1-GTP, total Lyn-Rac1, and endogenous PIPKI-α levels were detected by Western blot analysis using antibodies against GFP and Rac1. A representative example (n = 3) is shown. (C) The extent of integrin-induced Rac1-GTP loading correlates with Rac1 membrane association. PIPKI-α–depleted HT1080 cells transfected with plasmids expressing EGFP-tagged Lyn-Rac1, wild-type (wt) Rac1, or Rac1ΔCAAX were plated on FN substrate, serum starved, and Rac1-GTP was isolated by pull-down assay from cell lysates. EGFP–Rac1-GTP and total EGFP-Rac1 protein were detected by Western blot (WB) analysis of cell lysates using an anti-GFP antibody. To detect EGFP-Rac1 protein associated with membranes, aliquots of the same cell lysates were fractionated into particulate (P100) and soluble (S100) fractions, and EGFP-Rac1 in the P100 fraction was detected by Western blot analysis. Representative examples of Western blots (n = 3) are shown. (D and E) Quantitation of total EGFP–Rac1-GTP levels present in the cell lysate (D) and of EGFP-Rac1 protein levels present in the P100 fraction (E) are shown as calculated from C. EGFP–Rac1-GTP levels in D are expressed relative to those of total EGFP-Rac1 and levels were normalized relative to those of Lyn-Rac1, which were arbitrarily set to 100%. EGFP-Rac1 levels in the P100 fraction (E) are expressed relative to those of total EGFP-Rac1 and levels were normalized relative to those of Lyn-Rac1, which were arbitrarily set to 100%. Values indicate SEM (n = 3). (F) Integrin engagement induces membrane translocation of PIPKI-α and Rac1. Cell lysates were generated from quiescent HT1080 cells that were either kept in suspension or plated on FN for 20 min in the absence of serum and fractionated into 100,000 g supernatant (cytosol) and pellet (P100; membrane) fractions. Levels of PIPKI-α, Rac1, RhoGDI (cytosol), EGFR (plasma membrane), and tubulin (loading control) in the cell lysate and the P100 fraction were analyzed by SDS-PAGE and Western blot analysis. A representative experiment (n = 3) is shown. (G) Rac1 forms a stable complex with PIPKI-α. Rac1 was immunoprecipitated (IP) from cells that were either kept in suspension in serum-free medium or plated on FN for 20 min. Rac1 and copurifying PIPKI-α were detected by Western blot analysis using appropriate antibodies. IgG immunoprecipitates were probed as a negative control. A representative experiment is shown (n = 3). (H) Enforced Rac1 membrane targeting restores PDGF-induced Rac1 activation to PIPKI-α knockdown cells. Quiescent control or PIPKI-α–depleted HT1080 cells expressing Lyn-EGFP-Rac1 (Lyn-Rac1) were stimulated with 10 ng/ml PDGF (13’) or left untreated (0’). GTP-bound endogenous Rac1 or ectopically expressed Lyn-EGFP-Rac1 was isolated by pull-down assay using PBD-PAK1-GST. Total and GTP-bound levels of Rac1 and Lyn-EGFP-Rac1 were analyzed by Western blotting using antibodies directed against Rac1 and GFP, respectively.
PIPKI-α has been reported to form a complex with Rac1 in the cytoplasm (Tolias et al., 1998, 2000; van Hennik et al., 2003). The question then arises whether integrins regulate the translocation of a PIPKI-α–Rac1 complex to the membrane. Consistent with such a notion, we observed a concomitant increase in PIPKI-α and Rac1 levels in the membrane fraction after cell adhesion to FN (Fig. 6 F). Because similar levels of PIPKI-α protein can be coimmunoprecipitated with Rac1 in suspended and adherent HT1080 cells (Fig. 6 G), these results are best explained by a model wherein PIPKI-α forms a stable complex with Rac1 in the cytosol that translocates to the plasma membrane in response to integrin activation.
Interestingly, we also observed GTP loading of Lyn-Rac1 but not of endogenously expressed Rac1 in response to PDGF stimulation of PIPKI-α–depleted cells (Fig. 6 H). This may indicate that PIPKI-α also regulates PDGF-induced activation of Rac1 by controlling its membrane targeting. Consistent with such a notion, Rac1 translocation to the membrane can also be induced in response to serum stimulation of cells (Hansen and Nelson, 2001). Whatever the precise mechanism, these data show that Rac1-GTP loading can occur in the absence of PIPKI-α once efficient Rac1 plasma membrane localization is restored.
PIPKI-α is necessary for directional cell migration
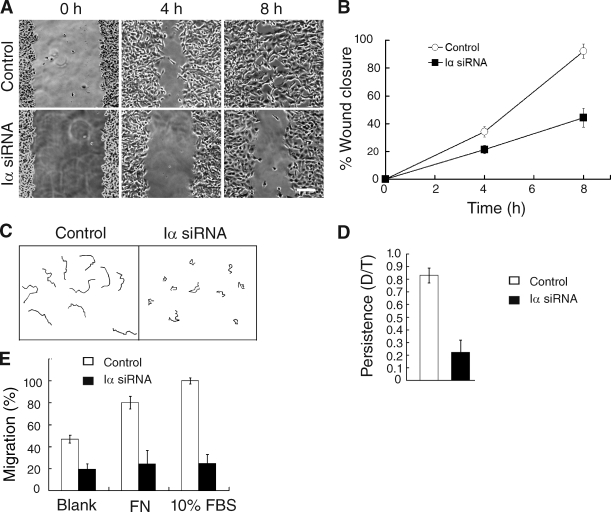
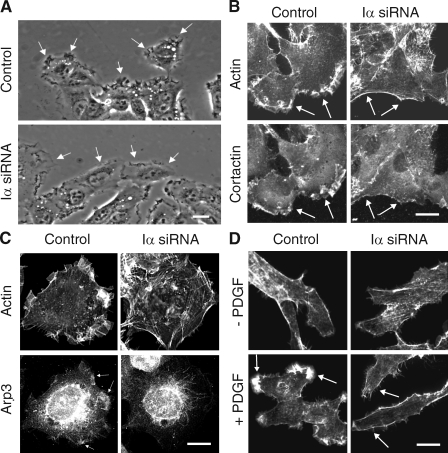
To establish the physiological significance of the Rac1–PIPKI-α interaction, we examined whether Rac1-regulated cellular processes are impaired in PIPKI-α–depleted cells. Previous studies have shown that Rac1 deficiency causes defects in the directional persistence of cell migration in many cell types (Glogauer et al., 2003; Sun et al., 2004; Pankov et al., 2005). Therefore, we examined directional cell migration in PIPKI-α–deficient HT1080 cells. Scratch wound assays showed that PIPKI-α–depleted HT1080 cells were significantly less motile than control siRNA–treated cells (58 ± 2% reduction; Fig. 7, A and B). Tracing of migration tracks of individual cells by time-lapse digital microscopy further revealed that the reduction in wound closure rates in PIPKI-α knockdown cells was mainly the result of a decrease in directional persistence (∼50% decrease; n = 20; P < 0.001), although a modest decrease in mean migration velocity was also observed (control cells, 36.0 ± 1.6 mm/h; PIPKI-α knockdown cells, 27.2 ± 2.3 mm/h; ∼25% decrease; n = 20; P < 0.001). The inhibition of directional persistence was even more pronounced when the migration of sparsely seeded, individually migrating PIPKI-α–depleted cells was analyzed (Fig. 7, C and D). Cells depleted for PIPKI-α also failed to respond to two chemoattractants, serum or FN (Fig. 7 E), in Transwell migration assays and were defective in chemotaxis toward a stable gradient of PDGF (Fig. 8). These results show that PIPKI-α is necessary for directional cell movement during random migration as well as during migration induced in response to chemotactic or haptotactic stimuli.
Figure 7.
PIPKI-α is required for directed cell migration. (A) Wound closure by PIPKI-α knockdown cells is shown. Control or PIPKI-α–depleted HT1080 cells were scratch wounded, and closure of the cell-free area was followed by time-lapse microscopy. Representative still photographs taken at the indicated time points after scratching are shown. Bar, 200 µm. (B) Quantification of the cell-free area during tissue culture wound closure from A, relative closure of the cell-free area, is shown. (C) Trajectories of sparsely plated and randomly migrating cells were tracked by time-lapse microscopy over a 6-h period. 10 trajectories of representative cells are shown. (D) Quantification of the directionality (persistence) of migration of individually migrating cells from C is shown. Persistence is calculated as the ratio (D/T) of the shortest distance from the initial to the last position (D) over the total length of the trajectory (T; n = 20). (E) PIPKI-α knockdown effects on chemotaxis and haptotaxis. Cell migration of control or PIPKI-α–depleted HT1080 cells across Transwell membranes was assayed in response to blank medium, serum (10% FBS), or 10 µg/ml FN present in the bottom chamber. Cells were incubated for 2 h at 37°C, and the total number of cells passing through the membrane was determined. Results shown are expressed as the percentage of migrated cells relative to control cells. Migration of control cells toward FBS was arbitrarily set to 100%. Error bars indicate SEM (n = 3).
Figure 8.
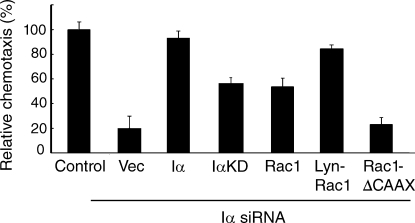
Membrane-targeted Rac1 suppresses the chemotaxis defect of PIPKI-α knockdown cells. Chemotaxis in response to a stable gradient of 10 µg/ml PDGF quiescent control or PIPKI-α–depleted HT1080 cells transfected with empty vector or vectors containing the indicated cDNAs. Chemotaxis was measured using µ-slide chemotaxis chambers. Results are shown as the percentage of migrated cells relative to control cells. Error bars indicate means ± SD (n = 3). Transfection efficiency was ∼85% based on GFP fluorescence.
Significantly, the chemotaxis defect of PIPKI-α–depleted cells toward a gradient of PDGF could be suppressed by expression of Lyn-Rac1 and, to a lesser extent, of wild-type Rac1 (Fig. 8). These findings suggest that the requirement for PIPKI-α in cell migration can be bypassed by increasing Rac1 plasma membrane levels. Consistent with such an idea, expression of a CAAX box–deleted Rac1 mutant that failed to target to the plasma membrane also failed to restore chemotaxis (Fig. 8). Notably, the migration defect associated with PIPKI-α depletion could also be partially rescued by expression of the kinase-dead PIPKI-α mutant IαKD compared with the wild-type protein (Fig. 8), a finding that is in line with our observation that PIPKI-α controls Rac1 membrane translocation through a kinase-independent mechanism. Collectively, these results suggest that the modulation of Rac1 membrane targeting by PIPKI-α is a key function through which PIPKI-α controls directional cell migration, but PI4,5P2-dependent mechanisms likely also contribute.
Depletion of PIPKI-α confers defects in membrane ruffling and actin organization
Rac1 plays a central role in directed cell migration by inducing lamellipodia extension and membrane ruffling (Nobes and Hall, 1999; Hall and Nobes, 2000). This is in part mediated by the Rac1-dependent recruitment of actin-binding proteins such as cortactin to the leading edge and by the stimulation of Arp2/3-mediated actin polymerization. We found that migrating HT1080 cells depleted for PIPKI-α showed markedly decreased membrane ruffling (Fig. 9 A) and appeared to produce multiple, unstable membrane protrusions in random directions when compared with control siRNA–treated cells (unpublished data). Phalloidin staining of F-actin further revealed that the majority of PIPKI-α knockdown cells (78 ± 5%; n = 100; P < 0.001) had lost actin-rich lamellae and showed a concomitant increase in actin stress fibers and cortical actin bundles (Fig. 9 B). Consistent with these alterations in actin morphology, cortactin and the Arp2/3 complex subunit Arp3 failed to localize to the plasma membrane in PIPKI-α–depleted cells, whereas >90% of control siRNA–treated cells contained prominent lamellipodial ruffles enriched in F-actin, cortactin, and Arp3 (Fig. 9, B and C). Notably, serum-starved PIPKI-α knockdown cells failed to induce actin-rich membrane ruffles in response to PDGF stimulation, whereas such structures were readily observed in control cells (Fig. 9 D). Collectively, these results suggest that PIPKI-α plays a central role in actin cytoskeletal reorganization and membrane ruffling. These data therefore support the notion that Rac1 activity is impaired in PIPKI-α–depleted cells.
Figure 9.
Knockdown of PIPKI-α leads to defects in membrane ruffling and F-actin organization. (A) Leading edge membrane ruffles are decreased in migrating PIPKI-α–depleted cells but not in control siRNA–treated cells. HT1080 cells, transfected with the indicated siRNAs, were stimulated to migrate by scratch wounding and monitored by video microscopy. Representative still pictures are shown. Arrows identify leading edge membrane ruffles. (B and C) Distribution of F-actin, cortactin, and Arp3 in PIPKI-α–depleted HT1080 cells. (top) Cells were fixed and stained with Alexa Fluor 594 phalloidin to visualize F-actin. Endogenous cortactin (B) or Arp3 (C) was visualized by indirect immunofluorescence using mouse anti-cortactin and anti-Arp3 antibodies followed by Cy2-conjugated goat anti–mouse antibodies (bottom). (D) PDGF induced F-actin distribution in PIPKI-α knockdown cells. Quiescent control or PIPKI-α–depleted HT1080 cells were stimulated with 10 ng/ml PDGF (bottom; +) or left untreated (top; −). Cells were fixed and stained for F-actin with Alexa Fluor 594 phalloidin. (B–D) Representative examples (n = 3) of cells are shown. Arrows indicate plasma membrane areas containing F-actin, cortactin, or Arp3 signal. Bars, 10 µm.
Depletion of PIPKI-α confers defects in focal adhesion formation and cell spreading
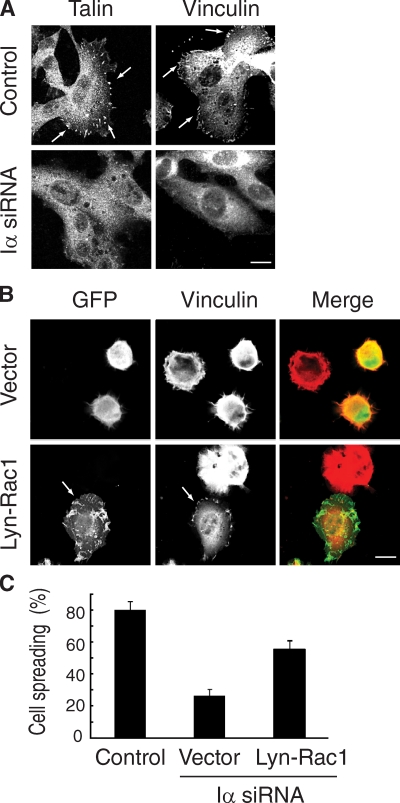
Rac1 is also important for cell spreading and the formation of focal adhesion contacts (Nobes and Hall, 1999; Hall and Nobes, 2000). Analysis of the distribution of focal adhesion markers, including vinculin and talin, in PIPKI-α–depleted cells showed that the number of adhesion plaques was significantly reduced in these cells compared with control siRNA–treated cells (Fig. 10 A). This suggests that focal adhesion formation or stability is impaired in PIPKI-α–depleted cells.
Figure 10.
PIPKI-α depletion impairs focal adhesion formation and cell spreading. (A) Control or PIPKI-α–depleted HT1080 cells were plated on FN substrate, fixed, and stained for vinculin or talin using specific monoclonal antibodies to reveal focal adhesions (arrows). (B) Expression of membrane-targeted Rac1 rescues the focal adhesion and spreading defects of PIPKI-α–depleted cells. Control or PIPKI-α–depleted HT1080 cells expressing Lyn-EGFP-Rac1 (Lyn-Rac1) or containing empty vector were resuspended and plated on FN substrate. After 45 min, cells were fixed and stained for vinculin to reveal focal adhesions. F-actin was visualized with Alexa Fluor phalloidin, and GFP fluorescence was recorded directly. Transfection efficiency was ∼80% based on GFP fluorescence. (C) Cells were treated as in B and assayed for cell spreading. A quantification of the results is shown in which spreading is expressed as the percentage of adherent cells to total cells. Error bars indicate means ± SD (n = 3). Bars, 10 µm.
Moreover, although PIPKI-α knockdown cells adhered to FN within a 10-min period in a manner similar to control RNAi–treated cells, these cells failed to extend lamellipodia, and only a small fraction of cells spread within a 1-h period, whereas control RNAi–treated cells were fully spread within 30 min after plating (Fig. 10 B). Together, these data suggest that PIPKI-α regulates the kinetics of cell spreading and integrin-mediated formation of cell–matrix contacts. Because focal complexes in the extending membrane protrusion are important for the formation of a stable lamellipodium, the impaired assembly of focal contacts may contribute to the cell migration defects of PIPKI-α knockdown cells.
Importantly, the focal adhesion phenotype of PIPKI-α–depleted cells was rescued by expression of Lyn-Rac1 because 60 ± 2% of cells expressing the plasma membrane–anchored Lyn-Rac1 variant were fully spread compared with 22 ± 2.5% of cells expressing vector alone (Fig. 10, B and C). Notably, Lyn-Rac1 also restored membrane ruffling to PIPKI-α knockdown cells (Fig. 10 B). Together, these data suggest that PIPKI-α–depleted cells have impaired membrane ruffling and defects in cell spreading and focal adhesion formation because of defective Rac1 signaling. Collectively, these results support the notion that PIPKI-α is an important regulator of Rac1 activity during directed cell migration.
Discussion
Integrin receptors have been shown to spatially regulate Rac1 signaling by controlling Rac1 membrane targeting (Price et al., 1998; del Pozo et al., 2000, 2002; Kraynov et al., 2000); however, the mechanism by which this occurs and the resulting implications for cell migration have so far not been described. The data presented in this study identify PIPKI-α as a mediator of Rac1 membrane targeting and show that PIPKI-α thereby regulates Rac1 activation and downstream signaling. This conclusion is based on (a) the characterization of PIPKI-α knockdown phenotypes, which shows that PIPKI-α modulates Rac1-dependent cellular processes such as actin and focal adhesion dynamics and the directional persistence of cell migration, and (b) the finding that all PIPKI-α–associated phenotypes can be suppressed by plasma membrane–anchored Rac1.
Notably, the role of PIPKI-α in Rac1 targeting is isoform specific, appears to require physical interaction with the polybasic domain of Rac1, and is independent of catalytic activity. Therefore, PIPKI-α appears to have two major signaling roles: a scaffolding role that serves to target Rac1 and a role as a lipid kinase to generate PI4,5P2. The finding that enforced targeting of Rac1 to the plasma membrane is able to bypass the requirement for PIPKI-α and suppresses the chemotaxis defect of PIPKI-α knockdown cells, further suggesting that the scaffolding role of PIPKI-α plays an important role in cell migration. These data therefore provide evidence for a new mechanism for the spatial regulation of Rac1 activity. This mechanism can operate independently and in addition to PI4,5P2 and PI3,4,5P3 gradients and modulate directed cell migration.
Our data show that PIPKI-α accumulates at plasma membrane sites where local activation of integrin receptors occurs. Because activated integrins are more prominent at the front of migrating cells (Moissoglu and Schwartz, 2006), integrin activation may lead to the preferential recruitment of PIPKI-α and, consequently, Rac1 to this site. Therefore, a key question is how activated integrins regulate the translocation of PIPKI-α to the plasma membrane. Previous studies have implicated Ajuba, a LIM domain–containing protein necessary for Rac1 activation and directed cell migration, in the targeting of PIPKI-α to lamellipodia and membrane ruffles (Doughman et al., 2003; Kisseleva et al., 2005). Thus, Ajuba is a plausible candidate for mediating integrin-dependent membrane translocation of PIPKI-α.
The finding that PIPKI-α in an upstream regulator of Rac1 in cell migration appears to contradict earlier reports, which identified PIPKI-α as an effector of small GTPases in growth factor– and G protein–coupled receptor pathways (Chatah and Abrams, 2001; Doughman et al., 2003). However, earlier studies were performed by overexpression of wild-type and dominant-negative PIPKI variants in conjunction with expression of constitutively active or dominant-negative Rac1 mutants (Chatah and Abrams, 2001; Doughman et al., 2003). Although these types of approaches have revealed roles of PIPKI isoforms in the regulation of many cellular processes, they have limitations and may not accurately reflect the function or localization of the endogenous PIPKI proteins and their relationship with members of the small GTPase family. In contrast, RNAi has emerged as a specific and effective means to test the physiological roles of individual PIPKI isoforms, although these approaches produce hypomorph and not complete loss of function phenotypes.
In summary, our study describes a novel, kinase-independent role of PIPKI-α in directed cell migration and suggests that PIPKI-α orchestrates this process by spatially regulating Rac1 activity. Interestingly, recent knockout and RNAi studies have demonstrated that PIPKI-α and PIPKI-γ play distinct roles during Fcγ receptor–mediated phagocytosis: PIPKI-γ acts upstream of Rac1 and RhoA, whereas PIPKI-α promotes N-WASP (neural Wiskott-Aldrich syndrome protein) activation and N-WASP–dependent de novo actin polymerization (Mao et al., 2009). Moreover, additional isoform-specific roles for PIPKI-β and the PIPKI-γ661 splice variant in the regulation of cell polarization and directed cell migration have been reported previously (Lacalle et al., 2007; Lokuta et al., 2007; Sun et al., 2007). Thus, all PIPK isoforms appear to contribute to the regulation of actin dynamics and cell migration but display remarkable specificity in their signaling roles, and a major focus of future research should be to understand the basis for this specificity.
Materials and methods
Cell culture
Human cervical carcinoma and human fibrosarcoma HT1080 cell lines (American Type Culture Collection) were grown in DME supplemented with 10% calf serum, penicillin, and streptomycin (Invitrogen) with 5% CO2 at 37°C.
Antibodies and reagents
Isoform-specific polyclonal rabbit anti–PIPKI-α and anti–PIPKI-β antibodies were generated against the C-terminal PIPKI tails and were affinity purified before use. Anti–PIPKI-γ–pan antibodies were provided by P. de Camilli (Yale University, New Haven, CT). Rabbit anti-HA antibodies were obtained from BabCo, mouse anti-Rac1 clone 102, mouse anti-cortactin, mouse anti-EGFR, and mouse anti-RhoGDI antibodies were obtained from BD, and mouse anti-Arp3 antibodies were obtained from Abcam. Cy2- and Cy3-conjugated goat anti–mouse and Cy2-conjugated goat anti–rabbit IgGs were obtained from Jackson ImmunoResearch Laboratories, Inc. Alexa Fluor–conjugated phalloidin was obtained from Invitrogen. LY294002 and neomycin were obtained from Enzo Life Sciences, Inc.
Plasmid constructs
We used the human PIPKI nomenclature throughout the manuscript, which is different from the mouse designation, as human and mouse PIPKI-α and PIPKI-β isoforms were named in a reciprocal manner; i.e., human PIPKI-α corresponds to mouse PIPKI-β and vice versa (Ishihara et al., 1996; Loijens and Anderson, 1996). Plasmids containing epitope-tagged wild-type PIPKI-α, PIPKI-β, PIPKI-γ635, PIPKI-γ661, and the kinase-dead IαKD mutant (PIPK Iα; D309N and R427Q) were previously described (Coppolino et al., 2002). GST-PAK-BD was obtained from G. Bokoch (The Scripps Research Institute, La Jolla, CA). Rac1 and Rac1V12 cDNAs in pRK5 were provided by A. Hall (Memorial Sloan-Kettering Institute, New York, NY). K-Ras4B, Rac2, and Rac3 cDNAs were obtained from E. Chang (Baylor College of Medicine, Houston, TX), D.A. Williams (Cincinnati Children’s Foundation, Cincinnati, OH), and N. Heisterkamp (Childrens Hospital of Los Angeles, Los Angeles, CA), respectively. pEGFP-WAVE2 was generated by PCR amplification of WAVE2 from plasmid pEF-BOS-FLAG-WAVE2 (provided by T. Takenawa, University of Tokyo, Tokyo, Japan). Plasmids EGFP-OMP25 (Nemoto and De Camilli, 1999) and EGFP-OMP25 containing human HA-tagged PIPKI-γ661 were obtained from P. de Camilli. The Lyn-EGFP plasmid was provided by T. Meyer (Stanford University, Palo Alto, CA). Point mutations and chimeras were created using the QuikChange site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s protocol and cloned into Escherichia coli and mammalian expression vectors. For construction of Iα-Mito and IαKD-Mito, the open reading frames of PIPKI-α and PIPKI-αKD were amplified by PCR, digested with appropriate restriction enzymes, and cloned in frame into the polylinker between EGFP and OMP25 in EGFP-OMP25. For construction of Lyn-Rac1, the open reading frame encompassing wild-type Rac1 was obtained by PCR and ligated into the polylinker of the pcDNA3-Lyn-EGFP vector. All constructs and mutations were confirmed by DNA sequencing.
RNAi
siRNA SMARTpool reagents (a mixture of four oligonucleotides specific for each targeted gene) were purchased from Thermo Fisher Scientific. The following siGENOME SMARTpool reagents were used: PIPKI-α, M004780-02-0010; PIPKI-β, M004058-00-0005; Rac1, M003560-02-0002; and scrambled siCont, D-001810-10-05. siRNA pools were transfected at a concentration of 20 nM into 30–50% confluent HeLa or HT1080 cells grown on 24-well plates in normal growth medium without antibiotics using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. siRNA-treated cells were analyzed 48 h after transfection for knockdown efficiency by Western blot analysis and for specific phenotypes.
Immunofluorescence and pixel intensity profiles
Cells grown on glass coverslips were fixed with 3.7% formaldehyde, permeabilized in 0.2% Triton X-100, and incubated with primary (1:50–1:100 dilution in PBS/0.1% Triton X-100) and appropriate secondary antibodies (1:200 dilution in PBS/0.1% Triton X-100). Coverslips were washed in PBS and mounted with gel mount aqueous mounting medium (Sigma-Aldrich). Images were acquired at room temperature using a confocal microscope (LSM 510 META; Carl Zeiss, Inc.) with a 40× 1.0 NA oil objective (Carl Zeiss, Inc.) and 2× zoom in magnification. Images were imported into Photoshop (version 8.0; Adobe) for processing. The degree of colocalization in immunostainings was determined by superposition of images acquired in the different channels (488, 594, and 647 nm). Line scans of fluorescence intensity were performed using the intensity profile function of the software (LSM 510; Carl Zeiss, Inc.).
Determination of Rac1-GTP levels
Rac1-GTP levels were analyzed 48 h after siRNA transfection from cell lysates using pull-down assays with a GST fusion protein containing the PBD domain of human PAK exactly as described previously (Benard and Bokoch, 2002). For determination of Rac1-GTP levels in PDGF-treated cells, cells were starved for 18 h in serum-free DME and treated with 10 ng/ml PDGF for 13 min before analysis. Adhesion-induced Rac1-GTP levels were measured exactly as described previously (del Pozo et al., 2002). Bound Rac1-GTP and total Rac1 present in cell lysates were analyzed by SDS-PAGE and Western blotting with a mouse anti-Rac1 antibody followed by HRP-conjugated anti–mouse secondary antibodies. For determination of Lyn-Rac1-GTP levels, cells were transfected with the Lyn-EGFP-Rac1 expression vector 24 h after siRNA treatment, and GTP levels were determined 24 h by GST-PAK pull-down from lysates. Precipitates were analyzed by SDS-PAGE and Western blotting with mouse anti-EGFP antibody followed by HRP-conjugated anti–mouse secondary antibodies. Blots were developed using an enhanced chemiluminescence system. Rac1 levels of at least three independent experiments were quantified by scanning densitometry using the Image Gauge program (version 4.0; Fujifilm), and multiple exposures of each gel were analyzed. In each experiment, Rac1-GTP levels were normalized against total Rac1 in the cell lysate.
Bead recruitment assays
HeLa cells were grown on coverslips and either left untreated or transfected with control siRNAs or siRNAs directed at PIPKIs or Rac1. 48 h after transfection, cells were incubated with 5 µm polystyrene divinylbenzene beads at 37°C for 20 min at a cell to bead ratio of 1:40 as described previously (del Pozo et al., 2002). Beads were coated with heat-treated 10 mg/ml BSA or 10 µg/ml FN at 4°C overnight exactly as described previously (del Pozo et al., 2002). Pixel intensity profiles across lines were used to analyze the extent of colocalization.
Subcellular fractionation
Cells grown on 10 µg/ml FN were transfected with control siRNAs or siRNAs targeting Iα or Rac1. 48 h after transfection, cells were serum starved for 2 h to eliminate contributions from growth factors. Cell lysates were generated and subfractionated into 100,000 g pellet (P100) or supernatant (S100) fractions exactly as described previously (del Pozo et al., 2002). Typically, 20 µg total protein from each fraction, representing 5% of cytosol fraction and 20–50% of the membrane fraction, was analyzed by Western blotting. Blots were quantified by scanning densitometry as described above.
Mitochondrial targeting assay
HeLa cells were cotransfected with either EGFP-OMP25–PIPKI-α, EGFP-OMP25–PIPKI-αKD, or EGFP-OMP25–PIPKI-γ and plasmids containing Flag-tagged wild-type and mutant Rac1 variants as indicated. 24 h after transfection, cells were fixed and analyzed by indirect immunofluorescence using mouse monoclonal anti-Flag antibodies followed by Cy3-conjugated goat anti–mouse IgGs. Actin was visualized by Alexa Fluor 647–conjugated phalloidin. EGFP-OMP25 fusions were visualized by 488-nm excitation. Pixel intensity profiles were used to analyze the extent of colocalization in cells expressing moderate levels of EGFP-OMP25 and Flag-tagged fusion proteins; high and low expressing cells were avoided. Mitotracker (Invitrogen) was used to visualize mitochondria.
Purification of recombinant proteins and pull-down assays
GST fusion proteins were expressed in E. coli BL21(λDE3) cells and purified using GST-bound resin (EMD) following the manufacturer’s protocol. In vitro transcription and translation and labeling of proteins with [35S] Easy Tag Express Protein Labeling Mix (NEN Life Science Products) was performed using a quick-coupled transcription–translation system (TNT T7; Promega). Pull-downs using radioactively labeled PIPKI-α, PIPKI-β, and PIPKI-γ661 and purified GST fusion proteins were performed essentially as described previously (Fadri et al., 2005). In brief, 6 µl of product from in vitro transcription–translation reactions was incubated for 1 h at 4°C with 1 µg different GST fusion proteins bound to agarose beads in binding buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.1% Tween 20, and 0.1% SDS containing protease inhibitors [1 mM PMSF, 1 µg/ml leupeptin, and 1 µg/ml pepstatin A]). Agarose beads were collected by centrifugation and washed five times with 1 ml of cold binding buffer. Bound proteins were eluted, denatured in SDS sample buffer by incubation at 68°C for 15 min, and separated by SDS-PAGE. Gels were dried and exposed to film (BiomaxMR; Kodak).
Scratch wound assays and cell tracking
HT1080 cells grown on 10 µg/ml FN-coated glass coverslips were transfected with the indicated siRNA pools using Lipofectamine 2000 (Invitrogen). Transfected cells were cultured in medium containing 10% FBS for 36 h, serum starved for an additional 12 h, and cell monolayers were transferred to medium containing 10% FBS at 37°C under 5% CO2, wounded by scraping with a 20 µl pipet tip, and washed several times to remove dislodged cells. Time-lapse sequences of migrating monolayers were acquired using an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.) with a 10× objective (0.25Ph1; Carl Zeiss, Inc.) for lower magnification and a 20× objective (0.30Ph1 var1; Carl Zeiss, Inc.) for higher magnification at 37°C under 5% CO2. Video frames were taken with a camera (Axiocam MRm; Carl Zeiss, Inc.) every 5 min for 8 h. Cell paths were generated and quantified using Axiovision software (LE Rel 4.5; Carl Zeiss, Inc.). Ratios of the shortest direct distance from the starting point of each recording to the end point (D), to the total distance traversed by the cell (T), were compared as a measure of directional persistance.
Transwell and chemotaxis assays
siRNA-treated HT1080 cells were plated in the top chamber of a Transwell chamber (8.0-µm pore size; BD), and chemotaxis assays were performed with 10% FBS (vol/vol) medium added to the bottom chamber. For haptotaxis assays, filters were coated on the lower side with 10 µg/ml FN or 10 mg/ml heat-denatured BSA. Cultures were incubated for 2 h at 37°C, nonmigrating cells were removed from the top chamber with cotton swabs, and the remaining cells were fixed for 10 min in 3.7% formaldehyde/0.5% Triton X-100 in PBS, stained with DAPI, mounted, and cells were counted by fluorescence microscopy. To analyze the chemotactical response of HT1080 cells toward a linear and stable concentration gradient of PDGF, we used µ-slide chambers (ibidi). siRNA-treated HT1080 cells were loaded into the chambers, and chemotaxis toward 10 ng/ml PDGF was analyzed as recommended by the manufacturer. Cell migration was quantitated by counting the number of cells that migrated toward the PDGF gradient into a defined circular area of the slide during the 2-h time observation period relative to the total number of cells loaded.
Rescue experiments
HT1080 cells were treated with the indicated siRNA pools and incubated in 10% FBS medium. 12 h before bead recruitment or Transwell migration assays, cells were retransfected with cDNAs encoding the EYFP-tagged murine orthologues of the indicated human PIPKI isoforms or mutant variants thereof, which are resistant to siRNA treatment. Transfection with empty vector was used as a control. Alternatively, siRNA-treated cells were retransfected with EGFP-tagged Rac1 or Lyn-Rac1. Cells were incubated in serum-free medium, and Transwell assays were performed as described above. Transfection efficiency as judged by the number of YFP- or-GFP positive cells was similar in all cases and typically >85%.
Online supplemental material
Fig. S1 shows efficiency and specificity of mRNA and PIPKI-α protein knockdown. Fig. S2 shows that PIPKI-α and Rac1 colocalize at the plasma membrane. Fig. S3 shows that PIPKI-α specifically and directly binds Rac1 via the polybasic tail of Rac1. Fig. S4 shows that mitochondrial targeting of PIPKI-α affects Rac1 subcellular localization and chemotaxis. Fig. S5 shows targeting of GFP-tagged Rac1 variants in PIPKI-α–depleted cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200911110/DC1.
Acknowledgments
We thank Gary Bokoch, Pietro de Camilli, Eric Chang, Alan Hall, Nora Heisterkamp, Tobias Meyer, Tadaomi Takenawa, Ralph Isberg, and David A. Williams for their generous gifts of antibodies and plasmids, Tegy J. Vadakkan for assistance with live cell tracking, and Joseph Henri Bayle for comments on the manuscript.
This work was supported by grants from the National Institutes of Health (GM068098 and R03CA139545) and by a Dan L. Duncan Cancer Center Pilot Project Grant (all to J. Kunz).
Footnotes
Abbreviations used in this paper:
- FN
- fibronectin
- GEF
- GTP exchange factor
- PH
- pleckstrin homology
- PI4
- 5P2, phosphatidylinositol-4,5-bisphosphate
- PIPKI
- type I phosphatidylinositol-4-phosphate 5-kinase
- RhoGDI
- Rho guanine nucleotide dissociation inhibitor
References
- Balla T., Várnai P. 2002. Visualizing cellular phosphoinositide pools with GFP-fused protein-modules. Sci. STKE. 2002:pl3 10.1126/stke.2002.125.pl3 [DOI] [PubMed] [Google Scholar]
- Baumeister M.A., Martinu L., Rossman K.L., Sondek J., Lemmon M.A., Chou M.M. 2003. Loss of phosphatidylinositol 3-phosphate binding by the C-terminal Tiam-1 pleckstrin homology domain prevents in vivo Rac1 activation without affecting membrane targeting. J. Biol. Chem. 278:11457–11464 10.1074/jbc.M211901200 [DOI] [PubMed] [Google Scholar]
- Benard V., Bokoch G.M. 2002. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 345:349–359 10.1016/S0076-6879(02)45028-8 [DOI] [PubMed] [Google Scholar]
- Chatah N.E., Abrams C.S. 2001. G-protein-coupled receptor activation induces the membrane translocation and activation of phosphatidylinositol-4-phosphate 5-kinase I alpha by a Rac- and Rho-dependent pathway. J. Biol. Chem. 276:34059–34065 10.1074/jbc.M104917200 [DOI] [PubMed] [Google Scholar]
- Coppolino M.G., Dierckman R., Loijens J., Collins R.F., Pouladi M., Jongstra-Bilen J., Schreiber A.D., Trimble W.S., Anderson R., Grinstein S. 2002. Inhibition of phosphatidylinositol-4-phosphate 5-kinase Ialpha impairs localized actin remodeling and suppresses phagocytosis. J. Biol. Chem. 277:43849–43857 10.1074/jbc.M209046200 [DOI] [PubMed] [Google Scholar]
- Côté J.F., Motoyama A.B., Bush J.A., Vuori K. 2005. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 7:797–807 10.1038/ncb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo M.A., Price L.S., Alderson N.B., Ren X.D., Schwartz M.A. 2000. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19:2008–2014 10.1093/emboj/19.9.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo M.A., Kiosses W.B., Alderson N.B., Meller N., Hahn K.M., Schwartz M.A. 2002. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 4:232–239 10.1038/ncb759 [DOI] [PubMed] [Google Scholar]
- Doughman R.L., Firestone A.J., Wojtasiak M.L., Bunce M.W., Anderson R.A. 2003. Membrane ruffling requires coordination between type Ialpha phosphatidylinositol phosphate kinase and Rac signaling. J. Biol. Chem. 278:23036–23045 10.1074/jbc.M211397200 [DOI] [PubMed] [Google Scholar]
- Dransart E., Olofsson B., Cherfils J. 2005. RhoGDIs revisited: novel roles in Rho regulation. Traffic. 6:957–966 10.1111/j.1600-0854.2005.00335.x [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2002. Rho GTPases in cell biology. Nature. 420:629–635 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Fadri M., Daquinag A., Wang S., Xue T., Kunz J. 2005. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell. 16:1883–1900 10.1091/mbc.E04-07-0564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glogauer M., Marchal C.C., Zhu F., Worku A., Clausen B.E., Foerster I., Marks P., Downey G.P., Dinauer M., Kwiatkowski D.J. 2003. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J. Immunol. 170:5652–5657 [DOI] [PubMed] [Google Scholar]
- Guo F., Debidda M., Yang L., Williams D.A., Zheng Y. 2006. Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J. Biol. Chem. 281:18652–18659 10.1074/jbc.M603508200 [DOI] [PubMed] [Google Scholar]
- Hall A., Nobes C.D. 2000. Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:965–970 10.1098/rstb.2000.0632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Luby-Phelps K., Das B., Shu X., Xia Y., Mosteller R.D., Krishna U.M., Falck J.R., White M.A., Broek D. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 279:558–560 10.1126/science.279.5350.558 [DOI] [PubMed] [Google Scholar]
- Hansen M.D., Nelson W.J. 2001. Serum-activated assembly and membrane translocation of an endogenous Rac1:effector complex. Curr. Biol. 11:356–360 10.1016/S0960-9822(01)00091-4 [DOI] [PubMed] [Google Scholar]
- Heo W.D., Inoue T., Park W.S., Kim M.L., Park B.O., Wandless T.J., Meyer T. 2006. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 314:1458–1461 10.1126/science.1134389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Shibasaki Y., Kizuki N., Katagiri H., Yazaki Y., Asano T., Oka Y. 1996. Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J. Biol. Chem. 271:23611–23614 10.1074/jbc.271.39.23611 [DOI] [PubMed] [Google Scholar]
- Kisseleva M., Feng Y., Ward M., Song C., Anderson R.A., Longmore G.D. 2005. The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKI alpha. Mol. Cell. Biol. 25:3956–3966 10.1128/MCB.25.10.3956-3966.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovárová M., Tolar P., Arudchandran R., Dráberová L., Rivera J., Dráber P. 2001. Structure-function analysis of Lyn kinase association with lipid rafts and initiation of early signaling events after Fcepsilon receptor I aggregation. Mol. Cell. Biol. 21:8318–8328 10.1128/MCB.21.24.8318-8328.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov V.S., Chamberlain C., Bokoch G.M., Schwartz M.A., Slabaugh S., Hahn K.M. 2000. Localized Rac activation dynamics visualized in living cells. Science. 290:333–337 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Lacalle R.A., Peregil R.M., Albar J.P., Merino E., Martínez-A C., Mérida I., Mañes S. 2007. Type I phosphatidylinositol 4-phosphate 5-kinase controls neutrophil polarity and directional movement. J. Cell Biol. 179:1539–1553 10.1083/jcb.200705044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K., Schill N.J., Wagoner M.P., Sun Y., Anderson R.A. 2006. Movin’ on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 16:276–284 10.1016/j.tcb.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Loijens J.C., Anderson R.A. 1996. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J. Biol. Chem. 271:32937–32943 10.1074/jbc.271.51.32937 [DOI] [PubMed] [Google Scholar]
- Lokuta M.A., Senetar M.A., Bennin D.A., Nuzzi P.A., Chan K.T., Ott V.L., Huttenlocher A. 2007. Type Igamma PIP kinase is a novel uropod component that regulates rear retraction during neutrophil chemotaxis. Mol. Biol. Cell. 18:5069–5080 10.1091/mbc.E07-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Yamaga M., Zhu X., Wei Y., Sun H.-Q., Wang J., Yun M., Wang Y., Di Paolo G., Bennett M., et al. 2009. Essential and unique roles of PIP5K-γ and -α in Fcγ receptor-mediated phagocytosis. J. Cell Biol. 184:281–296 10.1083/jcb.200806121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissoglu K., Schwartz M.A. 2006. Integrin signalling in directed cell migration. Biol. Cell. 98:547–555 10.1042/BC20060025 [DOI] [PubMed] [Google Scholar]
- Moissoglu K., Slepchenko B.M., Meller N., Horwitz A.F., Schwartz M.A. 2006. In vivo dynamics of Rac-membrane interactions. Mol. Biol. Cell. 17:2770–2779 10.1091/mbc.E06-01-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., De Camilli P. 1999. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18:2991–3006 10.1093/emboj/18.11.2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes C.D., Hall A. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144:1235–1244 10.1083/jcb.144.6.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K.M. 2005. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 170:793–802 10.1083/jcb.200503152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Price L.S., Leng J., Schwartz M.A., Bokoch G.M. 1998. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell. 9:1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Small J.V., Stradal T., Vignal E., Rottner K. 2002. The lamellipodium: where motility begins. Trends Cell Biol. 12:112–120 10.1016/S0962-8924(01)02237-1 [DOI] [PubMed] [Google Scholar]
- Sun C.X., Downey G.P., Zhu F., Koh A.L., Thang H., Glogauer M. 2004. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood. 104:3758–3765 10.1182/blood-2004-03-0781 [DOI] [PubMed] [Google Scholar]
- Sun Y., Ling K., Wagoner M.P., Anderson R.A. 2007. Type Iγ phosphatidylinositol phosphate kinase is required for EGF-stimulated directional cell migration. J. Cell Biol. 178:297–308 10.1083/jcb.200701078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T., Miki H. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801–1809 [DOI] [PubMed] [Google Scholar]
- ten Klooster J.P., Jaffer Z.M., Chernoff J., Hordijk P.L. 2006. Targeting and activation of Rac1 are mediated by the exchange factor β-Pix. J. Cell Biol. 172:759–769 10.1083/jcb.200509096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias K.F., Couvillon A.D., Cantley L.C., Carpenter C.L. 1998. Characterization of a Rac1- and RhoGDI-associated lipid kinase signaling complex. Mol. Cell. Biol. 18:762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias K.F., Hartwig J.H., Ishihara H., Shibasaki Y., Cantley L.C., Carpenter C.L. 2000. Type Ialpha phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr. Biol. 10:153–156 10.1016/S0960-9822(00)00315-8 [DOI] [PubMed] [Google Scholar]
- Ueyama T., Eto M., Kami K., Tatsuno T., Kobayashi T., Shirai Y., Lennartz M.R., Takeya R., Sumimoto H., Saito N. 2005. Isoform-specific membrane targeting mechanism of Rac during Fc gamma R-mediated phagocytosis: positive charge-dependent and independent targeting mechanism of Rac to the phagosome. J. Immunol. 175:2381–2390 [DOI] [PubMed] [Google Scholar]
- van Hennik P.B., ten Klooster J.P., Halstead J.R., Voermans C., Anthony E.C., Divecha N., Hordijk P.L. 2003. The C-terminal domain of Rac1 contains two motifs that control targeting and signaling specificity. J. Biol. Chem. 278:39166–39175 10.1074/jbc.M307001200 [DOI] [PubMed] [Google Scholar]
- Vidali L., Chen F., Cicchetti G., Ohta Y., Kwiatkowski D.J. 2006. Rac1-null mouse embryonic fibroblasts are motile and respond to platelet-derived growth factor. Mol. Biol. Cell. 17:2377–2390 10.1091/mbc.E05-10-0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler A.P., Wells C.M., Smith S.D., Vega F.M., Henderson R.B., Tybulewicz V.L., Ridley A.J. 2006. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J. Cell Sci. 119:2749–2757 10.1242/jcs.03024 [DOI] [PubMed] [Google Scholar]
- Yin H.L., Janmey P.A. 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65:761–789 10.1146/annurev.physiol.65.092101.142517 [DOI] [PubMed] [Google Scholar]