Dividing cells can sense DNA damage and initiate a primary response, but repair isn’t completed until the cells enter G1.
Abstract
The signaling cascade initiated in response to DNA double-strand breaks (DSBs) has been extensively investigated in interphase cells. Here, we show that mitotic cells treated with DSB-inducing agents activate a “primary” DNA damage response (DDR) comprised of early signaling events, including activation of the protein kinases ataxia telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK), histone H2AX phosphorylation together with recruitment of mediator of DNA damage checkpoint 1 (MDC1), and the Mre11–Rad50–Nbs1 (MRN) complex to damage sites. However, mitotic cells display no detectable recruitment of the E3 ubiquitin ligases RNF8 and RNF168, or accumulation of 53BP1 and BRCA1, at DSB sites. Accordingly, we found that DNA-damage signaling is attenuated in mitotic cells, with full DDR activation only ensuing when a DSB-containing mitotic cell enters G1. Finally, we present data suggesting that induction of a primary DDR in mitosis is important because transient inactivation of ATM and DNA-PK renders mitotic cells hypersensitive to DSB-inducing agents.
Introduction
The maintenance of an intact genome is crucial for cellular homeostasis. DNA double-strand breaks (DSBs), generated by ionizing radiation (IR) and radiomimetic drugs, are the most cytotoxic lesions. Failure to repair DSBs causes genomic instability and can lead to tumorigenesis and other age-related diseases (Jackson and Bartek, 2009). Upon DSB induction, cells activate a DNA damage response (DDR) that comprises two major stages: initial sensing of DNA breaks followed by downstream events leading to cell cycle arrest, DNA damage repair, and subsequent cell cycle resumption.
Numerous factors involved in DSB processing, signaling, and repair accumulate at damaged sites in focal structures termed IR-induced foci (IRIF). Within seconds, DSBs are detected by the Mre11–Rad50–Nbs1 (MRN) and Ku70–Ku80 complexes, which in turn recruit the apical PI3-kinase–like kinases (PIKKs), ataxia telangiectasia mutated (ATM), and DNA-dependent protein kinase catalytic subunit (DNA-PKcs), respectively (Falck et al., 2005). A prime PIKK target is the C terminus of the histone variant H2AX, whose derivative phosphorylated on serine 139 (S139) is referred to as γH2AX (Rogakou et al., 1998). Phospho-S139 of γH2AX is then bound by the tandem BRCA1 C-terminal domain (BRCT) domains of the DDR-mediator protein MDC1 (mediator of DNA damage checkpoint 1; Stucki et al., 2005). ATM-mediated phosphorylations near DSB sites are propagated via phospho-dependent recruitment of MRN-ATM by MDC1, thus helping to create megabase-sized γH2AX-MDC1 foci (for review see van Attikum and Gasser, 2009). MDC1 phosphorylated by ATM also recruits the RING-finger ubiquitin E3-ligase RNF8, which, together with another ubiquitin E3-ligase, RNF168, produces DSB-associated ubiquitylations on histones H2A and H2AX that, in turn, promote accumulation of p53-binding protein 1 (53BP1) and breast cancer gene 1 (BRCA1) proteins (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Doil et al., 2009; Pinato et al., 2009; Stewart et al., 2009). These ubiquitylation events are thought to contribute to local changes in the chromatin structure near break sites to facilitate DSB signaling and repair.
Although DDR has been extensively studied in interphase cells, its precise mechanisms and functions in mitotic cells are still poorly understood. The onset of mitosis is characterized by nuclear envelope disassembly and the regulated compaction of chromatin into mitotic chromosomes, which is essential for the subsequent separation of sister chromatids in anaphase. Notably, vertebrate cells can delay mitosis, or even reverse mitotic progression if exposed to IR during antephase (late G2 to mid prophase) when chromatin condensation is actively taking place (Pines and Rieder, 2001; Chin and Yeong, 2009). However, once cells have passed a “point-of-no-return,” they are committed to completing mitosis even in the presence of DSBs (Rieder and Cole, 1998). The rate of mitotic progression can nevertheless be affected by the amount of DNA damage (Mikhailov et al., 2002). DNA breaks do not hinder mitotic progression per se, and do not appear to induce activation of a DNA damage checkpoint (Rieder and Salmon, 1998). Nevertheless, γH2AX foci do form in mitotic cells treated with IR (Nakamura et al., 2006; Kato et al., 2008), which suggests that DSBs generated during mitosis are not left unnoticed by the DDR machinery. Here, we show that mitotic cells treated with DSB-inducing agents exhibit apical aspects of the DDR but not a full DDR. We also provide evidence that marking of DSBs generated in mitosis with γH2AX enhances cell viability, which suggests that it acts to facilitate full DDR induction in the more favorable chromatin environment of the G1 cell.
Results and discussion
Mitotic DSBs are marked by PIKK-dependent γH2AX, MDC1, and MRN foci
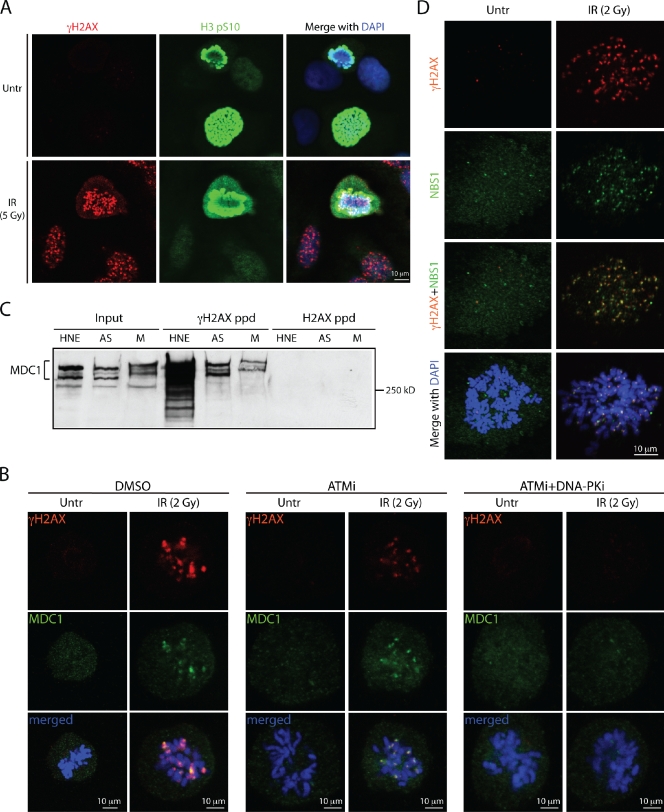
γH2AX is a hallmark of unrepaired DSBs in interphase cells (Rogakou et al., 1998; Paull et al., 2000). Several studies have described focal or pan-nuclear γH2AX staining in mitotic cells that were either untreated or treated with DNA-damaging agents (Ichijima et al., 2005; McManus and Hendzel, 2005; Kato et al., 2008). To obtain additional insights into γH2AX production during mitosis, we examined γH2AX focus formation in mitotic cells arising from asynchronously growing cultures of human U2OS, HeLa, BJ, and MRC5 cells (Figs. 1 A and S1 A). Multiple discrete γH2AX foci were detected only in mitotic cells that had been exposed to IR or the radiomimetic drug phleomycin, but were not readily observed in untreated mitotic cells. Some mitotic cells did occasionally display γH2AX foci under untreated conditions, which could reflect DNA damage arising from endogenous sources (Deckbar et al., 2007; Kato et al., 2009). As we observed that nocodazole caused an overall increase in γH2AX levels as detected by immunoblotting (Fig. 3 F), we did not use nocodazole to enrich for M-phase cells in our subsequent immunofluorescence analyses.
Figure 1.
ATM and DNA-PK mediate IRIF formation in mitosis. All experiments were performed with U2OS cells. (A) IR induces γH2AX focus formation on mitotic chromosomes costained for histone H3pS10. (B) ATM inhibitor or a combination of ATM and DNA-PK inhibitors affect γH2AX and MDC1 IRIF formation in mitosis. (C) Immunoblot of MDC1 after peptide pull-downs with unmodified and phosphorylated H2AX C-terminal peptides. Inputs represent 10% of the total protein in the whole cell extracts prepared from asynchronous (AS) and mitotic (M) cells. HNE, HeLa nuclear extract. (D) NBS1 and γH2AX colocalize on mitotic chromosomes after IR.
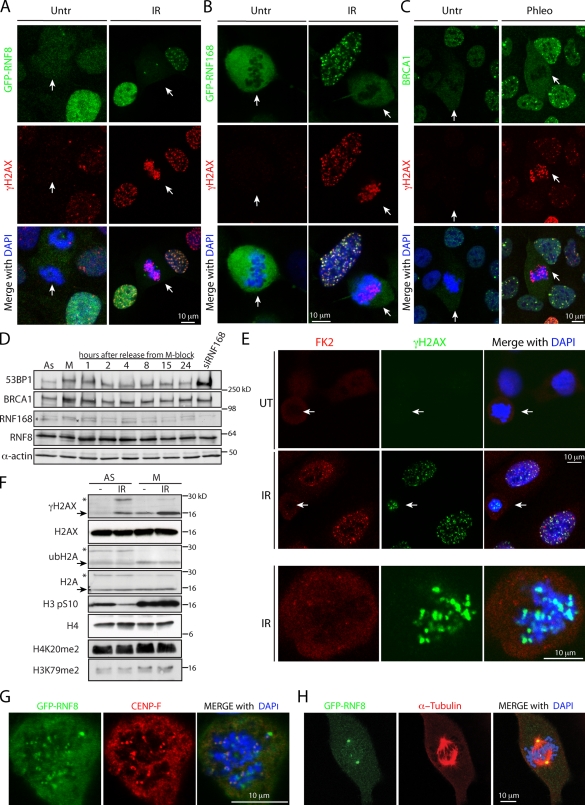
Figure 3.
Lack of recruitment of the ubiquitin E3 ligases RNF8, RNF168, and BRCA1 to DSB sites in mitosis. (A–C) GFP-RNF8 (A), GFP-RNF168 (B), and BRCA1 (C) are excluded from chromatin and are not recruited to IRIF upon DSB induction during mitosis. White arrows indicate mitotic cells. (D) Levels of indicated proteins throughout the cell cycle. M-phase cells, obtained by a shake-off procedure after thymidine-nocodazole arrest, were released into fresh medium and collected at specified times. (E) FK2 antibody staining does not detect ubiquitin conjugates on mitotic chromosomes upon IR treatment. (F) Histones H2A and H2AX are deubiquitylated in mitosis. Arrows point to nonubiquitylated forms and asterisks mark ubiquitylated forms of the proteins. Note that histone H4K20me2 and H3K79me2 marks are present in mitosis. (G) GFP-RNF8 colocalizes with CENP-F at kinetochores. (H) GFP-RNF8 localizes to centrosomes.
Because γH2AX provides a docking site for the DDR-mediator protein MDC1 (Stucki et al., 2005), we assessed whether MDC1 was recruited to DSB sites in mitosis. Using two different anti-MDC1 antibodies, as well as cells stably expressing a GFP-MDC1 fusion, we found that, in various cell lines, MDC1 colocalized with γH2AX foci in mitotic cells that had been treated with IR or phleomycin (Figs. 1 B and S1, B–D). More detailed analyses revealed that γH2AX and MDC1 IRIF were present at all mitotic stages (Fig. S1 B). Consistent with these observations, we found that the hyperphosphorylated, slower migrating forms of MDC1 derived from mitotically arrested cells (Xu and Stern, 2003) were capable of binding to the γH2AX phospho-epitope in peptide pull-down experiments, as was MDC1 from extracts of asynchronous cells (Fig. 1 C).
We next assessed whether IRIF formation in mitotic cells required ATM and DNA-PK kinase activities. By using small-molecule inhibitors specific to each kinase (Hickson et al., 2004; Leahy et al., 2004), we observed partial redundancy between the two kinases in mediating H2AX phosphorylation and IRIF formation (Figs. 1 B and S2, A and B). Consistent with previous studies (Burma et al., 2001; Hickson et al., 2004; Stiff et al., 2004; Wang et al., 2005), ATM inhibition reduced IRIF intensity, whereas combined ATM and DNA-PK inhibition almost completely abrogated IRIF in both M-phase and interphase cells. DNA-PK inhibition alone did not visibly affect the DDR markers (Fig. S2 B and not depicted). In line with DNA damage leading to ATM activation in mitotic cells, we detected the S1981-phosphorylated form of ATM (Bakkenist and Kastan, 2003) in extracts of IR-treated mitotic cells (Fig. S2 C; van Vugt et al., 2010). Furthermore, in accord with ATM activation being mediated by the MRN complex (Williams et al., 2007), NBS1 colocalized with γH2AX in mitotic cells treated with IR (Fig. 1 D).
Exclusion of 53BP1 from mitotic IRIF precedes its association with IRIF in G1
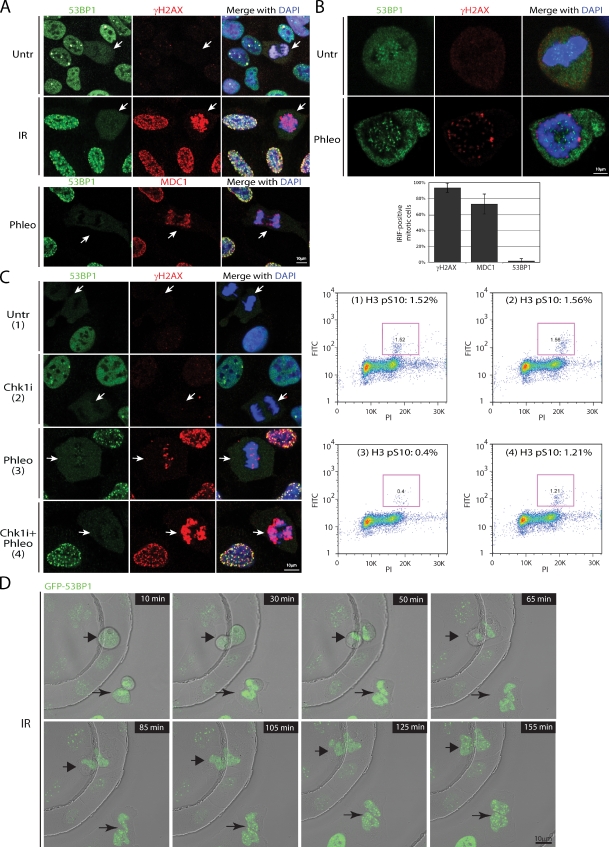
Having established that mitotic cells respond to DSB induction by phosphorylation of γH2AX and by the recruitment of MDC1 and MRN to DSB sites, we next examined the behavior of another mediator protein, 53BP1, which in interphase cells associates with IRIF within 5 min of IR exposure (Schultz et al., 2000). By using an anti-53BP1 antibody or U2OS cells stably expressing GFP-53BP1, we found that, in marked contrast to interphase cells, during mitosis, 53BP1 was mostly excluded from chromatin and was not recruited to IRIF (Fig. 2, A–D; and Fig. S3, A–C). The punctate staining of 53BP1 on condensed pro-metaphase chromosomes in both untreated and irradiated cells, which never colocalized with γH2AX (Fig. 2, A and B), is consistent with previously reported 53BP1 association with kinetochores (Jullien et al., 2002). Importantly, 53BP1 protein levels were similar during the cell cycle, and, in agreement with an earlier study (Jullien et al., 2002), 53BP1 in nocodazole-arrested cells displayed slower gel mobility due to hyperphosphorylation in mitosis (Fig. 3 D). The finding that 53BP1 also did not form IRIF in mitotic HeLa, BJ, or MRC5 cells (Fig. S3 B) demonstrates that 53BP1 exclusion from mitotic chromatin is not cell-line specific.
Figure 2.
53BP1 is excluded from IRIF during mitosis. White arrows point to mitotic cells. (A) Costaining of either mock- or IR-treated asynchronously growing U2OS cells with 53BP1 and γH2AX antibodies. Phleomycin-treated U2OS cells costained for 53BP1 and MDC1. (B) Enlarged images of mitotic cells show exclusion of 53BP1 from chromatin and a lack of colocalization between the punctate 53BP1 staining and γH2AX foci. The histogram represents a quantification of γH2AX, MDC1, and 53BP1 focus-positive mitotic cells after IR treatment from three independent experiments (n > 200). Error bars indicate standard deviation. (C) Active exclusion of 53BP1 from mitotic IRIF. After a 3-h incubation with 50 nM of Chk1 inhibitor (Chk1i), cells were treated with phleomycin for 2 h, fixed and costained for 53BP1 and γH2AX, or collected for FACS analyses to determine mitotic indices by H3 pS10 immunofluorescence (purple boxes). (D) Time-lapse frames of U2OS cells stably expressing EGFP-53BP1. After irradiation with 0.5 Gy, two mitotic cells marked with arrows were imaged for up to 3 h.
To determine whether 53BP1 is actively excluded from IRIF upon mitotic entry, we irradiated asynchronously growing U2OS cells in the presence or absence of AZD7762, an inhibitor of the DNA-damage checkpoint kinases Chk1 and Chk2 (Zabludoff et al., 2008), then measured mitotic indices by FACS with an antibody against histone H3 phosphorylated on serine 10 (H3pS10; Juan and Darzynkiewicz, 2004). In the undamaged samples, both mock- and Chk1 inhibitor–treated, ∼1.5% of cells were in mitosis (Fig. 2 C). After a 2-h phleomycin treatment, this dropped to 0.4% because the G2/M checkpoint prevents damaged interphase cells from proceeding into mitosis, whereas a large proportion of cells damaged in mitosis progress into G1 within the 2-h time frame. In contrast, when the G2/M checkpoint was inactivated by preincubating cells with Chk1 inhibitor before phleomycin treatment, the mitotic index was restored to 1.2%, as had been observed previously in combination with other DNA-damaging agents (Zabludoff et al., 2008). This result thereby implied that the majority of such mitotic cells would have arisen from damaged G2 cells progressing into mitosis within the 2-h phleomycin treatment. Under these experimental settings, where damaged cells with an inhibited G2/M checkpoint entered mitosis, we still did not observe 53BP1 foci in mitotic cells (Fig. 2 C). These data are in agreement with a recent report demonstrating that 53BP1 dissociates from endogenously arising DSBs at the G2/M boundary (Nelson et al., 2009), and suggest that 53BP1 is actively removed from mitotic chromatin even though the known 53BP1-binding histone marks H3K79me2 and H4K20me2 (FitzGerald et al., 2009) are retained in mitosis (Fig. 3 F).
In addition, we used live-cell imaging of asynchronous U2OS cells stably expressing GFP-53BP1 to assess 53BP1 localization after cells irradiated during mitosis subsequently entered G1 (Fig. 2 D). Interphase cells present in the field next to M-phase cells formed 53BP1 foci within several minutes after irradiation. In contrast, 53BP1 signal remained diffuse in mitotic cells until division was complete and nuclei reformed as cells entered G1, at which stage 53BP1 started accumulating within IRIF (Figs. 2 D and 4 B). 53BP1 focus induction in the G1 cells was not caused by the imaging process itself, as it did not occur after prolonged imaging of unirradiated cells (Fig. S3 C). Collectively, these findings suggest that inhibition of 53BP1 recruitment to DSBs is limited to mitosis, and that association of 53BP1 with IRIF is concomitant with nuclear envelope formation and chromatin decompaction in G1.
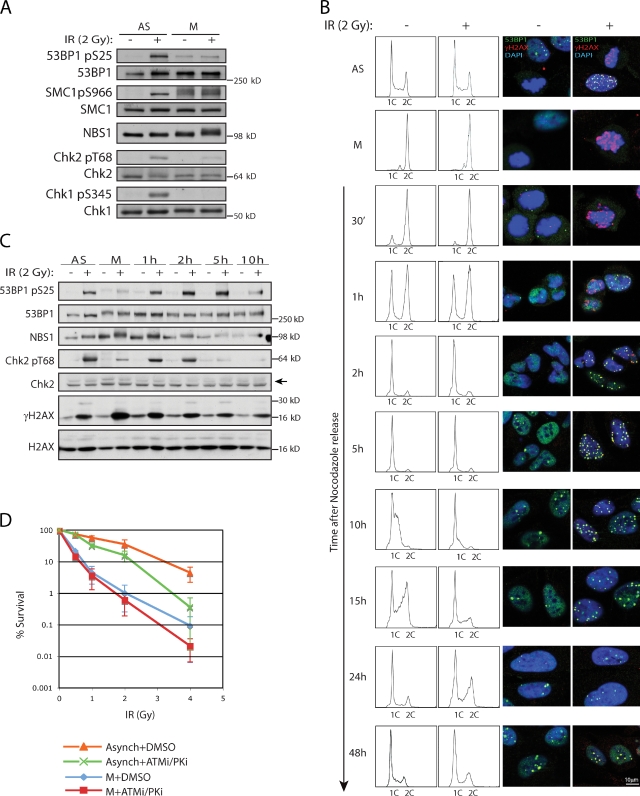
Figure 4.
Marking of DSBs in mitosis precedes full DDR activation in G1 and affects cell survival. Mitotic cells were obtained as in Fig. 3 D. (A) Diminished phosphorylation of ATM targets in IR-treated mitotic cells compared with asynchronous cells. (B) Untreated or irradiated mitotic cells were released from nocodazole and monitored for cell cycle progression and 53BP1 recruitment to IRIF (see text for details). (C) Chk2 T68 and 53BP1 S25 phosphorylation in cells released into G1 after IR treatment in mitosis. (D) Radiosensitivity of asynchronous and mitotic cells pretreated for 1 h with DMSO or a combination of ATM and DNA-PK inhibitors and then exposed to various doses of IR. 30 min after irradiation, inhibitors were washed away and cells were plated in fresh medium. Error bars represent standard deviations of the mean from six experiments. P-values were calculated at the standard 0.05 threshold. Treatment with PIKK inhibitors had statistically significant effects on radiosensitivity of both asynchronous (P = 0.00015) and mitotic (P = 0.041) cells.
E3 ubiquitin ligases RNF8 and RNF168 are not recruited to mitotic IRIF
Recent work has demonstrated that the ubiquitin E3 ligases RNF8 and RNF168 are needed for the productive association of 53BP1 and BRCA1 with IRIF (Stewart, 2009). Strikingly, by using U2OS cells stably expressing GFP-RNF8 or GFP-RNF168, we found that both proteins were excluded from mitotic chromatin and did not colocalize with γH2AX foci upon IR or phleomycin treatment (Fig. 3, A and B), even though the RNF8 docking site provided by MDC1 phosphorylated on the TQXF motifs (Kolas et al., 2007) appears to be constitutively present in mitotic cells (Fig. S2 D). Further examination revealed that in addition to the previously reported midbody localization (Tuttle et al., 2007; Plans et al., 2008), GFP-RNF8 associated with other mitotic structures: kinetochores and centrosomes (Fig. 3, G and H). Our data, together with the proposed function of RNF8 in mitotic exit control (Tuttle et al., 2007; Plans et al., 2008), suggest that RNF8 plays important roles in the regulation of mitosis that might be independent of its involvement in the DDR. Consistent with the exclusion of RNF8 and RNF168 from IRIF during mitosis, the ubiquitin E3 ligase BRCA1 was also undetectable in mitotic γH2AX foci (Fig. 3 C). Importantly, there were no marked changes in 53BP1, BRCA1, RNF8, or RNF168 protein levels throughout the cell cycle (Fig. 3 D), although all these proteins have altered gel migration properties during M phase, likely reflecting hyperphosphorylation in mitosis that is characteristic of many proteins (Poon, 2007). Furthermore, γH2AX foci in mitotic cells did not display costaining with the FK2 antibody that detects protein–ubiquitin conjugates, which implies that ubiquitin conjugates are not effectively formed in DSB foci during mitosis (Fig. 3 E).
RNF8 and RNF168 ubiquitylate nucleosomal histones H2A and H2AX at DSB sites (Stewart, 2009). Previous work has shown that ubiquitylated H2A (ubH2A) is essentially absent from mitotic chromatin and that active H2A deubiquitylation at the G2/M transition is required for chromatin condensation (Matsui et al., 1979; Cai et al., 1999; Joo et al., 2007). In agreement with these findings, we observed a drastic reduction in ubiquitylated H2A and H2AX in mitotic cells, before or after irradiation (Fig. 3 F), despite ubiquitin levels being similar throughout the cell cycle (unpublished data). Collectively, these data suggest that ubiquitylation at DSB sites does not take place in mitosis; this is possibly to prevent changes in chromatin conformation in the context of highly condensed mitotic chromosomes, which might otherwise perturb mitotic progression. The lack of RNF8 recruitment to DSB sites in mitosis could be caused by a combination of chromatin structure and mitosis-specific posttranslational modifications (Fig. S3 D), as well as sequestration of RNF8 in mitotic structures (Fig. 3, G and H).
Mitotic cells mark DSBs before full DDR activation in G1
The progression of mitotic cells into G1 is not apparently delayed by DSBs (Fig. S3 E), which supports the idea of a lack of the DNA damage checkpoint during mitosis (Rieder and Cole, 1998; Mikhailov et al., 2002). Indeed, we found that DSB-induced phosphorylations of ATM substrates, including the critical DDR effectors Chk1 and Chk2, occurred to a lesser extent in mitotic cells than in interphase cells (Fig. 4 A), despite ATM autophosphorylation on S1981 and formation of γH2AX-containing IRIF. Interestingly, in line with the lack of 53BP1 recruitment, ATM-mediated 53BP1 S25 phosphorylation was not induced by DNA damage in mitotic cells (Fig. 4 A). Attenuation of certain ATM-mediated phosphorylations in mitosis is in agreement with a recent study by van Vugt et al. (2010).
To see whether DSBs carried over from mitosis can trigger full DDR activation in G1, we collected mitotic cells from a presynchronized culture, treated them with IR, released them from nocodazole block, and then analyzed samples at different times for cell cycle distribution, presence of 53BP1 in IRIF, and Chk2 phosphorylation (Fig. 4, B and C). Cells damaged in M phase progressed into G1 with kinetics similar to those of untreated cells for up to 5 h after nocodazole release (Fig. 4 B). Consistent with full DDR activation occurring when cells entered G1, irradiated cells exhibited slower entry and/or progression through S-phase, as well as a delay in G2 (Fig. 4 B; 10–24 h time-points). Similar to our findings with EGFP-53BP1 in live-cell imaging experiments (Fig. 2 D), endogenous 53BP1 started forming foci 1–2 h after G1 entry (Fig. 4 B). Furthermore, IRIF were still detectable 24 h after irradiation, although fewer foci remained, which is indicative of ongoing DSB repair. Parallel examination of IR-induced Chk2 phosphorylation on threonine 68 (T68) revealed that this modification was reduced in M phase in comparison with asynchronous cells (Fig. 4 C). Nevertheless, once cells progressed into G1, T68 became highly phosphorylated, which supports the notion that an attenuated DDR during mitosis becomes fully activated in the following interphase.
To assess the potential physiological significance of DSB marking in mitosis, we performed clonogenic survival assays on asynchronous and mitotic cells that had been irradiated either in the absence or presence of ATM and DNA-PK inhibitors (Fig. 4 D). In agreement with earlier studies (Stobbe et al., 2002), mitotic cells displayed much higher radiosensitivity compared with asynchronous cells. Furthermore, acute PIKK inhibition, at a dose sufficient to ablate γH2AX IRIF formation, further enhanced the killing of mitotic and asynchronous cells.
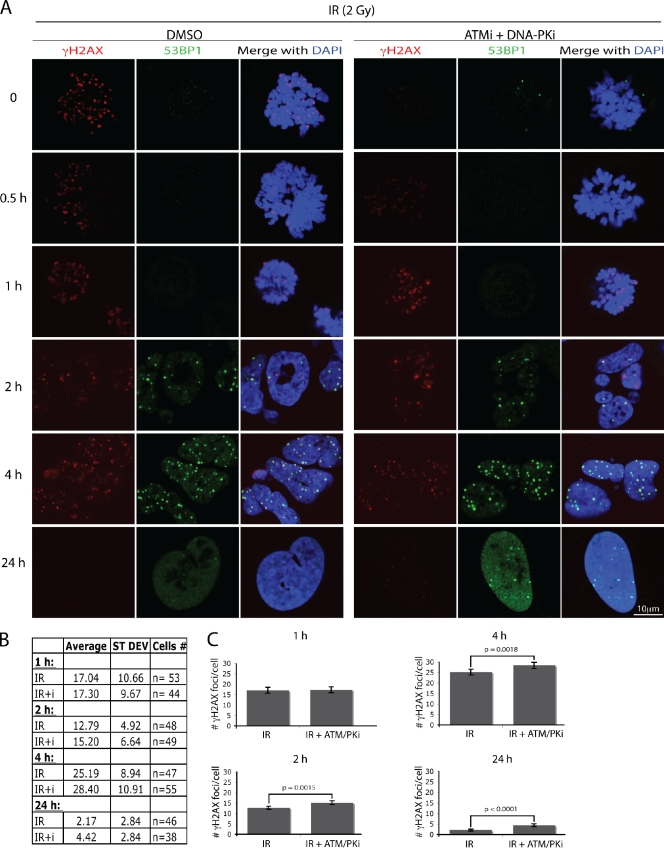
To assess how PIKK inhibition during mitosis leads to radiosensitization, we quantified γH2AX foci at various time points after release from inhibitor treatment and nocodazole block. The inhibitors were removed 30 min after irradiation, and the effect of PIKK inhibition on γH2AX was readily reversible within 1 h (Fig. 5 A). Notably, at the 1 h time point, similar numbers of foci were present in cells that had been mock- or PIKK inhibitor–treated (Fig. 5 A), which indicates that similar amounts of damage were generated under the two conditions, and that the majority of DSBs formed in mitosis were not repaired before G1 entry. Lack of ongoing DSB repair in mitosis is also supported by the difference in PIKK inhibitor–mediated radiosensitization between asynchronous and mitotic cells: the effect of the PIKK inhibitors on mitotic cells is approximately threefold lower than on asynchronous cells (Fig. 4 D). At later time points, however, there were significant differences in IRIF numbers between mock- and inhibitor-treated cells, with the inhibitor-treated cells showing twofold more residual foci at the 24 h time point (Fig. 5, B and C). One possibility is that the IRIF that persist after 24 h in cells derived from mitotic cells pretreated with PIKK inhibitors correspond to more complex lesions that might otherwise have benefited from the immediate marking after DNA damage induction.
Figure 5.
Transient inhibition of ATM and DNA-PK during mitosis before irradiation results in the increase of persistent IRIF 24 h after treatment. U2OS cells were treated as in Fig. 4 D. (A) Representative images of the cells at different times after nocodazole and PIKK inhibitor release. The table (B) and the graph (C) summarize γH2AX focus quantification. Error bars represent standard errors (SE = SD/√n).
Collectively, our data support a model in which mitotic cells treated with DSB-inducing agents prioritize timely passage through mitosis over activation of a full DDR. The latter is likely to involve substantial changes in chromatin structure proximal to DSB sites that would presumably not be feasible in the context of highly condensed mitotic chromosomes. Nevertheless, mitotic cells do activate early DDR events, including the marking of DSBs by γH2AX, MDC1, and MRN, which may facilitate recognition of DNA damage and its repair during the following cell cycle.
Materials and methods
Cell culture
U2OS, HeLa, BJ, and MRC5 cells were cultured in standard Dulbecco’s modified Eagle’s minimal essential medium (Sigma-Aldrich) supplemented with 10% FBS (BioSera), 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (Sigma-Aldrich). The medium for U2OS cells stably expressing GFP-MDC1 (Kolas et al., 2007), GFP-53BP1, GFP-RNF8 (Mailand et al., 2007) and GFP-RNF168 was supplemented with 0.5 mg/ml G418 (Invitrogen).
Treatment with small-molecule inhibitors and DNA-damaging agents
ATM KU-55933 and DNA-PK NU-7441 inhibitors were obtained from KuDOS Pharmaceuticals, and Chk1/2 inhibitor AZD7762 was obtained from AstraZeneca (Zabludoff et al., 2008). IR treatment was performed with an x-ray machine (Faxitron X-Ray LLC). Phleomycin (Duchefa Biochemie) was added at 30 µg/ml. Where appropriate, ATM and DNA-PK inhibitors (20 µM and 2 µM, respectively) were applied to the culture medium 1 h before DSB induction. Chk1 inhibitor was added (50 nM final concentration) 3 h before phleomycin treatment. Cells were processed for analyses 30 min after IR or phleomycin treatment.
Cell synchronization
After a 20–24-h presynchronization with 2.5 mM thymidine, cells were extensively washed and released into fresh medium. After 8–10 h, nocodazole was added (final concentration 40 ng/ml) for 3–4 h to accumulate cells in early mitosis.
Immunofluorescence analyses
Cells were either grown on coverslips or harvested by mitotic shake-off and attached to poly-l-lysine–coated coverslips using a cytospin centrifuge at 500 rpm for 5 min. All the following procedures were performed at room temperature. Cells were fixed with 2% paraformaldehyde for 20 min, washed three times with PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and blocked with 5% BSA in PBS/0.1% Tween 20 for 10 min. Primary antibodies used are listed in Table S1. Incubation with primary antibodies was for 45 min followed by three washes with PBS and a 30-min incubation with the corresponding secondary antibodies: Alexa Fluor 488 (green), 594 (red), and 647 (far red) at 1:1,000 (Invitrogen). Coverslips were washed three times with PBS and mounted with Vectashield mounting medium containing DAPI (Vector Laboratories). Images were acquired with a confocal microscope (Radiance 2100; BioRad Laboratories) with a 40× or 60× objective and processed by Photoshop (Adobe).
Immunoblotting
Standard procedures were used. Cells were lysed in 2× Laemmli buffer (Laemmli, 1970), and samples were resolved by SDS-PAGE, transferred onto polyvinylidene fluoride or nitrocellulose membranes, and probed with primary antibodies (Table S1). Secondary antibodies were obtained from Dako.
Peptide pull-downs
Peptide pull-downs were performed as described previously (Stucki et al., 2005). To prepare whole-cell extracts (WCEs), U2OS cells were harvested by mitotic shake-off or scraped in PBS, washed with PBS, and lysed in buffer containing 420 mM NaCl, 20 mM Hepes, pH 7.9, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 0.2% NP-40 supplemented with protease (Roche), and phosphatase (Sigma-Aldrich) inhibitors for 30 min on ice. Lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4°C. 1 mg of U2OS WCE or HeLa nuclear extract (CilBiotech) was incubated with peptide-coupled beads. Beads were washed extensively with Tris-buffered saline (pH 7.5) containing 0.1% Tween 20, and bound proteins were eluted in SDS sample buffer.
FACS
Cells were fixed with 70% ethanol at 4°C followed by incubation for 30 min with 250 µg/ml RNase A and 10 µg/ml propidium iodide at 37°C. Cells were analyzed by FACS on a CyAn flow cytometer (Beckman Coulter). Data were analyzed with FlowJo software (Tree Star, Inc.).
Live cell imaging
Cells were cultured in 35 mm Ibidi dishes (Thistle Scientific) in phenol red-free medium (Invitrogen). Images were acquired with an inverted microscope (Axiovert 200 M; Carl Zeiss, Inc.) equipped with a 37°C heated stage and CO2 chamber (PeCon GmbH) and LSM510 software (Carl Zeiss, Inc.).
Cell survival assays
Mitotic cells obtained by shake-off of the cultures presynchronized by thymidine-nocodazole were treated with ATM and DNA-PKcs inhibitors for 1 h and irradiated with 0.5, 1, 2, or 4 Gy. 30 min later, cells were extensively washed to remove inhibitors and/or nocodazole, counted, and plated. Asynchronous cells were plated 24 h before the treatments. Cells were left for 10–14 d at 37°C to allow colony formation. Colonies were stained with 0.5% crystal violet/20% ethanol and counted. Results were normalized to plating efficiencies. Statistical analysis was performed using the R Language for Statistical Computing. Coefficients and corresponding p-values were calculated separately for mitotic and asynchronous cells using the linear modeling function in R by the following model: log(y) = β0 + β1IR + β2IR2 + β3t + β4IR × t, where y is the percentage of cells surviving, IR is the radiation level in Gy, and t is the treatment (encoded as 0 for DMSO and 1 for ATMi/DNA-PKi). The quadratic term was included because the response curve is nonlinear.
Immunoprecipitation
For ATM immunoprecipitation (Fig. S2 C), cells were lysed for 30 min on ice in buffer containing 20 mM Hepes, pH 7.9, 450 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 0.1% Tween 20, 10% glycerol, Ser-Thr phosphatase inhibitor cocktail (Sigma-Aldrich), and protease inhibitor cocktail (Roche). Extracts were cleared by centrifugation at 14,000 rpm for 30 min at 4°C and diluted with the same buffer lacking NaCl to a final concentration of 150 mM NaCl. Extracts were subsequently incubated with Protein G–coated Dynabeads (Invitrogen) prebound with antibodies for 2 h at 4°C. After five washes with the immunoprecipitation buffer, beads were boiled for 5 min in SDS sample buffer. Eluted proteins were resolved by 4–8% gradient SDS-PAGE and transferred to polyvinylidene fluoride membrane (GE Healthcare). Immunoblotting was performed with the appropriate antibodies (Table S1).
Online supplemental material
Fig. S1 supplements the data in Fig. 1 (A and B), demonstrating formation of γH2AX- and MDC1-containing IRIF in mitotic HeLa, BJ, and MRC5 cells upon DSB induction. Fig. S2 contains controls for the data presented in Fig. 1 (C and D): the effect of the ATM and DNA-PK inhibitors on IRIF formation and DDR markers in the asynchronously growing U2OS cells, IR-induced ATM phosphorylation on S1981 in mitosis, and MDC1 phosphorylation on TQXF motif during mitosis. Fig. S3 supplements Fig. 2 by providing additional examples of 53BP1 exclusion from IRIF in mitotic cells derived from various cell lines and the “untreated” control for the time-lapse images; Fig. S3 also shows altered gel mobility of RNF8 in mitosis and lack of delay in mitotic exit following irradiation of the mitotic cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200911156/DC1.
Acknowledgments
We thank all members of the Jackson laboratory for advice, particularly Jeanine Harrigan and Kate Dry. We are grateful to Jonathon Pines, for his valuable comments and critical reading of the manuscript, Rachael Walker for help with flow cytometry and David Knowles for help with statistical analyses. We also thank Thanos Halazonetis and Grant Stewart for the kind gift of anti-RNF8 and anti-RNF168 antibodies, respectively, Sonya Zabludoff (AstraZeneca) for providing AZD7762, and Yaron Galanty for providing the GFP-RNA168 cell line.
S. Giunta is supported by a U.K. Biotechnology and Biological Sciences Research Council (BBSRC) Cooperative Award in Science and Engineering (CASE) studentship with KuDOS Pharmaceuticals/AstraZeneca (BB/D526 129). R. Belotserkovskaya is funded by a Cancer Research UK Programme Grant C6/A5290. Research in the Jackson laboratory is made possible by core infrastructure funding from Cancer Research UK and the Wellcome Trust, and by funding from the European Community (EU Projects DNA Repair [LSHG-CT-2005-512113] and GENICA).
Footnotes
Abbreviations used in this paper:
- 53BP1
- p53-binding protein 1
- ATM
- ataxia telangiectasia mutated
- BRCA1
- breast cancer gene 1
- DDR
- DNA damage response
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- DSB
- double-strand break
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- MDC1
- mediator of DNA damage checkpoint 1
- MRN
- Mre11–Rad50–Nbs1
- PIKK
- phosphatidylinositol 3-kinase–like kinase
References
- Bakkenist C.J., Kastan M.B. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467 10.1074/jbc.C100466200 [DOI] [PubMed] [Google Scholar]
- Cai S.Y., Babbitt R.W., Marchesi V.T. 1999. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc. Natl. Acad. Sci. USA. 96:2828–2833 10.1073/pnas.96.6.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.F., Yeong F.M. 2010. Safeguarding entry into mitosis: the antephase checkpoint. Mol. Cell. Biol. 30:22–32 10.1128/MCB.00687-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar D., Birraux J., Krempler A., Tchouandong L., Beucher A., Walker S., Stiff T., Jeggo P., Löbrich M. 2007. Chromosome breakage after G2 checkpoint release. J. Cell Biol. 176:749–755 10.1083/jcb.200612047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S.P. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 434:605–611 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- FitzGerald J.E., Grenon M., Lowndes N.F. 2009. 53BP1: function and mechanisms of focal recruitment. Biochem. Soc. Trans. 37:897–904 10.1042/BST0370897 [DOI] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159 10.1158/0008-5472.CAN-04-2727 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y., Sakasai R., Okita N., Asahina K., Mizutani S., Teraoka H. 2005. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem. Biophys. Res. Commun. 336:807–812 10.1016/j.bbrc.2005.08.164 [DOI] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H.Y., Zhai L., Yang C., Nie S., Erdjument-Bromage H., Tempst P., Chang C., Wang H. 2007. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 449:1068–1072 10.1038/nature06256 [DOI] [PubMed] [Google Scholar]
- Juan G., Darzynkiewicz Z. 2004. Detection of mitotic cells. Curr Protoc Cytom. Chapter 7:Unit 7.24 [DOI] [PubMed] [Google Scholar]
- Jullien D., Vagnarelli P., Earnshaw W.C., Adachi Y. 2002. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci. 115:71–79 [DOI] [PubMed] [Google Scholar]
- Kato T.A., Okayasu R., Bedford J.S. 2008. Comparison of the induction and disappearance of DNA double strand breaks and gamma-H2AX foci after irradiation of chromosomes in G1-phase or in condensed metaphase cells. Mutat. Res. 639:108–112 [DOI] [PubMed] [Google Scholar]
- Kato T.A., Okayasu R., Bedford J.S. 2009. Signatures of DNA double strand breaks produced in irradiated G1 and G2 cells persist into mitosis. J. Cell. Physiol. 219:760–765 10.1002/jcp.21726 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Leahy J.J., Golding B.T., Griffin R.J., Hardcastle I.R., Richardson C., Rigoreau L., Smith G.C. 2004. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 14:6083–6087 10.1016/j.bmcl.2004.09.060 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Matsui S.I., Seon B.K., Sandberg A.A. 1979. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc. Natl. Acad. Sci. USA. 76:6386–6390 10.1073/pnas.76.12.6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus K.J., Hendzel M.J. 2005. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell. 16:5013–5025 10.1091/mbc.E05-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov A., Cole R.W., Rieder C.L. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797–1806 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Sedelnikova O.A., Redon C., Pilch D.R., Sinogeeva N.I., Shroff R., Lichten M., Bonner W.M. 2006. Techniques for gamma-H2AX detection. Methods Enzymol. 409:236–250 10.1016/S0076-6879(05)09014-2 [DOI] [PubMed] [Google Scholar]
- Nelson G., Buhmann M., von Zglinicki T. 2009. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle. 8:3379–3383 [DOI] [PubMed] [Google Scholar]
- Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895 10.1016/S0960-9822(00)00610-2 [DOI] [PubMed] [Google Scholar]
- Pinato S., Scandiuzzi C., Arnaudo N., Citterio E., Gaudino G., Penengo L. 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 10:55–69 10.1186/1471-2199-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Rieder C.L. 2001. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 3:E3–E6 10.1038/35050676 [DOI] [PubMed] [Google Scholar]
- Plans V., Guerra-Rebollo M., Thomson T.M. 2008. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 27:1355–1365 10.1038/sj.onc.1210782 [DOI] [PubMed] [Google Scholar]
- Poon R.Y. 2007. Mitotic phosphorylation: breaking the balance of power by a tactical retreat. Biochem. J. 403:e5–e7 10.1042/BJ20070290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W. 1998. Entry into mitosis in vertebrate somatic cells is guarded by a chromosome damage checkpoint that reverses the cell cycle when triggered during early but not late prophase. J. Cell Biol. 142:1013–1022 10.1083/jcb.142.4.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310–318 10.1016/S0962-8924(98)01299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390 10.1083/jcb.151.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S. 2009. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle. 8:1532–1538 [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Stiff T., O’Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P.A. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64:2390–2396 10.1158/0008-5472.CAN-03-3207 [DOI] [PubMed] [Google Scholar]
- Stobbe C.C., Park S.J., Chapman J.D. 2002. The radiation hypersensitivity of cells at mitosis. Int. J. Radiat. Biol. 78:1149–1157 10.1080/09553000210166570 [DOI] [PubMed] [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 123:1213–1226 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19:207–217 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- van Vugt M.A., Gardino A.K., Linding R., Ostheimer G.J., Reinhardt H.C., Ong S.E., Tan C.S., Miao H., Keezer S.M., Li J., et al. 2010. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 8:e1000287 10.1371/journal.pbio.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang M., Wang H., Böcker W., Iliakis G. 2005. Complex H2AX phosphorylation patterns by multiple kinases including ATM and DNA-PK in human cells exposed to ionizing radiation and treated with kinase inhibitors. J. Cell. Physiol. 202:492–502 10.1002/jcp.20141 [DOI] [PubMed] [Google Scholar]
- Williams R.S., Williams J.S., Tainer J.A. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85:509–520 10.1139/O07-069 [DOI] [PubMed] [Google Scholar]
- Xu X., Stern D.F. 2003. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 278:8795–8803 10.1074/jbc.M211392200 [DOI] [PubMed] [Google Scholar]
- Zabludoff S.D., Deng C., Grondine M.R., Sheehy A.M., Ashwell S., Caleb B.L., Green S., Haye H.R., Horn C.L., Janetka J.W., et al. 2008. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol. Cancer Ther. 7:2955–2966 10.1158/1535-7163.MCT-08-0492 [DOI] [PubMed] [Google Scholar]