Abstract
Polymorphisms of the gene encoding the regulator of G protein signaling, subtype 4 (RGS4), may be associated with schizophrenia. Among first-episode schizophrenia patients, they are also associated with dorsolateral prefrontal cortex (DLPFC) volume. The DLPFC is a key region that regulates heritable cognitive functions implicated in schizophrenia pathogenesis. To further understand the relationship of RGS4 variants to schizophrenia, we examined their associations with cognitive functions among schizophrenia patients and their relatives. We analyzed 31 multiplex, multigenerational Caucasian families with schizophrenia recruited on the basis of 2 affected first-degree relatives. All participants underwent a computerized neurocognitive battery that evaluates accuracy and speed (response time) of performance on abstraction/mental flexibility; attention; verbal, spatial, and face memory; and spatial ability. “Tag” single-nucleotide polymorphisms (SNPs) representing common polymorphisms were genotyped. Measured genotype analyses accounting for family relationships were performed using Sequential Oligogenic Linkage Analysis Routines. SNPs rs10917670 (“SNP1”) and rs951439 (“SNP7”) were associated with face memory speed (P = .0003) at a significance level that survived Bonferroni correction (P = .039). The same SNPs have earlier been reported to be associated with schizophrenia. There also were uncorrected associations with rs10917670 (“SNP1”) and rs951439 (“SNP7”) on face memory efficiency (P = .03) and verbal memory efficiency (P = 0.02), rs28757217 on abstraction/mental flexibility speed (P = .02) and verbal memory efficiency (P = .03), SNP18 (rs2661319) on spatial memory accuracy (P = 0.02) and face memory speed (P = .03). RGS4 polymorphisms are associated with variations in cognitive functions and contribute a small but statistically significant proportion of variance in a family-based sample.
Keywords: schizophrenia, genetics, cognition, memory
Introduction
The regulator of G protein signaling, subtype 4 (RGS4), has been implicated in the etiology and pathogenesis of schizophrenia. RGS4 is localized to chromosome 1q23.3 in close proximity to a region that showed linkage to schizophrenia in a meta-analysis.1 Convergent evidence from microarray studies,2 genetic association studies,3–8 as well as imaging studies on first-episode, antipsychotic-naive schizophrenia subjects9 and healthy subjects10 also implicate certain RGS4 variations in the etiology of schizophrenia. On the other hand, some studies on RGS4 polymorphisms11,12 have not yielded significant associations. A meta-analysis of 13 studies13 observed an association of RGS4 variants with schizophrenia, whereas 2 meta-analyses14,15 involving smaller subsets of the samples used in the initial meta-analysis did not show an association. Subsequently, an association study involving a comprehensive list of common RGS4 polymorphisms has been published. It suggests that RGS4 haplotypes associated with schizophrenia may have promoter activity in vitro, providing a possible functional basis for the genetic associations.3 The association of some RGS4 variants with baseline symptom severity and treatment response suggests clinical significance of these variations.16
RGS4 negatively regulates the G protein activation by accelerating the hydrolysis of guanosine triphosphate from Gα to produce guanosine diphosphate and Pi at the G protein–coupled receptors. This reaction shortens the duration of signal transduction of many neurotransmitters of interest to schizophrenia, such as dopamine, glutamate, serotonin, and γ-aminobutyric acid (GABA). Although it is known that RGS4 participates in neuronal signal transduction, the precise mechanisms by which RGS4 variations may confer risk for schizophrenia are unclear.
A potentially useful approach to clarify such mechanisms is to examine the association of RGS4 variants with putative intermediate phenotypes of schizophrenia. Cognitive deficits have been consistently demonstrated among individuals with schizophrenia and their first-degree relatives. A meta-analysis of cognitive deficits in schizophrenia found that intelligence, memory, language, executive function, and attention were impaired in patients compared with healthy controls.17 Some cognitive measures have been suggested as “endophenotypes” or intermediate phenotypes, ie, heritable variables that occur proximal to the clinical syndrome in the chain of pathogenic events.18 Although there are questions regarding the effect size of genetic variations on endophenotypes relative to the clinical syndrome, they are useful markers to investigate the pathways linking the genotype and the phenotype. Significant heritability of cognitive domains among healthy individuals19–21 and among schizophrenia patients22,23 has been documented in several studies. These domains include working memory, accuracy of verbal memory, face memory, spatial processing and emotion identification, and speed of attention and abstraction. Some of these cognitive functions are regulated by the dorsolateral prefrontal cortex (DLPFC), eg, working memory24 and executive function.25 Our group has reported an association between decreased volume of the DLPFC among first-episode, antipsychotic-naive schizophrenia patients and specific RGS4 polymorphisms.9 These observations were replicated and extended by a subsequent study.10 Therefore, a systematic examination of association of RGS4 variants with cognitive performance could provide insight into the impact of underlying candidate gene variations and provide clues to elucidate the molecular pathways associated with such variations.
Variations in schizophrenia susceptibility genes have been associated with cognitive deficits frequently observed in schizophrenia. An exonic polymorphism (rs4380) on the catechol-O-methyltransferase gene (COMT) has been associated with executive subprocesses of working memory.26 This polymorphism leads to the substitution of valine by methionine in the COMT enzyme that results in altered metabolism of synaptic dopamine. Similarly, dysbindin (dystrobrevin binding protein; DTNBP1) and disrupted in schizophrenia-1 (DISC1) have also been associated with variations in cognitive functions in schizophrenia. A 6-SNP haplotype (CTCTAC) of DTNBP1 has been observed to correlate with cognitive decline in schizophrenia.27 DISC1 variations were associated with performance in verbal working memory and rapid visual search.28 While these studies have examined the variance contributed by specific variants of a susceptibility gene, none have been comprehensively interrogated for the common polymorphisms at those genes.
We hypothesized that the RGS4 polymorphisms contribute to the variability of cognitive functions in schizophrenia in line with suggested links between RGS4 variants and different aspects of schizophrenia pathology outlined above. In this study, we used the quantitative trait approach among multiplex, multigenerational families comprising schizophrenia probands. We reasoned that the quantitative approach among related individuals may enhance the prospects of detecting associations. Thus, we examined the association of common polymorphisms of RGS4 with variability in cognitive functions in such families and estimated the proportion of variance in selected cognitive functions attributable to polymorphisms in or near RGS4.
Methods
Clinical Evaluation
The sample consisted of European Americans from 37 multiplex, multigenerational families, recruited at 2 sites. Details of clinical evaluation are given in our previous publication.23 Briefly, the Diagnostic Interview for Genetic Studies (version 2.0), the Family Interview for Genetic Studies, and medical records review were used for the psychiatric evaluation. Trained interviewers with established reliability conducted interviews under the supervision of the investigators. Two investigators blind to the subject group reviewed each case independently and provided Diagnostic and Statistical Manual of Mental Disorders (Fourth Edition) multiaxial lifetime diagnoses. Consensus diagnosis was reached in diagnostic meetings attended by board-certified psychiatrists/psychologists. Complex cases were discussed between the 2 ascertainment sites. At each site, interrater reliability among investigators and interviewers was tested at regular intervals using videotaped interviews and bimonthly joint interviews. Kappa values > 0.8 were maintained. After fully explaining the study procedures, we obtained informed consent from all subjects.
Genetic Analysis
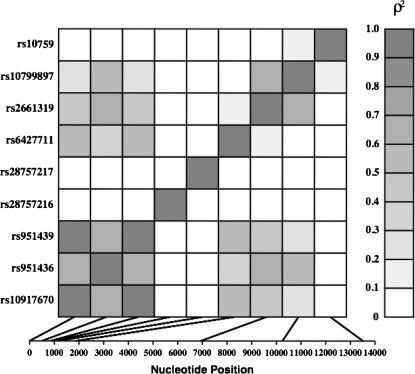
Genomic DNA was collected from all consenting subjects. Based on sequencing, linkage disequilibrium (LD) analysis, and our previous observations, we selected 9 representative “tag” SNPs at a correlation threshold of r2 < 0.80 between loci. In addition, we included the 4 SNPs that were found to be associated with schizophrenia in our previous report.29 The final list of genotyped SNPs were rs10917670 (“SNP1”), rs951436 (“SNP4”), rs951439 (“SNP7”), rs28757216 (“SNP8”), rs28757217 (“SNP9”), rs6427711, rs2661319 (“SNP18”), rs10799897, and rs10759. The SNPs were genotyped using the SNaPshot assay (SNaPshot, ABI Biosystems Inc, Carlsbad, CA)30 and were read independently by 2 investigators. To assure quality control, positive controls (determined by sequencing) were included in all assays. No discrepancies were detected between the positive controls across assays. All SNPs could be genotyped reliably except for SNP4 (rs951436). Therefore, this SNP was not included in the statistical analyses. Two common risk haplotypes have been reported by us and others,29 namely A-T-A-A and G-G-G-G on SNPs 1, 4, 7, and 18, respectively. SNP4 could not be genotyped reliably in this sample; we encountered failures in the assay in a substantial proportion of subjects. SNP1 (rs10917670) and SNP4 (rs951436) are in virtually complete LD with SNP 7 (rs951439) and SNP 18 (rs2661319), respectively (figure 1). Therefore, haplotypes were constructed using SNP7 and SNP18 only.
Fig. 1.
Linkage Disequilibrium Structure of Regulator of G Protein Signaling, Subtype 4, in the Multiplex, Multigenerational Families.
Neuropsychological Methods
A computerized neurocognitive battery (CNB)31,32 validated in both subjects with schizophrenia and healthy comparison subjects was used. The CNB includes a training module and automated scoring with direct data downloading. It evaluates 8 domains: abstraction and mental flexibility, spatial memory, attention, spatial processing, verbal memory, sensorimotor dexterity, face memory, emotion processing.
Two performance indices were calculated for each domain: accuracy and speed. In addition, we derived the efficiency of performance in each domain as a ratio between the accuracy and response time (speed). A brief description of each of these tests is given in table 1. Details regarding the test administration and a description of individual tests are provided in our earlier publication.23
Table 1.
| Cognitive Domain | Test | Description |
| Abstraction and mental flexibility | Penn Conditional Exclusion Test | Four objects are presented at a time, subject selects the object that does not belong with the other 3 based on a sorting principle, sorting principle changes, and the feedback guides further performance |
| Attention | Penn Continuous Performance Test | Subject responds to a set of vertical and horizontal lines in 7-segment displays represent a digit; uses the continuous performance test paradigm |
| Verbal memory | Penn Word Memory Test | Twenty target words are presented followed by forced-choice recognition of these words when mixed with 20 distracters. Words are matched for frequency, length, concreteness, and imageability |
| Face memory | Penn Face Memory Test | Twenty black and white pictures of faces of neutral emotional expression and 40 foils are presented. Immediate and delayed forced-choice recognition of faces |
| Spatial memory | Visual Object Learning Test | Twenty euclidean shapes are presented followed by immediate and delayed recognition when these shapes are presented along with 40 distracters |
| Spatial processing | Judgment of Line Orientation | Two lines at an angle are shown, and subjects are asked to identify the corresponding lines in a simultaneously presented array of lines |
| Emotion processing | Emotion Intensity Discrimination Test | A 40-item test where 2 faces of same emotion are presented. Subject is asked to pick the one with more intense emotion expression |
| Sensorimotor dexterity | — | Subject is shown squares that progressively get smaller on the computer monitor and asked to click on the squares |
Total number of true positive responses and median reaction times are the performance measures.
Statistical Analyses
Variance component–based quantitative trait analyses were conducted using Sequential Oligogenic Linkage Analysis Routines (SOLAR).33 SNP genotypes were coded as 0 for heterozygotes, −1 for one homozygote, and 1 for the other homozygote, and maximum likelihood methods, implemented in SOLAR, were used to test whether the trait mean varies by genotype. A model in which a regression parameter for the SNP is estimated was compared with a model in which the parameter is fixed at 0 for a simple 1 df likelihood ratio test. Differences in trait value by RGS4 genotype were assessed with a fixed-effects model, while accounting for nonindependence of family members by modeling covariance among related individuals. Age and sex were included as covariates. The multivariate t distribution was used in all analyses because the neurocognitive scores showed significant kurtosis. Heritability for each trait was estimated using maximum likelihood variance component–based methods, implemented in SOLAR, and whether heritability was significantly different from 0 was assessed using a likelihood ratio test. We, first, examined the efficiency of cognitive performance in each domain and then the accuracy and speed measures separately.
LD was estimated as ρ2 values using unrelated members of the sample (figure 1). In order to examine the haplotype associations, haplotypes were estimated using Simwalk2.34 These haplotypes were binned such that haplotypes A-A and G-G were considered high risk and haplotypes A-G and G-A low risk, consistent with earlier publications.3,13,29 We tested for a difference in mean trait values by number of high-risk haplotypes carried. Nominal significance of P < .05 (uncorrected) is reported for most comparisons. Where feasible, corrections for multiple testing were made. The corrections accounted for the number of SNPs and the number of cognitive traits tested.
Results
The mean age of the sample was 44.56 ± 18.16 years (range 11–85 years). About 46% were males, and 54% were females. Average family size was 10.51 ± 8.46 members per family with a range of 3–32. Table 2 shows the demographic variables of the study sample. The genotypes and complete neuropsychological data together were available for all subjects on 7 SNPs. The genotypes for 2 SNPs (rs951436 and rs10759) were not available for all subjects; therefore, these 2 SNPs were not included in the analyses. Complete data were available for 327 individuals from 31 families for 7 SNPs (rs10917670 [“SNP1”], rs951439 [“SNP7”], rs28757216 [“SNP8”], rs28757217 [“SNP9”], rs6427711, rs2661319 [“SNP18”], rs10799897).
Table 2.
Demographic Characteristics of the Study Sample
| Demographic Variable | Study Group |
Statistics | |
| Schizophrenia | Relatives | ||
| N | 45 | 282 | χ2 = 4.38, df = 1, P = .036 |
| Sex (male:female) | 27:18 | 122:160 | |
| Age, mean (SD) | 45.58 (11.88) | 44.40 (18.99) | t = 0.56, P = .58 |
| Education, mean (SD) | 12.09 (2.30) | 12.68 (2.36) | t = 1.56, P = .12 |
| Parental education, mean (SD) | 11.51 (4.65) | 11.07 (3.86) | t = 0.68, P = .5 |
Among the cognitive functions examined, variability in face memory efficiency and speed, verbal memory efficiency, spatial memory accuracy, and abstraction and mental flexibility speed were associated with RGS4 polymorphisms. Subjects with schizophrenia who comprised less than 16% of the total sample were included in the analyses. We did not include sensorimotor dexterity in this study because it was not found to be significantly different between the patients and comparison subjects and was not heritable in our previous study on the same dataset.23 Similarly, verbal memory speed and efficiency and accuracy on abstraction and mental flexibility were not heritable in our samples; therefore, these measures were not included in the analyses.23
Among the SNPs examined, SNPs rs10917670 (“SNP1”), rs951439 (“SNP7”), rs28757217 (“SNP9”), rs6427711 and rs2661319 (“SNP18”) showed nominally significant associations with variability in cognitive functions. Table 3 shows significant associations between the SNPs and the cognitive domains. SNPs rs10917670 (“SNP1”) and rs951439 (“SNP7”) were significantly associated with face memory speed (P = .0003). The associations with face memory speed survived Bonferroni correction (P = .039) (19 traits with 7 markers, total tests 133). Whereas the efficiency measures showed nominally significant associations with these SNPs, the accuracy measures did not. Mean trait values for face memory speed for probands, first-degree relatives, and other relatives were similar across the genotypes of these 2 SNPs (rs10917670 [“SNP1”] and rs951439 [“SNP7”]), as expected (CC 0.81 ± 0.46, CT 0.60 ± 1.09, and TT 0.60 ± 0.88 for probands; CC 0.34 ± 0.87, CT 0.55 ± 1.13, and TT 0.88 ± 0.99 for first-degree relatives; and CC 0.49 ± 1.46, CT 0.51 ± 1.05, and TT 0.61 ± 1.02 for second- and third-degree relatives). We also reanalyzed our data based on the ascertainment status, which was based on another affected first-degree relative of the proband. We observed that the face memory speed remained significant (P = .01).
Table 3.
Association of ‘Tag’ SNPs With Cognitive Functions
| ‘Tag’ SNP | Cognitive Measure | Significance Values for Cognitive Measures |
||
| Efficiency | Accuracy | Speed | ||
| rs10917670 (SNP1) | Face memory | .03 | .14 | .0003a |
| Verbal memory | .02 | .09 | b | |
| rs951439 (SNP7) | Face memory | .03 | .14 (NS) | .0003a |
| Verbal memory | .02 | .09 | b | |
| rs28757217 (SNP9) | Abstraction/mental flexibility | b | b | .02 |
| Verbal memory | .03 | .14 (NS) | b | |
| rs6427711 | Face memory | 0.27 (NS) | 0.59 (NS) | .005 |
| rs2661319 (SNP18) | Face memory | 0.11 (NS) | 0.29 (NS) | .03 |
| Spatial memory | .06 | .02 | .92 (NS) | |
Note: Traits that showed significant heritability in our previous report (Gur et al23) were examined. Note that SNP1 and SNP7 show identical results because the SNPs are in complete linkage disequilibrium. SNP, single-nucleotide polymorphism; NS, not significant.
Survived Bonferroni correction for multiple testing.
These measures were not included in the analysis because these measures were not significantly heritable in our report.
Besides, rs6427711 was nominally associated with face memory speed (P = .005). The efficiency of verbal memory showed nominal associations with SNPs rs10917670 (“SNP1”) (P = .02), rs951439 (“SNP7”) (P = .02), and rs28757217 (“SNP9”) (P = .03). Furthermore, rs28757217 (“SNP9”) was also associated with the speed of abstraction/mental flexibility (P = .02). We observed similar association with spatial memory accuracy (P = .02) for rs2661319 (“SNP18”). These associations did not survive Bonferroni correction for multiple tests.
Haplotype frequencies in this sample were as follows: A-A 35%, G-G 41%, G-A 15%, and A-G 9%. Face memory speed and accuracy were not significantly associated with any haplotype. Verbal memory efficiency and accuracy were negatively correlated with the number of risk haplotypes carried (P = .005 for efficiency and .015 for accuracy). Other cognitive traits did not show significant associations.
We estimated the variance contributed by the associated SNPs to heritability of the traits. After accounting for the effects of age and sex, SNPs 1 and 7 explained an estimated 3.5% of the residual trait variance for face memory speed and 1.4% for face memory accuracy and verbal memory scores. Because SNPs 1 and 7 are in complete LD,3 the trait variance may be the joint effect of these SNPs or of an unknown functional variant with which these SNPs may be in LD.
Discussion
We have evaluated a comprehensive set of common RGS4 polymorphisms in moderately sized, multiplex, multigenerational families with schizophrenia for their associations with selected domains of cognitive function that are heritable and known to be impaired among patients and unaffected relatives. The SNPs included those previously reported to be associated with schizophrenia3,4,13 and prefrontal structure and function.9,10 Using an efficient computerized battery, the accuracy, speed, and efficiency of performance were estimated. Two RGS4 variants, denoted SNP1 and SNP7, are involved in most of the associations in this study, which include the speed (response time) for face memory, along with suggestive associations for the efficiency of face memory and verbal memory. The findings are notable because these SNPs are also associated with schizophrenia,3,4,13 and SNP1 showed suggestive associations with the DLPFC volume in a previous study.9 Because SNP1 (rs10917670) and SNP7 (rs951439) are in complete LD (r2 = 1.0) in this sample (figure 1), identical associations with cognitive performance scores for these SNPs provides internal validation for our observations. A previous study also showed that these 2 SNPs are in strong LD (r2 = 0.96).3
Individual SNPs appear to be associated with either accuracy or speed of cognitive traits such as face and verbal memory. It is difficult to say whether the differences reflect LD or whether, as the haplotype analyses suggest, these cognitive traits are influenced by different functional variants in the region. At the individual SNP level, the variants affecting the face memory appear to be in LD with rs951439 (SNP7) alone, whereas verbal memory measures may be affected by putative functional variant(s) in LD with the haplotype comprising rs951439 (“SNP7”) and rs2661319 (“SNP18”). Similarly, the associations of rs6427711 and rs2661319 (SNP18) with face memory may be explained by the LD between these 2 SNPs and SNPs1/7 (r2 between rs6427711 and SNPs1/7 is 0.53, r2 between rs2661319 and SNPs1/7 is 0.50).
At present, molecular mechanisms to explain the associations are unknown, but studies such as ours may help elucidate the chain of events leading from RGS4 variants, through structural changes in certain regions of the brain and cognitive function, and possibly culminating in the diagnostic entity. The order of the events in our model is speculative at present. For example, it is uncertain if and how the cognitive variations are linked with gray matter volume, though prior studies have suggested associations between these variables.35 When such relationships are observed, the nature of association between brain structure and cognitive domain is complex.36 However, functional imaging studies could provide further clues; a recent imaging study showed that rs951436 (“SNP18”) of RGS4 was associated with blood oxygen level–dependent responses in the frontotemporal and frontoparietal networks during a working memory (n-back) task performance.10 It is unclear whether such relationships would hold for facial memory, too. Furthermore, the specificity of the association of RGS4 variations with facial memory requires replication and validation with imaging and electrophysiological studies.
The expression of RGS4 messenger RNA (mRNA) and protein in the brain is region specific.37 For example, the prefrontal and some of the medial temporal regions express RGS4 more densely. Mirnics et al2 reported underexpression of RGS4 in the neocortical regions, including the prefrontal cortex among schizophrenia patients. The associated SNPs in the present study contribute to haplotypes that may alter transcription.3 If prior in vitro results mirror an effect of these SNP/s on mRNA levels in vivo, the associated RGS4 polymorphisms could impact signal transduction at dopamine, glutamate, serotonin, or some GABA neurons. Abnormalities in these neurotransmitters have been associated with cognitive variations.38,39 It is unclear whether such relationships would hold for facial memory, too. Furthermore, the specificity of the association of RGS4 variations with facial memory requires replication and validation with imaging and electrophysiological studies.
The domains of memory and executive functions that showed significant associations in this study have been consistently reported to be impaired among patients with schizophrenia.17 Previous studies have demonstrated that memory and executive functions are the central components of higher order information processing40 that may be one of the core deficits observed in schizophrenia. Face and verbal working memory deficits have been consistently identified in patients with schizophrenia and their relatives.41 For this reason, it is important to investigate the variance contributed by a putative schizophrenia susceptibility gene to variations in cognitive domains. Such efforts could assist in delineating the pathway between these cognitive traits and genetic variations.
The variance contributed to the heritability of cognitive variations by the RGS4 SNP that is relatively (approximately 3.5%) consistent with a multifactorial causation for these traits. This is a lower bound, and the exact variance depends on the strength of the LD between the associated SNP and the presumably ungenotyped functional variant(s). Though Bonferroni corrections may be overly conservative, corrections based on effective number of markers did not alter our findings. Replicate studies using adequately powered samples to examine genome-wide associations are needed to not only replicate the present findings with RGS4 but also account for the rest of the trait variance attributable to genetic factors.
A comprehensive LD map of common SNPs at RGS4 is now available.3 We selected the “tag” SNPs for the current studies, based on the LD patterns among Caucasians3 and in our earlier reported association study among schizophrenia patients.29 However, reliable genotype data could not be obtained for 2 SNPs in the present sample (rs951436, aka “SNP4”) and rs10759. Of these, rs951436 is in strong LD with rs2661319 (aka SNP18, r2 = 0.88),3 so rs2661319 was a suitable surrogate for rs951436 using our chosen correlation threshold for tag SNPs and the loss of information is probably not substantial. On the other hand, rs10759 is not in significant LD with any of the other SNPs assayed here. This SNP is localized to the 3′ untranslated region (3′ UTR), and its function is unknown. It was not associated with schizophrenia in our prior analyses.3
In summary, polymorphisms of RGS4 reported to be associated with schizophrenia were also found to be associated with selected heritable cognitive measures known to be impaired among schizophrenia patients. Our results show that the RGS4 variations were associated with the speed (response time) and accuracy of memory and executive functions. Such findings can help build a model for pathogenesis linking genetic variation, cognitive traits, and the diagnosis. To investigate this model further, multimodal imaging studies on an extended sample are planned. Replication and further molecular analyses of the associations are also warranted.
Funding
National Institute of Mental Health (MH63420, MH56242, MH66263 to V.L.N., MH72995 to K.M.P., MH61622, MH59490 to L.A., MH42191 to R.E.G., MH60722 to R.C.G.).
Acknowledgments
Presented as a “Hot Topic” at the 45th Annual Meeting of the American College of Neuropsychopharmacology, Hollywood, FL, December 3–7, 2006. Nothing to disclose and no conflicts of interest for Prasad, Almasy, Pogue-Geile, Chowdari, and Talkowski). Nimgaonkar has received a research grant from Lundbeck Research USA for unrelated research. R.C. Gur and R.E. Gur have received research grants from Astra Zeneca and Merck for unrelated studies.
References
- 1.Lewis CM, Levinson DF, Wise LH, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- 3.Chowdari KV, Bamne M, Joel W, et al. Linkage disequilibrium patterns and functional analysis of RGS4 polymorphisms in relation to schizophrenia. Schizophr Bull. 2007;34:118–126. doi: 10.1093/schbul/sbm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams N, Preece A, Spurlock G, et al. Support for RGS4 as a susceptibility gene for schizophrenia. Biol Psychiatry. 2004;55:192–195. doi: 10.1016/j.biopsych.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Morris DW, Rodgers A, McGhee KA, et al. Confirming RGS4 as a susceptibility gene for schizophrenia. Am J Med Genet. 2004;125B:50–53. doi: 10.1002/ajmg.b.20109. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Dunham C, Kendler S, et al. Regulator of G-protein signaling 4 (RGS4) gene is associated with schizophrenia in Irish high density families. Am J Med Genet. 2004;129B:23–26. doi: 10.1002/ajmg.b.30078. [DOI] [PubMed] [Google Scholar]
- 7.Cordeiro Q, Talkowshi ME, Chowdari KV, Wood J, Nimgaonkar VL, Vallada H. Association and linkage analysis of RGS4 polymorphisms with schizophrenia and bipolar disorder in Brazil. Genes Brain Behav. 2005;4:45–50. doi: 10.1111/j.1601-183x.2004.00096.x. [DOI] [PubMed] [Google Scholar]
- 8.Fallin MD, Lasseter VK, Avramopoulos D, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad KMR, Chowdari KV, Nimgaonkar VL, Talkowski ME, Lewis DA, Keshavan MS. Polymorphism of the RGS4 gene and the DLPFC morphometry in first episode schizophrenia: a ROI study using the structural magnetic resonance imaging. Mol Psychiatry. 2005;10:213–219. doi: 10.1038/sj.mp.4001562. [DOI] [PubMed] [Google Scholar]
- 10.Buckholtz JW, Meyer-Lindenberg A, Honea RA, et al. Allelic variation in RGS4 impacts functional and structural connectivity in the human brain. J Neurosci. 2007;27:1584–1593. doi: 10.1523/JNEUROSCI.5112-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiguro H, Horiuchi Y, Koga M, et al. RGS4 is not a susceptibility gene for schizophrenia in Japanese: association study in a large case-control population. Schizophr Res. 2007;89:161–164. doi: 10.1016/j.schres.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Sobell JL, Richard C, Wirshing DA, Heston LL. Failure to confirm association between RGS4 haplotypes and schizophrenia in Caucasians. Am J Med Genet B Neuropsychiatr Genet. 2005;139:23–27. doi: 10.1002/ajmg.b.30221. [DOI] [PubMed] [Google Scholar]
- 13.Talkowski ME, Seltman H, Bassett AS, et al. Evaluation of a susceptibility gene for schizophrenia: genotype based metaanalysis of RGS4 polymorphisms from thirteen independent samples. Biol Psychiatry. 2006;60:152–162. doi: 10.1016/j.biopsych.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo S, Tang W, Shi Y, et al. RGS4 polymorphisms and risk of schizophrenia: an association study in Han Chinese plus meta-analysis. Neurosci Lett. 2006;406:122–127. doi: 10.1016/j.neulet.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 15.Li D, He L. Association study of the G-protein signaling 4 (RGS4) and proline dehydrogenase (PRODH) genes with schizophrenia: a meta-analysis. Eur J Hum Genet. 2006;14:1130–1135. doi: 10.1038/sj.ejhg.5201680. [DOI] [PubMed] [Google Scholar]
- 16.Campbell DB, Ebert PJ, Skelly T, et al. Ethnic stratification of the association of RGS4 variants with antipsychotic treatment response in schizophrenia. Biol Psychiatry. 2007;63:32–41. doi: 10.1016/j.biopsych.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fioravanti M, Carlone O, Vitale B, Cinti ME, Clare L. A meta-analysis of cognitive deficits in adults with a diagnosis of schizophrenia. Neuropsychol Rev. 2005;15:73–95. doi: 10.1007/s11065-005-6254-9. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 19.Finkel D, Pedersen NL, Plomin R, McClearn GE. Longitudinal and cross-sectional twin data on cognitive abilities in adulthood: the Swedish Adoption/Twin Study of Aging. Dev Psychol. 1998;34:1400–1413. doi: 10.1037//0012-1649.34.6.1400. [DOI] [PubMed] [Google Scholar]
- 20.McClearn GE, Johansson B, Berg S, et al. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 1997;276:1560–1563. doi: 10.1126/science.276.5318.1560. [DOI] [PubMed] [Google Scholar]
- 21.Tuulio-Henriksson A, Haukka J, Partonen T, et al. Heritability and number of quantitative trait loci of neurocognitive functions in families with schizophrenia. Am J Med Genet. 2002;114:483–490. doi: 10.1002/ajmg.10480. [DOI] [PubMed] [Google Scholar]
- 22.Alfimova M, Uvarova L. Cognitive peculiarities in relatives of schizophrenic and schizoaffective patients: heritability and resting EEG-correlates. Int J Psychophysiol. 2003;49:201–216. doi: 10.1016/s0167-8760(03)00133-8. [DOI] [PubMed] [Google Scholar]
- 23.Gur RE, Nimgaonkar VL, Almasy L, et al. Neurocogntive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164:813–819. doi: 10.1176/ajp.2007.164.5.813. [DOI] [PubMed] [Google Scholar]
- 24.Callicott JH, Mattay VS, Verchinski BA, Marenco S, Egan MF, Weinberger DR. Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry. 2003;160:2209–2215. doi: 10.1176/appi.ajp.160.12.2209. [DOI] [PubMed] [Google Scholar]
- 25.Krawczyk DC. Contributions of the prefrontal cortex to the neural basis of human decision making. Neurosci Biobehav Rev. 2002;26:631–664. doi: 10.1016/s0149-7634(02)00021-0. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg TE, Egan MF, Gscheidle T, et al. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003;60:889–896. doi: 10.1001/archpsyc.60.9.889. [DOI] [PubMed] [Google Scholar]
- 27.Burdick KE, Goldberg TE, Funke B, et al. DTNBP1 genotype influences cognitive decline in schizophrenia. Schizophr Res. 2007;89:169–172. doi: 10.1016/j.schres.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burdick KE, Hodgkinson CA, Szeszko PR, et al. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- 29.Chowdari KV, Mirnics K, Semwal P, et al. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- 30.Mansour HA, Wood J, Logue T, et al. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5:150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- 31.Gur RC, Ragland JD, Moberg PJ, et al. Computerized neurocognitive scanning: II. The profile of schizophrenia. Neuropsychopharmacology. 2001;25:777–788. doi: 10.1016/S0893-133X(01)00279-2. [DOI] [PubMed] [Google Scholar]
- 32.Gur RC, Ragland JD, Moberg PJ, et al. Computerized neurocognitive scanning: I. methodology and validation in healthy people. Neuropsychopharmacology. 2001;25:766–776. doi: 10.1016/S0893-133X(01)00278-0. [DOI] [PubMed] [Google Scholar]
- 33.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 35.Persson J, Nyberg L, Lind J, et al. Structure-function correlates of cognitive decline in aging. Cereb Cortex. 2006;16:907–915. doi: 10.1093/cercor/bhj036. [DOI] [PubMed] [Google Scholar]
- 36.Demakis GJ. Frontal lobe damage and tests of executive processing: a meta-analysis of the category test, Stroop test, and trail-making test. J Clin Exp Neuropsychol. 2004;26:441–450. doi: 10.1080/13803390490510149. [DOI] [PubMed] [Google Scholar]
- 37.Erdely HA, Lahti RA, Lopez MB, et al. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci. 2004;19:3125–3128. doi: 10.1111/j.0953-816X.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- 38.Braver TS, Cohen JD. Dopamine, cognitive control, and schizophrenia: the gating model. Prog Brain Res. 1999;121:327–349. doi: 10.1016/s0079-6123(08)63082-4. [DOI] [PubMed] [Google Scholar]
- 39.Williams GV, Goldman-Rakic PS. Modulation of memory fields by dopamine D1 receptors in prefrontal cortex. Nature. 1995;376:572–575. doi: 10.1038/376572a0. [DOI] [PubMed] [Google Scholar]
- 40.Engle RW, Tuholski SW, Laughlin JE, Conway AR. Working memory, short-term memory, and general fluid intelligence: a latent-variable approach. J Exp Psychol Gen. 1999;128:309–331. doi: 10.1037//0096-3445.128.3.309. [DOI] [PubMed] [Google Scholar]
- 41.Snitz BE, MacDonald AW, III, Carter CS. Cognitive deficits in unaffected first-degree relatives of schizophrenia patients: a meta-analytic review of putative endophenotypes. Schizophr Bull. 2006;32:179–194. doi: 10.1093/schbul/sbi048. [DOI] [PMC free article] [PubMed] [Google Scholar]