Abstract
Previously, we reported the design and properties of alkyne C-lysine conjugates, a powerful and tunable family of DNA cleaving reagents. We also reported that, upon photoactivation, these molecules are capable of inducing cancer cells death. To prove that the cell death stems from DNA cleavage by the conjugates, we investigated intracellular DNA damage induced by these molecules in LNCap cancer cells using single cell gel electrophoresis (SCGE) assays. The observation of highly efficient DNA damage confirmed that lysine acetylene conjugate is capable of cleaving the densely compacted intracellular DNA. This result provides a key mechanistic link between efficient DNA cleavage and cytotoxicity towards cancer cells for this family of light-activated anticancer agents.
1. Introduction
Because double stranded (ds) DNA cleavage is much harder to repair than single stranded (ss) DNA cleavage, ds damage is particularly efficient in inducing self-programmed cell death or apoptosis [1]. A particularly striking example of this efficiency is provided by natural enediyne antibiotics [2]. These compounds, often hailed as the most potent family of anticancer agents [3], produce cleavage of both strands of DNA duplex via two hydrogen abstractions from two opposite strands of DNA backbone by a reactive biradical, p-benzyne, generated from the enediyne core via a process, called the Bergman cyclization [4–6]. However, natural enediynes not only lack selectivity towards cancer cells, but also do not cause the ds cleavage with 100% efficiency. Even the best of them, calicheamicin leads to only 25% cleavage [7]. Thus, design of compounds which are capable of more efficient ds DNA cleavage and combine this efficiency with selectivity towards cancer cells remains the focal point of the anticancer therapeutic agents targeting DNA.
We have found that DNA damaging potential of enediynes can be increased if their reactivity is tuned towards C1–C5 photocyclizations, a new reaction discovered in our lab which leads to incorporation of four rather than two hydrogen atoms from the environment [8, 9].
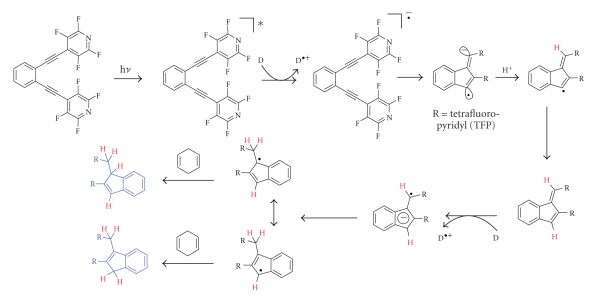
Because C1–C5 cyclization proceeds under photochemical conditions for thermal C1–C5 cyclization, see [10, 11], it takes advantage of the high degree of spatial and temporal controls over reactivity inherent to the photochemical activation. The use of tissue-penetrating light allows for efficient, and selective, spatial and temporal control over prodrug activation as light can be delivered directly to the tumor when it contains a high concentration of the prodrug. Skin cancer is the most obvious target for this therapy and, in 2006, the UK National Institute of Health and Clinical Excellence (NICE) recommended PDT for basal cell carcinoma. However, PDT can be also used to treat tumors on the lining of internal organs or cavities. Other tumors can be targeted with low-energy tissue penetrating photons, especially if the three-dimensional control of activation is provided by the two-photon excitation mode. For two photon excitation of enediynes, see [12–14]. In addition, this radical-anionic C1–C5 cyclization of enediynes is triggered by photoinduced electron transfer (PET). This mechanistic feature increases cellular selectivity because activation is possible only in the direct vicinity of a suitable electron donor such as DNA to occur. In the absence of such a donor, TFP-substituted enediynes (Scheme 1) are unreactive, both thermally and photochemically.
Scheme 1.
C1–C5 photocyclization of bis-TFP-enediyne and proposed mechanism in the proximity to DNA (four abstracted hydrogens are shown in red, TFP = tetrafluoropyridine).
We have also found that related TFP-substituted monoacetylenes are capable of photochemical alkylation of electron rich π-systems [15–17] and investigated whether this reaction can be also used for controlled DNA-modification. A priori, efficient DNA-cleavage by monoalkynes incapable of the Bergman or C1–C5 cyclizations can involve several possible mechanisms like base alkylation, hydrogen abstraction, generation of reactive oxygen species as well as PET.
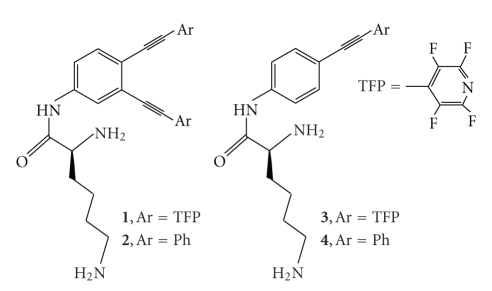
In order to increase solubility of TFP-warheads in water and their affinity to DNA, we combined them with lysine via carboxyl moiety of the amino acid, Figure 1 [18].
Figure 1.
Structures of C-lysine conjugate.
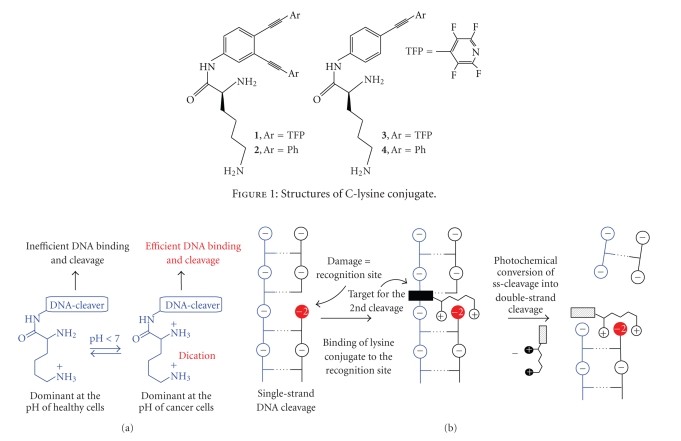
Importantly, this mode of attachment leaves both amino groups of lysine available for an acid-base reaction which converts them into cationic ammonium groups. We found that DNA-damaging ability of such hybrid molecules can be fine-tuned in the narrow range of physiological pH conditions which results in a dramatic increase in reactivity at the lower pH of hypoxic tumor cells [19]. Less basic α-amino group is protonated at the lower pH than 7 and this protonation not only prevents quenching the excited state of the chromophore but also provides tighter binding to negatively charged DNA. Remarkably, the change in reactivity occurs at a relatively narrow and predefined pH point (~pH 6). These DNA-photocleavers provide the DNA cleavage ratios of up to the 1 : 2 ds : ss at pH 5.5 at concentrations and irradiation times where almost no ds cleavage is observed at the pH of healthy cells. This dramatic increase of ds DNA cleavage at the lower pH renders these molecules more efficient ds DNA cleavers than calicheamicine under the conditions suitable for selective targeting of acidic cancer tissues (Figure 2(a)). We also found that the C-lysine conjugates bind selectively to nicks and gaps in a DNA duplex and, upon photochemical activation, transform the easily repairable ss-DNA damage into much more therapeutically important ds-DNA damage [20] (Figure 2(b)).
Figure 2.
(a) Design of pH-dependent DNA-cleavers based on different stages of protonation of the lysine side chain, (b) photochemical conversion of ss-DNA cleavage into more therapeutically important ds-DNA cleavage through lysine-phosphate monoester recognition of the initial damage site.
The medicinal potential of these molecules has been illustrated by a >90% LNCap cancer cell death induced by photochemically activated TFP-acetylene-lysine conjugate 3 in one treatment at concentrations as low as 10 nM. Notably, at these concentrations, toxicity without light is negligible. Similar increases in reactivity upon activation with light were observed in parallel experiments with UMRC3, UMRC6, and 786-O cancer cell lines [19].
In summary, our previous work led to the development of a family of powerful and tunable DNA cleaving reagents which have been shown to cleave both plasmid DNA and DNA oligomers outside of cells [15, 18]. We have also proven that these reagents can induce cancer cells death at the low concentrations. However, our previous work offered no evidence for DNA-damage by TFP-enediynes and acetylenes inside of cells. Such evidence is important because cell death can result from mechanisms other than DNA cleavage and because DNA-cleavage of intracellular DNA should be more difficult since this DNA is compactly organized around histone proteins. The aim of this work is to test the efficiency of our light-activated ds-DNA-cleavers towards intracellular DNA using single cell gel electrophoresis assay which can measure DNA damage in individual eukaryote cells [21–25]. This assay has been used as a standard technique for evaluation of DNA damage/repair, biomonitoring, and genotoxicity testing [26–33]. The alkaline SCGE assay detects both ss and ds DNA damages. The cleaved DNA fragments are able to migrate out of the cell under an electric field after lysis and alkali treatments while undamaged DNA moves slower and remains with the confines of the nucleoid.
2. Materials and Methods
2.1. General Information
Reagent kit for single cell gel electrophoresis assay kit, CometAssay, and control cells containing different levels of DNA damage, CometAssay Control Cell, were purchased from Trevigen, Inc. The CC0 sample corresponds to cells with undamaged DNA whereas CC1, CC2, and CC3 have different levels of DNA-cleavage induced with Etoposide [34]. Miligel FisherBiotech Horizontal Electrophoresis System was used for electrophoresis. Olympus BX61 microscope attached with the DP71 color digital camera was used to take fluorescence images of SCGE assay. The images were qualified by Comet Score 1.5 software (Tritec). Tail moment, the ratio of tail length to head diameter (L/H), DNA percentage in tail, and tail length were used to estimate DNA damage. The tail moment has been regarded as an appropriate index of induced DNA damage by computerized image analysis. It represents both the amount of damaged DNA and the distance of migration by a single number. The tail moment was calculated by multiplying the percentage of DNA in the tail by the tail length; see [35].
2.2. Preparation of LNCap Cells and Their Treatment with Conjugate 3
LNCap cells (P.35) were plated in 6 (100 mm) plates at density of 250,000 cells/well and were maintained in RPMI 1640 medium supplemented with 10% FBS, sodium bicarbonate (2 g/L). When they reach 70% confluence, compound 3 was dissolved in serum-free RPMI 1640 medium supplemented with sodium bicarbonate (2 g/L). After the RPMI 1640, medium containing the compound 3 (0, 10, and 50 μM) were added to the cells and the cells were placed in the incubator for 4 hours. The cells were exposed to UV with cover removed for maximum exposure for 10 minutes and were trypsinized and counted. Solutions in ice cold 1× PBS (Ca2+ and Mg2+ free), with 1 × 105 cells/mL, were prepared based on CometAssay instruction from Trevigen, Inc.
2.3. Alkaline Single Cell Gel Electrophoresis Assay
LMAgarose was melt in boiling water bath for 5 minutes and placed in 37°C water bath for at least 20 minutes to cool. Cells at 1 × 105/mL were combined with molten LMAgarose at a ratio of 1 : 10 (v/v) and 50 μL of the mixture was transferred on CometSlides. The slides were placed at 4°C in the dark for 30 minutes and they were immersed in prechilled lysis solution. After 30-minute immersion at 4°C, the slides were immersed in alkaline solution prepared freshly with NaOH (0.6 g), 200 mM EDTA (250 μL), and dH2O (49.75 mL) for 20 minutes at room temperature, in the dark. Then, the slides were removed from alkaline solution and washed by immersing in 1× TBE buffer for 5 minutes twice. After adding 1× TBE buffer not to exceed 0.5 cm above slides in electrophoresis tank, the voltage at 1 volt per cm was applied for 10 minutes. The slides were immersed in dH2O twice for 10 minutes, then in 70% ethanol for 5 minutes. The samples were dried at ≤45°C for 15 minutes and 100 μL of diluted SYBR Green I was placed on the gels and the slides were stored at refrigerator. After 5 minutes, excess SYBR solution was removed by gentle tapping and the slides were completely dried at room temperature in the dark. The fluorescence images were taken by epifluorescence microscopy.
3. Results and Discussion
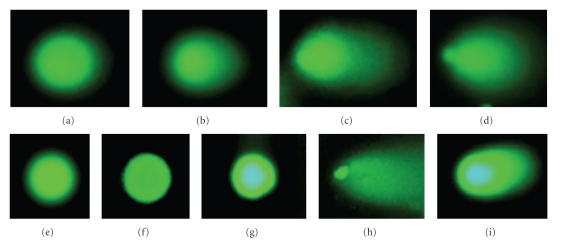
The control SCGE assay results for undamaged cells (CC0) and commercially obtained cells with variable amount of DNA damage (CC1–3) are summarized in the top part of Figure 3 (entries (a)–(d)). As expected, while SCGE assay with healthy cells showed no tails indicative of DNA damage, the assays with the damaged cells produced characteristic tails, the size of which correlates with the extent of DNA damage in these cells. Qualified data of the assays are given in Table 1. With the pretreated control cells (Table 1, (b)–(d)), 33, 47, and 98% of DNA were detected in tails, respectively. Tail moment values are also consistent with different levels of DNA damage.
Figure 3.
Images of SCGE assays. Controls: (a) Undamaged control cell, (b)–(d) Control cells with variable amount of DNA damage. LNCap Cells: (e) No compound + No UV; (f) No compound + UV; (g) 3 (50 μM) + No UV; (h) 3 (50 μM) + UV; (i) 3 (10 μM) + UV. All UV irradiations were carried out for 10 minutes.
Table 1.
Qualified data from SCGE assays.
| Exp. | Tail moment | L/H | %DNA in tail | Tail length (px) |
|---|---|---|---|---|
| (a) | 0 | 0 | 0.3 ± 0.3 | 0 |
| (b) | 9.9 ± 0.9 | 0.4 ± 0.1 | 33.2 ± 2.9 | 29.7 ± 1.5 |
| (c) | 22.1 ± 2.0 | 0.8 ± 0.1 | 47.0 ± 5.5 | 47.7 ± 9.3 |
| (d) | 107.3 ± 7.8 | 6.8 ± 1.4 | 96.8 ± 1.3 | 110.8 ± 6.9 |
| (e) | 0 | 0 | 0.9 ± 1.2 | 0.2 ± 0.4 |
| (f) | 0.1 ± 0.1 | 0 | 2.0 ± 2.3 | 2.2 ± 2.0 |
| (g) | 0 | 0 | 1.3 | 0 |
| (h) | 155.3 ± 62.6 | 4.6 ± 2.0 | 92.0 ± 4.1 | 167.0 ± 62.2 |
| (i) | 26.1 ± 10.1 | 0.7 ± 0.1 | 41.4 ± 9.0 | 61.2 ± 12.8 |
After confirming that assay conditions work in the control cells, we proceed to investigate DNA damage induced by conjugate 3 in LNCap cancer cells. To find whether UV itself or thermal reactions of compound 3 may be responsible for the DNA cleavage in cancer, we included two control experiments with cells exposed to UV for 10 minutes in the absence of a DNA-cleaver (Figure 3(f)) and with cells treated with 50 μM of compound 3 for 4 hours without photochemical activation (Figure 3(g)). No DNA damage is observed in the control cases. This result confirms that neither UV nor compound 3 in the dark can damage DNA under these experimental conditions. In contrast, photochemical activation of 50 μM of compound 3 produced very efficient DNA damage (more than 90% DNA in the tail, Table 1) in individual cells (Figure 3(h)). Irradiation in the presence of 10 μM of compound 3 also showed significant DNA damage (~40% DNA in the tail, Figure 3(i)). These results confirm that compound 3 can penetrate into the nucleus of the cancer cell and damage highly compacted DNA photochemically.
The concentrations of lysine conjugates used in our comet experiments are significantly higher than >0.01 mM concentrations sufficient to cause significant photocytotoxicity to several cancer cells lines. This difference is not limited to the comet assay—our earlier experiments with pure DNA also required micromolar concentrations of the conjugate to observe the cleavage [18, 19]. The observation has two consequences. First, it suggests (somewhat surprisingly) that the efficiency of cleavage for isolated plasmid DNA and compacted cellular DNA is not drastically different, thus indicating that our compounds should accumulate in the cell nucleus rather efficiently.
Second, this observation may indicate the presence of an additional, even more efficient, mechanism for cytotoxicity which may not be based on DNA cleavage. Alternatively, it may also mean that even small amount of DNA cleavage (which is not detected by the conventional, relatively insensitive assays) is still sufficient for causing apoptosis. Although we cannot distinguish between these two mechanisms at this point, this mechanistic ambiguity renders important the observation that lysine-acetylene conjugate can indeed target and damage cellular DNA.
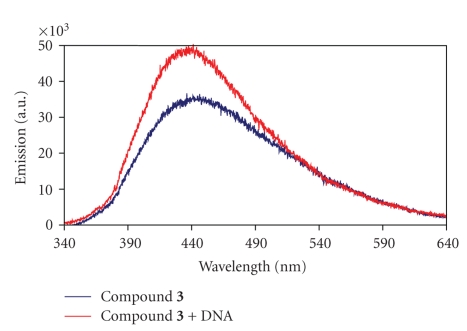
Interestingly, the fluorescence images of cells treated with compound 3 (Figures 3(g) and 3(i)) showed blue fluorescence in the nucleus region on top of the green fluorescence from the DNA-staining dye, SYBR Green I. Because this blue fluorescence is not observed in control cells without the conjugate, the emission is likely to result either from compound 3 itself which has the maximum emission at 440 nm (Figure 4) or from one of the respective photoproducts derived from the DNA-photocleaver. This observation provides additional evidence that conjugate 3 can be uptaken into the nucleus of cancer cells. It is also interesting that there is no residual blue fluorescence in Figure 3(h), where the DNA is broken completely.
Figure 4.
Emission spectrum of 3 (10 μM) in phosphate buffer at pH 7. Blue: without DNA, Red: with 30 μM/b.p of Calf thymus DNA.
4. Conclusions
SCGE assays confirm the occurrence of efficient cleavage of highly compacted intracellular DNA by a light-activated C-lysine acetylene conjugate. This result provides a key mechanistic link between efficient DNA cleavage and significant cytotoxicity in cell proliferation assays.
Acknowledgments
The authors are grateful to the National Science Foundation (CHE-0848686) and TTCP Grant from the James and Ester King Florida Biomedical Research Program to IVA, and DOD Grant W81XWH-07-1-0225 from US Congressionally Directed Medical Research Programs to QXS for partial support of this research.
References
- 1.Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Molecular and Cellular Biology. 1997;17(7):4070–4079. doi: 10.1128/mcb.17.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolaou KC, Smith AL, Yue EW. Chemistry and biology of natural and designed enediynes. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(13):5881–5888. doi: 10.1073/pnas.90.13.5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galm U, Hager MH, Van Lanen SG, Ju J, Thorson JS, Shen B. Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chemical Reviews. 2005;105(2):739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 4.Jones RR, Bergman RG. p-Benzyne. Generation as an intermediate in a thermal isomerization reaction and trapping evidence for the 1,4-benzenediyl structure. Journal of the American Chemical Society. 1972;94(2):660–661. [Google Scholar]
- 5.Bergman RG. Reactive 1,4-dehydroaromatics. Accounts of Chemical Research. 1973;6(1):25–31. [Google Scholar]
- 6.Alabugin I, Breiner B, Manoharan M. Cycloaromatization reactions: the testing ground for theory and experiment. Advances in Physical Organic Chemistry. 2007;42:1–33. [Google Scholar]
- 7.Elmroth K, Nygren J, Mårtensson S, Ismail IH, Hammarsten O. Cleavage of cellular DNA by calicheamicin γ 1 . DNA Repair. 2003;2(4):363–374. doi: 10.1016/s1568-7864(02)00235-5. [DOI] [PubMed] [Google Scholar]
- 8.Alabugin IV, Kovalenko SV. C1-C5 photochemical cyclization of enediynes. Journal of the American Chemical Society. 2002;124(31):9052–9053. doi: 10.1021/ja026630d. [DOI] [PubMed] [Google Scholar]
- 9.Alabugin IV, Manoharan M. Radical-anionic cyclizations of enediynes: remarkable effects of benzannelation and remote substituents on cyclorearomatization reactions. Journal of the American Chemical Society. 2003;125(15):4495–4509. doi: 10.1021/ja029664u. [DOI] [PubMed] [Google Scholar]
- 10.Prall M, Wittkopp A, Schreiner PR. Can fulvenes form from enediynes? A systematic high-level computational study on parent and benzannelated enediyne and enyne-allene cyclizations. Journal of Physical Chemistry A. 2001;105(40):9265–9274. [Google Scholar]
- 11.Vavilala C, Byrne N, Kraml CM, Ho DM, Pascal Jr. RA. Thermal C1-C5 diradical cyclization of enediynes. Journal of the American Chemical Society. 2008;130(41):13549–13551. doi: 10.1021/ja803413f. [DOI] [PubMed] [Google Scholar]
- 12.Kauffman JF, Turner JM, Alabugin IV, et al. Two-photon excitation of substituted enediynes. Journal of Physical Chemistry A. 2006;110(1):241–251. doi: 10.1021/jp056127y. [DOI] [PubMed] [Google Scholar]
- 13.Turro NJ, Evenzahav A, Nicolaou KC. Photochemical analogue of the Bergmancycloaromatization reaction. Tetrahedron Letters. 1994;35(44):8089–8092. [Google Scholar]
- 14.Evenzahav A, Turro NJ. Photochemical rearrangement of enediynes: is a “photo-Bergman” cyclization a possibility? Journal of the American Chemical Society. 1998;120(8):1835–1841. [Google Scholar]
- 15.Breiner B, Schlatterer JC, Kovalenko SV, Greenbaum NL, Alabugin IV. Protected 32P-labels in deoxyribonucleotides: investigation of sequence selectivity of DNA photocleavage by enediyne-, fulvene-, and acetylene-lysine conjugates. Angewandte Chemie. International Edition. 2006;45(22):3666–3670. doi: 10.1002/anie.200504479. [DOI] [PubMed] [Google Scholar]
- 16.Zeidan TA, Kovalenko SV, Manoharan M, Clark RJ, Ghiviriga I, Alabugin IV. Triplet acetylenes as synthetic equivalents of 1,2-bicarbenes: phantom n, π ∗ state controls reactivity in triplet photocycloaddition. Journal of the American Chemical Society. 2005;127(12):4270–4285. doi: 10.1021/ja043803l. [DOI] [PubMed] [Google Scholar]
- 17.Zeidan TA, Clark RJ, Kovalenko SV, Ghiviriga I, Alabugin IV. Triplet acetylenes as synthetic equivalents of 1,2-bicarbenes, part II: new supramolecular scaffolds from photochemical cycloaddition of diarylacetylenes to 1,4-cyclohexadienes. Chemistry—A European Journal. 2005;11(17):4953–4960. doi: 10.1002/chem.200500180. [DOI] [PubMed] [Google Scholar]
- 18.Kovalenko SV, Alabugin IV. Lysine-enediyne conjugates as photochemically triggered DNA double-strand cleavage agents. Chemical Communications. 2005;(11):1444–1446. doi: 10.1039/b417012a. [DOI] [PubMed] [Google Scholar]
- 19.Yang W-Y, Breiner B, Kovalenko SV, et al. C-lysine conjugates: pH-controlled light-activated reagents for efficient double-stranded DNA cleavage with implications for cancer therapy. Journal of the American Chemical Society. 2009;131(32):11458–11470. doi: 10.1021/ja902140m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breiner B, Schlatterer JC, Kovalenko SV, Greenbaum NL, Alabugin IV. DNA damage-site recognition by lysine conjugates. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13016–13021. doi: 10.1073/pnas.0705701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochemical and Biophysical Research Communications. 1984;123(1):291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 22.Sing NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Experimental Cell Research. 1988;175(1):184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 23.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiation Research. 1990;122(1):86–94. [PubMed] [Google Scholar]
- 24.McKelvey-Martin VJ, Green MHL, Schmezer P, Pool-Zobel BL, De Meo MP, Collins A. The single cell gel electrophoresis assay (comet assay): a European review. Mutation Research. 1993;288(1):47–63. doi: 10.1016/0027-5107(93)90207-v. [DOI] [PubMed] [Google Scholar]
- 25.Rojas E, Lopez MC, Valverde M. Single cell gel electrophoresis assay: methodology and applications. Journal of Chromatography B. 1999;722(1-2):225–254. doi: 10.1016/s0378-4347(98)00313-2. [DOI] [PubMed] [Google Scholar]
- 26.Virag P, Brie I, Postescu ID, et al. Comparative study of two evaluation methods for the genotoxic effects of environmental heavy metals on normal cells. Toxicology and Industrial Health. 2009;25(4-5):253–258. doi: 10.1177/0748233709103411. [DOI] [PubMed] [Google Scholar]
- 27.Coronas MV, Pereira TS, Rocha JAV, et al. Genetic biomonitoring of an urban population exposed to mutagenic airborne pollutants. Environment International. 2009;35(7):1023–1029. doi: 10.1016/j.envint.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Azqueta A, Shaposhnikov S, Collins AR. DNA oxidation: investigating its key role in environmental mutagenesis with the comet assay. Mutation Research: Genetic Toxicology and Environmental Mutagenesis. 2009;674(1-2):101–108. doi: 10.1016/j.mrgentox.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Wirzinger G, Weltje L, Gercken J, Sordyl H. Genotoxic damage in field-collected three-spined sticklebacks (Gasterosteus aculeatus L.): a suitable biomonitoring tool? Mutation Research: Genetic Toxicology and Environmental Mutagenesis. 2007;628(1):19–30. doi: 10.1016/j.mrgentox.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Migliore L, Colognato R, Naccarati A, Bergamaschi E. Relationship between genotoxicity biomarkers in somatic and germ cells: findings from a biomonitoring study. Mutagenesis. 2006;21(2):149–152. doi: 10.1093/mutage/gel012. [DOI] [PubMed] [Google Scholar]
- 31.Kim B-S, Park JJ, Edler L, et al. New measure of DNA repair in the single-cell gel electrophoresis (comet) assay. Environmental and Molecular Mutagenesis. 2002;40(1):50–56. doi: 10.1002/em.10090. [DOI] [PubMed] [Google Scholar]
- 32.Kassie F, Parzefall W, Knasmüller S. Single cell gel electrophoresis assay: a new technique for human biomonitoring studies. Mutation Research: Reviews in Mutation Research. 2000;463(1):13–31. doi: 10.1016/s1383-5742(00)00041-7. [DOI] [PubMed] [Google Scholar]
- 33.Garaj-Vrhovac V, Kopjar N. The comet assay—a new technique for detection of DNA damage in genetic toxicology studies and human biomonitoring. Periodicum Biologorum. 1998;100(3):361–365. [Google Scholar]
- 34.Walles SAS, Zhou R, Liliemark E. DNA damage induced by etoposide; a comparison of two different methods for determination of strand breaks in DNA. Cancer Letters. 1996;105(2):153–159. doi: 10.1016/0304-3835(96)04266-8. [DOI] [PubMed] [Google Scholar]
- 35.Hellman B, Vaghef H, Bostrom B. The concepts of tail moment and tail inertia in the single cell gel electrophoresis assay. Mutation Research. 1995;336(2):123–131. doi: 10.1016/0921-8777(94)00047-a. [DOI] [PubMed] [Google Scholar]