Abstract
The Drosophila dumpy gene consists of seventy eight coding exons and encodes a huge extracellular matrix protein containing large numbers of epidermal growth factor-like (EGF) modules and a novel module called dumpy (DPY). A molecular analysis of forty five mutations in the dumpy gene of Drosophila melanogaster was carried out. Mutations in this gene affect three phenotypes: wing shape, thoracic cuticular defects, and lethality. Most of the mutations were chemically induced in a single dumpy allele and were analyzed using a nuclease that cleaves single base pair mismatches in reannealed duplexes followed by dHPLC. Additionally, several spontaneous mutations were analyzed. Virtually all of the chemically induced mutations, except for several in a single exon, either generate nonsense codons or lesions that result in downstream stop codons in the reading frame. The remaining chemically induced mutations remove splice sites in the nascent dumpy message. We propose that the vast majority of nonsense mutations that affect all three basic dumpy phenotypes are in constitutive exons, whereas nonsense mutants that remove only one or two of the basic functions are in alternatively spliced exons. Evolutionary comparisons of the dumpy gene from seven Drosophila species show strong conservation of the 5′ ends of exons where mutants with partial dumpy function are found. In addition, reverse transcription PCR analyses reveal transcripts in which exons marked by nonsense mutations with partial dumpy function are absent.
“All things considered, it is better to have a mutant than not to have a mutant.”
Gerry Finkca 1970
Introduction
The history of the dumpy gene in Drosophila melanogaster encompasses virtually the entire history of Drosophila genetics itself. Early last century, several mutants which initially seemed to have different phenotypes were recovered by the Morgan lab at Columbia University. Morgan himself noticed a fly in August of 1910 with shortened wings which he called Truncate [1]–[3]. A fly with pits on the thorax and whorls of the bristles was found in 1916 and termed vortex-II due to its location on the second chromosome [4]. In 1918 [5] a fly was discovered with both shortened wings and with whorls of bristles and hairs on the thorax. This mutant was called dumpy, the first time this term was used. These mutants, along with a second vortex mutant and another mutant named thoraxate showing thoracic vortices and homozygous lethality, were eventually combined by Bridges and Muller as a series of recessive allelomorphs possibly occurring in different parts of a single gene [6]–[8]. In the 1950s, Elof Carlson, then at UCLA, and his students generated a large number of dumpy mutant alleles, primarily with chemical mutagens [9]–[13]. A genetic fine structure map with discrete subloci was developed culminating in the map published by Dale Grace in 1980 [14]–[17].
Beginning in the middle of the last century, Drosophila geneticists defined and analyzed a number of complex loci. Like dumpy, these genes were characterized primarily by mutations with different and sometimes overlapping phenotypes, complex patterns of complementation, and genetic fine structure maps exhibiting separable clusters of mutant sites called subloci. With the advent of molecular cloning and sequencing, the underlying basis for the phenotypic complexity and the complementation patterns of many, if not most, of those loci could be explained. In addition, different functions could be assigned to groups of mutant alleles mapping at discrete subloci in fine structure maps of the genes. Two genes where cloning and sequencing provided explanations for their complexity are rudimentary, where complementing mutants affect distinct domains in the protein [18] and cut, where complementing mutants map either in the regulatory region or in the coding exons of the gene [19]. In contrast, the complexity of the dumpy gene in Drosophila melanogaster, despite being cloned and sequenced [20], has remained unexplained.
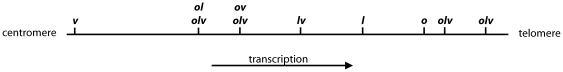
Recessive mutant alleles of dumpy have three primary effects: oblique (dp o) that affect the shape of the wing, vortex (dp v) that disrupt the attachments of indirect flight muscle to the dorsal thoracic cuticle causing pits and protrusions, and lethal (dp l) acting mostly at larval moults. The oblique and vortex phenotypes are shown in Figure 1, b and d respectively from Wilkin et al. [20].
Figure 1. Fine structure genetic map of the subloci in the dumpy gene based on the mutants examined in this study (adapted from Grace [17]).
The map is drawn approximately to scale in terms of recombinational distances. Classes of dumpy mutant alleles found at each sublocus are shown above the line, and the direction of transcription is shown below the map. Dp olv mutations are found at many sites throughout the gene.
Pleiotropic individual alleles of dumpy, shown in Table 1, can exhibit any combination of the three mutant phenes, and heteroallelic heterozygotes will show the phenotype of the homozygous “alleles”, e.g. dp olv /dp v flies will be viable with normal wings but mutant for vortex.
Table 1. Phenotypes and complementation patterns of the classes of dumpy mutant alleles.
| Allele | Phenotype | Phenotypes of heterozygotes | ||||||
| dp o | dp v | dp l | dp ov | dp ol | dp lv | dp olv | ||
| dp o | oblique | O | + | + | O | O | + | O |
| dp v | vortex | V | + | V | + | V | V | |
| dp l | lethal | L | + | L | L | L* | ||
| dp ov | oblique, vortex | OV | O | V | OV | |||
| dp ol | oblique, lethal | L | L* | L* | ||||
| dp lv | vortex, lethal | L | L* | |||||
| dp olv | oblique, lethal, vortex | L* | ||||||
+ = wild type, O = oblique wings, V = vortex, L = lethal.
*may show interallelic complementation for lethality.
Importantly, there are also cases of intragenic or interallelic complementation between some dp ol, dp lv, dp l, and dp olv alleles—marked with an asterisk in Table 1—revealing additional genetic complexity, presumably reflecting different biological roles for dumpy at different developmental stages.
The large size of the dumpy gene (the largest euchromatic gene in Drosophila) has made the construction of fine structure maps of the locus feasible. A detailed map—adapted from Grace's paper [17] to include just the mutants analyzed in this study —is shown in Figure 1. Note that dp ol, dp ov, dp v, dp lv, and dp l, alleles occupy recombinationally distinct subloci, whereas dp olv alleles are found throughout the locus. In Grace's original genetic map, dp o alleles also mapped at several places in the gene.
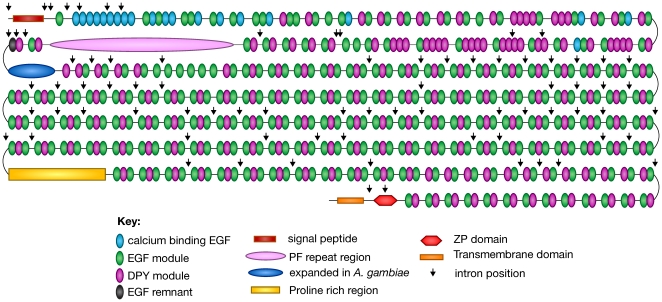
As shown in Figure 2, dumpy encodes a large protein comprised of more than 300 epidermal growth factor (EGF) repeats, a class of modules found in many extracellular matrix (ECM) proteins. Most of the EGF modules are interspersed with a novel repeat of 21 amino acids, which we have termed the DPY module, and much of the Dumpy protein is composed of contiguous repeats of a three-module EGF-DPY-EGF unit. The EGF-DPY-EGF repeats are interrupted by an insert of a repetitive, proline-rich sequence (PR) and by approximately 40 tandem, nearly identical copies of a novel 102 amino acid repeat which we call “PIGSFEAST” (PF) (since the single letter amino acid code of the repeat contains these two “words”). Our lab recently showed the PIGSFEAST region is evolving in a concerted fashion, most likely by unequal crossing over [21], [22]. Near Dumpy's N-terminus are found copies of a sub-class of Ca2+ binding EGF modules, and near its C-terminus there is a single Zona Pellucida (ZP) domain, found in a number of important ECM proteins where they mediate homotypic and heterotypic covalent crosslinking to other ZP domains.
Figure 2. Modular structure of the dumpy gene product.
Adapted from Wilkin et al. [20]. Modules are designated as shown in the key. Note that a large part of the protein is composed of EGF-DPY-EGF triads, with two repeated regions containing PIGSFEAST (PF) and Proline rich repeats respectively. The N-terminus is enriched in calcium binding EGF repeats and the C-terminus contains a transmembrane domain and a Zona Pellucida (ZP) domain. The arrowheads denote the positions of introns in the gene.
Dumpy, along with two other ZP domain proteins, Piopio and Papilotte, function in the adhesion of the apical surface of the Drosophila wing epithelia to the overlying cuticle, and loss of function of each of these three genes results in a blistering phenotype in the wing [23], [24]. A further role of Dumpy in cuticle adhesion is revealed by certain larval lethal dumpy mutations that fail to molt due to a failure of cuticle detachment rather than a failure of adhesion. Dumpy also plays an important role in the epithelial cells that mediate the attachment of the muscles to the overlying cuticle. As mentioned above, dumpy vortex (dp v) mutations result in depressions or pits in the cuticle where it overlies the muscle attachment sites. During embryogenesis, dumpy is expressed in many tube-forming structures that form an apical ECM that lines their internal lumens. These include the salivary gland, fore and hind-gut, and developing trachea [20]. Certain embryonic lethal dumpy mutations result in failure of tracheal cells in the small vessels to connect to form tubes [25]. The effect of dumpy mutations on the trachea may be responsible for the lethal phenotype of dp l, dp ol, dp lv, and dp olv mutations. Hence, Dumpy has functionally diverse roles including cell adhesion, ECM assembly and mechanical properties, morphogenesis and tube formation, and as a ZP domain containing protein may interact with and modulate developmental signaling pathways [25], [26].
In this paper, we identify the molecular lesions responsible for some 45 dumpy mutants including examples of each kind of mutant allele shown in Table 1, and those that either complement or fail to complement other alleles. We report in this paper that most dumpy mutants are directly due to or lead to downstream nonsense codons, even when the mutation disrupts only one or two of the three basic mutant phenotypes. We propose that such mutants mark alternatively spliced exons whereas mutants which affect all three phenotypes (dp olv) are located in constitutive exons. We provide some experimental evidence for this hypothesis using RT-PCR analyses. We also discuss the possibility that the complementation of certain dp olv mutations results from trans-splicing. Hence, alternative cis and trans-splicing events generating different and perhaps tissue specific Dumpy isoforms can provide a rationale for the complexity of this long studied Drosophila gene.
Results
Properties of chemically induced dumpy mutations
The crosses employed in the screens for EMS induced dumpy mutations in defined isoallelic backgrounds are outlined in Table 2. The distribution of mutations from crosses 1, 2A, and 2B is as follows: 60 dp olv, 32 dp ol, 7 dp lv, 2 dp ov, and 2 are dp o. Like Jenkins [13], the majority of our mutants were dp olv. However, in our case dp ol mutants outnumbered dp lv mutants. From the screens depicted in crosses 2A and 2B in Table 2, we recovered 90 transmitted dumpy mutants, 46 from cross 2A in the net chromosome and 44 from cross 2B in the clot chromosome. All of these mutations are in an identical dumpy allele derived from an isofemale line from Australia. The mutants, along with the flanking visible marker, their dumpy phenotypes, and the balancer chromosomes are listed in Table S1. The cross schemes followed in Table 2 also allowed us to detect dumpy lethal alleles which complement the dp lvI mutation in the CyO balancer chromosome.
Table 2. Crosses used to produce dumpy mutants in defined chromosomal backgrounds.
| Cross | Mutagenized males | Females | F1 phenotype |
| 1 | cn bw- 2nd chromosome isogenic | dp ov cl | oblique, vortex |
| 2A | net, dp + isoallele* | net dp ov cl | oblique, vortex |
| 2B | cl, dp + isoallele | dp ov cn bw | oblique, vortex |
| 3 | cl; e(dp v) dp + isoallele | dp v1; e(dp v) | vortex |
| 4 | net; dp + isoallele | In(2LR) Gla/dp lv cl; e(dp v) | vortex |
*A single dp + allele isolated from a wild type strain collected in Australia. See text for details.
We initiated the screens designated as crosses 3 and 4 in Table 2 to enrich for dumpy vortex mutants, since none was recovered from crosses 1 and 2. Cross 3, in which F1 males carrying the dp v1 mutant from the Bloomington stock collection in the presence of the e(dp v) mutation on the 3rd chromosome were scored, produced eleven dp olv mutations, one which complements dp lvI, and two new dp lv alleles. These are listed in Table S1 as dp olvRX or dp lvRX respectively. Since alleles with oblique phenotypes also came through this screen, we set up cross 4 in Table 2, this time examining F1 males carrying a previously generated dp lv allele and homozygous for e(dp v). 24,000 F1s were scored and four complementing dp olv mutants were obtained, along with a single dp ov mutant allele. Hence this screen appears to enrich for complementing dp olv mutations. We will discuss below how such mutations may help identify putative trans-splicing events in the dumpy gene.
Molecular basis of dumpy mutations
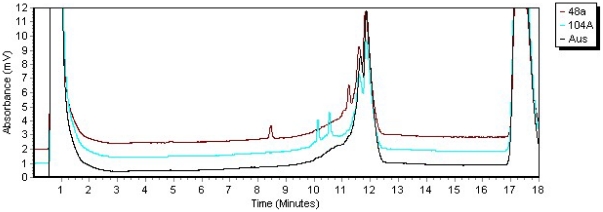
We outline in the Materials and Methods section the approaches we've taken to characterize preexisting dumpy mutations, those we generated in strains isoallelic for a wild type dumpy allele from Australia, and those recovered in a screen for spontaneous mutations. Most of these analyses relied on the generation of overlapping amplicons across the entire gene and the use of the WAVE dHPLC machine from Transgenomic, Inc. to detect cleavage fragments generated by the Surveyor nuclease at the sites of base pair mismatches. A typical dHPLC chromatogram is shown in Figure 3, where two mutations dp olv48a and dp olv104A are located in an amplicon from exon 11.
Figure 3. dHPLC patterns of two dumpy mutants, dp 48a and dp 104A, located in exon 11 and the wild type progenitor allele from Australia.
The elution of the two cleavage fragments generated by the Surveyor nuclease from each mutant are shown in dark red for dp 48a and light blue for dp 104A.
Table 3 and Figure 4 show our results to date. Clearly this approach is very effective in detecting and identifying mutations in the dumpy gene. The data are remarkable in that most of the mutations, including dp ol and dp lv mutants, result in stop codons either at the site of the mutation, are generated from a deletion, or cause the removal of a splice site. It is interesting that all missense mutations identified to date change cysteines in the protein. Given the repetitive nature of the Dumpy protein, i.e. all the EGF-DPY-EGF motifs, perhaps most missense mutations don't produce a visible phenotype.
Table 3. Results of molecular analyses of selected dumpy mutations.
| Mutant | Origin | Allele Class | Exon | Mutation or Deletion | Effect |
| v2 | SC | v | 5′ region | roo transposon | unknown |
| 1C5 | BM | olv | 3 | G->A | removes splice site |
| 36a | MG | ol | 5 | TGC->TAC | Cys->Tyr |
| 38a | MG | ol | 6 | 2bp deletion | frameshift and stops |
| 67b | MG | ol | 7 | TGT->TGA | Cys->STOP |
| 2P1 | BM | ol | 7 | CAA->TAA | Gln->STOP |
| 71a | MG | ol | 7 | TGG->TAG | Trp->STOP |
| 18b | MG | ol | 7 | TCG->TAG | Ser->STOP |
| D1311A | OG | ol | 7 | 15bp inversion | creates a STOP |
| 2G1A | BM | olv | 9 | CAA->TAA | Gln->STOP |
| 105A | MG | olv | 11 | CAA->TAA | Gln->STOP |
| 56a | MG | ov | 11 | TGT->TAT | Cys->Tyr |
| 7b | MG | ov | 11 | TGC->CGC | Cys->Arg |
| 104A | MG | olv | 11 | TGC->AGC | Cys->Ser |
| 61B | MG | olv | 11 | TGT->CGT | Cys->Arg |
| A12 | RM | ov | 11 | TGC->TAC | Cys->Tyr |
| 27B | MG | olv | 11 | TGC->TAC | Cys->Tyr |
| ovDG2 | SC | ov | 11 | TGT->TAT | Cys->Tyr |
| 48a | MG | olv | 11 | TGC->TCC | Cys->Ser |
| ov1 | SC | ov | Intron 11 | blood transposon | unknown |
| 6 | MG | olv | 15 | 4bp deletion | frameshift->STOP |
| R11 | RM | olv | 19 | CGA ->TGA | Arg->STOP |
| D2011A | OG | olv | 19 | 16bp deletion | frameshift->STOP |
| G8202B | OG | olv | Intron 21 | 6bp deletion | unknown |
| R4 | RM | olv | 33 | AGA->TGA | Arg->STOP |
| 2C1 | BM | olv | 34 | TGT->TGA | Cys->STOP |
| 89a | MG | olv | 34 | CAG->TAG | Gln->STOP |
| G3030B | OG | lv | 40 | 89bp deletion | frameshift ->STOP |
| L2311B | OG | lv | 40 | TAC->TAA | Tyr->STOP |
| 23b | MG | lv | 43 | 368bp deletion | frameshift |
| H1230B | OG | lv | 43 to 45 | 1140bp deletion | frameshift |
| lvR2 | RM | lv | 43 | G->A | removes splice site |
| 16 | MP | lv | 45 | 10bp deletion | frameshift and stops |
| 12 | MP | lv | 46 | 139bp deletion | removes splice site |
| 7a | MG | lv | 46 | CAG->TAG | Gln->STOP |
| D1191A | OG | lv | 47 | CAA->TAA | Gln->STOP |
| 65f | MG | lv | 48 | CAG->TAG | Gln->STOP |
| P1129B | OG | lv | 49 | 1bp deletion | frameshift |
| lDG82 | SC | l | 58 | GAG->TAG | Glu->STOP |
| o2 | SC | o | 72 | TGT->TAT | Cys->Tyr |
| o14b | MG | o | 72 | TGT->TAT | Cys->Tyr |
| 5B1 | BM | olv | 73 | CAA->TAA | Gln->STOP |
| 12B1 | BM | olv | 76 | 1482bp deletion | unknown |
| 21C2 | BM | olv | 76 | CAG->TAG | Gln->STOP |
| R3 | RM | olv | 76 | CAG->TAG | Gln->STOP |
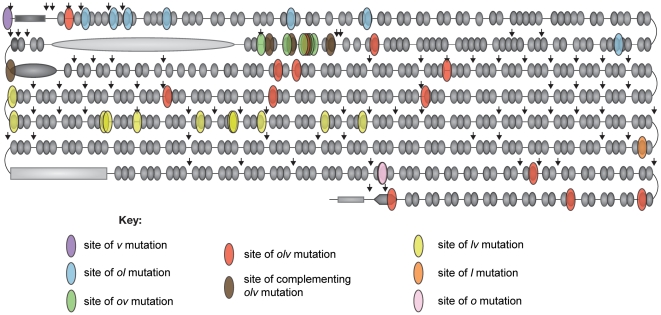
Figure 4. Positions of the sequenced dumpy mutations in the gene product as diagrammed in Figure 2 .
Note the regional localization of dp v (purple), dp ol (blue), dp ov (green), complementing dp olv (brown), dp lv (yellow), dp l (orange), and dp o (pink) mutations. Dp olv mutations (red) are located at several different sites in the protein.
The dp olv mutants are found scattered throughout the locus. Again, most identified so far introduce a stop codon or otherwise lead to a truncation of the protein via a frameshift, or remove a splice junction. It should be noted that this leads to the same severe class of phenotype whether the predicted molecular product is a short N-terminal region or includes the majority of the extracellular domain. It might be expected that generating such a long fragment of the protein would allow the mutant to act as a dominant negative. However, all of the dp olv mutants behave as straightforward recessive, loss of function alleles. This implies that, to retain any function, the product must translate through to the C-terminus.
In contrast to the dp olv mutant sites which are located at many different places in the gene, an observation that is consistent with their many sites in Grace's fine structure map [17], most of the other dumpy mutants are found clustered in discrete regions. We will discuss each of these clusters, proceeding from the 5′ to the 3′ end of the gene.
dp v mutants
To date, we've not recovered any EMS induced vortex mutants in the dumpy allele in 2nd chromosomes marked with the net and clot mutants. We did, however, examine pre-existing dp v alleles, dp v1 and dp v2 [4], [7], [8]. Both were originally recovered in the Morgan laboratory at Columbia University early last century. Using the primers listed in Table S2, we generated amplicons across the entire gene from each homozygous vortex mutant except with primers 5′19F and 5′19R. We then used long range PCR (see Materials and Methods) and recovered amplicons approximately 10kb in length from each mutant. Sequences from the ends of these amplicons indicated the presence of a roo element at bp 15448 upstream of the start codon, in the 5′ region of the gene in both alleles. If the roo element is responsible for the vortex mutant phenotype, its position in the gene is consistent with the position of the vortex sublocus in Grace's fine structure map. We will discuss the identity of the two supposedly independently isolated alleles below.
dp ol mutants
Note in Table 3 that these seven mutations localize to exons 5, 6, and 7 near the N-terminus of the protein, again consistent with the position of the ol sublocus in Grace's map. Except for dp ol36a, these mutations produce or result in a stop codon. Dp ol36a is a missense mutation in which a cysteine residue in a Ca2+ binding EGF motif is replaced with a tyrosine. The cysteine residues in these short motifs (ca 35 amino acids) are essential for their correct tertiary structure due to their participation in disulfide bonds [20]. The position of this missense mutation is in exon 5 and is the dp ol mutant closest to the 5′ end of the gene (see Figure 4).
dp ov mutants
All of these mutations are in exon 11 or in the adjacent intron. The mutant dp ov1, discovered by Morgan in 1918 [5], has been the canonical dumpy mutant used by many investigators in the last century. It is characterized by full penetrance and intermediate expressivity, e.g. virtually all females in dp ov1 containing stocks have oblique wings with an intermediate score of 3 on Dale Grace's scale [17], [27]. Males exhibit a lower penetrance and lower oblique scores on the Grace scale. We examined dp ov1 after finding the EMS induced dp ov mutations in the amplicon from exon 11 by sequencing. No nonsynonymous changes were found nor any changes affecting the canonical splice sites. Intron 10 and intron 11 were then sequenced. The primers used to amplify a region of intron 11 failed to produce a product from dp ov1 DNA. Long range PCR, however, did produce a product containing a blood transposon. This insertion is in the intron just preceding the large exon encoding the PIGSFEAST repeats that are undergoing concerted evolution [21], [22]. The EMS induced dp ov mutants, including dp ovDG2 from Dale Grace, are missense mutants affecting cysteine residues in the EGF-DPY diads that characterize exon 11. Curiously, except for dp ol36a which affects exon 5 and both dp o mutants affecting exon 72, exon 11 is the only other exon in which missense mutations have been recovered.
dp lv mutants
We have characterized 11 dp lv mutations and most either directly generate a stop codon or are out of frame deletions (see Table 3). The EMS generated mutant, dp lvR2, is a G to A transition removing a splice site between exons 43 and 44, and none of the dp lv mutants is due to an amino acid substitution, although the region of the protein affected, viz. exons 40–49 consists of consecutive repeats of EGF-DPY-EGF triad domains whose tertiary structures are surely stabilized by cysteines participating in disulfide bonds (see Figure 3c in Wilkin et al. [20]). Note also that there are no dp olv mutations located in the dp lv region, nor did Grace map any dp olv mutations in the lv sublocus. Once again the positions of the dp lv mutations in the protein are colinear with the position of the lv sublocus in Grace's map.
dp l mutant
We did not recover any dp l mutants in our screen for EMS induced mutants, nor did we expect to given the design of the cross scheme. We screened F1s for oblique and/or vortex phenotypes over dp ov1 or dp lvR1 alleles. Indeed, it is difficult to envision an F1 screen for mosaics which would allow for the recovery of dp l mutations. A search of the literature including Masters and PhD theses did not reveal how such mutants were recovered. Nevertheless, we were able to obtain two mutants, dp lDG82 and dp lDG83, from the Kyoto stock center induced by Dale Grace [15], [17]. Crosses with these mutually non-complementing mutants do indeed confirm their status as dp l mutants, i.e. they produce wild type F1 adults when crossed to dp o, dp v, or dp ov mutants and F1s from crosses to dp ol, dp lv, and most dp olv flies do not survive to adulthood (see Table 1). We determined that dp lDG82 is due to a nonsense mutation in exon 58. The dp l mutant, identified as distinct recombinationally from the lv sublocus, is also molecularly discrete from the exons marked by dp lv mutations.
dp olv mutants
Grace mapped dp olv mutations at many different sites in the gene, and we also find these mutants at many different places in the Dumpy protein. For example, dp olv1C5 affects the 3rd exon, dp olv2C1 and dp olv89a are both stop codons in exon 34, whereas dp olv21C2 and dp olvR3 are due to nonsense codons in exon 76 which encodes the ZP domain very near the C-terminus. Except for those mutations in exon 11 and dp olv1C5, which results in the removal of a splice site, dp olv mutants result from either stop codons or deletions, which generate frameshifts and downstream stop codons. Dp olv12B1 is a very large in frame deletion which removes a large portion of exon 76. Again, in agreement with Grace's genetic map, no dp olv mutations are found in the exons of the ol, lv, or l subloci. The ov sublocus, presumably encompassing only exon 11 and an adjacent intron, is another story. Here Grace mapped dp ov, dp olv, and dp o mutations at the same site, given the limited resolving power of recombination in a higher eukaryote such as Drosophila. We too find both dp olv and dp ov mutations in exon 11, but curiously, and except for dp olv105A, the dp olv mutations in this exon and only this exon are missense mutations, all four of which substitute a different amino acid for a cysteine residue. We will discuss below how dumpy mutations with several different phenotypes could be found in a single exon. We have also observed that certain dp olv mutations will complement other dumpy lethal alleles, particularly other dp lv and dp ol mutations. In these cases, the surviving F1s show good viability but will exhibit vortices or have oblique wings respectively. In these cases the complementing mutations result from a stop codon. We also find cases of complementation between different dp olv mutants for example, dp olv104A/dp olv6 F1s are fully viable but have vortices and oblique wings. Note in Figure 4 that complementing dp olv mutants we have analyzed appear to closely flank the highly repeated PIGSFEAST region, and indeed, all but dp olv6 are located in exon 11. Three of the complementing dp olv mutants in exon 11 are missense mutations but the complementing mutant dp olv105A is due to a nonsense mutation, and dp olv6 in exon 15 on the other side of the PIGSFEAST exon is a frameshift mutation which generates a stop codon.
dp o mutants
Our sample of sequenced mutants is deficient for oblique or dp o alleles. These unfortunately are only rarely recovered in EMS screens , although Grace found that, like dp olv mutants, they map at many places in the gene. We did analyze two dp o mutations. Both are missense mutations that, remarkably, are due to G to A transitions of the same nucleotide resulting in cysteine to tyrosine substitutions. We are certain these are different mutations since the SNP patterns and synonymous substitutions in the chromosomes surrounding the site are very different.
Alternative splicing in Dumpy: Evolutionary evidence
Our molecular analyses of the dumpy mutants indicates most are due to nonsense mutations. One might predict, if dumpy encodes a single transcript and translated message, that most, if not all, of these would affect all three basic functions, i.e. wing shape, tendon cell-cuticle attachment and ultimately viability. Hence they should have a dp olv phenotype. How then do we explain the observations that the dp ol, dp lv, and dp lmutants, i.e. those that have only partial dumpy function, are also due to the presence of stop codons in the dumpy message? We propose that these mutations producing partial functions will be found in alternatively spliced exons. For example, exons tagged by dp lv nonsense mutations will be expressed in certain tissues, e.g. in tendon cells and in the trachea, the latter presumably necessary for viability, but not be present in dumpy messages in the developing wing. Dp olv nonsense mutants would be found in so called constitutive exons expressed in most, if not all, tissues at all developmental stages. Other explanations of our results are discussed below.
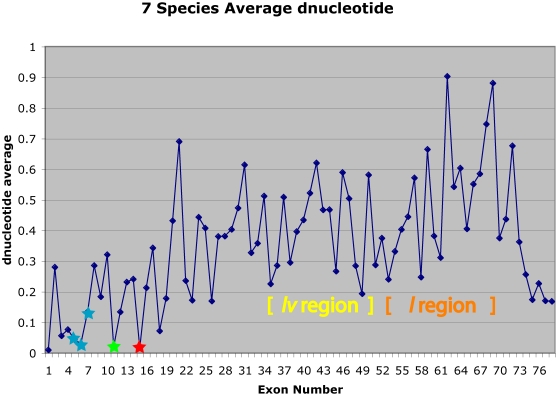
The hypothesis that the dumpy gene encodes both alternative and constitutive exons makes several predications. First, there are distinct differences between alternative and constitutive exons in other systems. Xing and Lee [28], [29] noted that RNA sequences from alternatively spliced exon/intron boundaries leads to selection pressure for nucleotide sequence conservation in these regions while there is significantly less conservation in constitutive exons. Thus, they noted that Ks, the number of synonymous substitutions per synonymous site, is much lower in human-mouse comparisons of alternatively spliced exons than in constitutive exons. To assess sequence divergence in dumpy's exons, we compared the first 30 nucleotides of the exons from seven Drosophila species (D. melanogaster, D. ananassae, D. pseudoobscura, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi), as shown in Figure 5.
Figure 5. Graph of average dnucleotide differences between the first 30 sites at the 5′ ends of dumpy exons from seven Drosophila species.
There is strong conservation of the sequences at the 5′ ends of ol exons (blue stars), exon 11 (green star), and exon 15 containing the complementing mutation dp olv6 (red star). In general, exons containing other dp olv mutations show higher levels of divergence. See Materials and Methods for details regarding alignment and computational procedures.
It should be noted that the seven species compared in a pairwise fashion (the averages are shown in Figure 5) are, in each case, from different subgenera or from different species groups. The conservation of the 5′ ends of exons marked by dp ol (blue stars) mutations is striking. Exon 11 (green star) and exon 15 (red star), both of which are affected by complementing dp olv mutations, are also striking in their conservation of 5′ nucleotides. The nearly complete sequence identity of the 5′ ends of these exons over 60 million years of evolutionary time indicates there is a highly conserved interaction between the dumpy message from these regions and proteins involved in the splicing process, possibly in a tissue specific manner [30]. The pattern of conservation in the lv and l regions is also very interesting (bracketed yellow and orange areas). There may be several different mechanisms creating various alternative transcripts in the lv and l region such as competing intronic RNA secondary structures [31], steric hinderance of multiple splicing factor binding sites, or major and minor splicesome usage [32]. In general, exons with non complementing dp olv mutations do not show marked 5′ end conservation. For example, exons 19 and 34 show higher levels of nucleotide divergence viz. 0.178 and 0.512 respectively and each is marked by two non complementing dp olv nonsense mutations, dp olvR11 and dp olvD2011A in exon 19, and dp olv2C1 and dp olv89a in exon 34. As in the case of the human-mouse comparisons [28], [29], there is marked conservation in some alternatively spliced exons.
Alternative splicing in Dumpy: Evidence from RT-PCR
We have evidence that alternatively spliced dumpy mRNAs can be detected by RT-PCR. We extracted mRNA from wild type 3rd instar larvae and from Drosophila S2 cell lines and used primers spanning the set of ol and lv exons. Primers spanning the two exons of the ZP domain were used as a positive control. We also chose primers located in exons marked by dp olv nonsense mutations that we believe are constitutive.
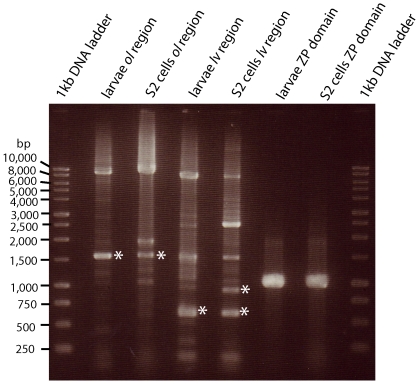
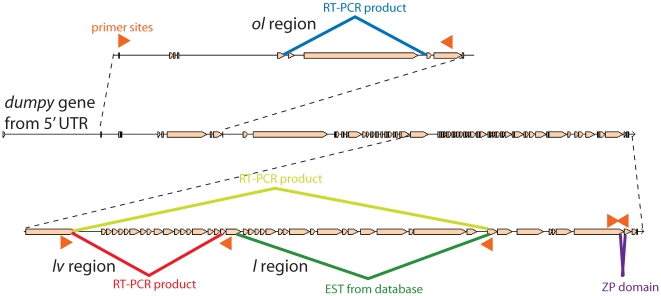
Figure 6 is a gel image of the RT-PCR products obtained from the two sources of mRNA. There are a number of shorter products, some of which were sequenced (identified by asterisks in Figure 6). Sequence data clearly indicate the presence of alternatively spliced mRNAs in both 3rd instar larvae and S2 cells. The primers spanning the ol region detected a mRNA lacking exons 6 and 7 diagrammed in Figure 7. Recall that 6 of the 7 dp ol nonsense mutations are in these two exons. At least two differently spliced messages were obtained using the primers spanning the lv exons, one is missing exons 35 to 50 where all of the dp lv mutations are located, whereas the other skips a larger number of exons, 35 to 69. The largest band in each is consistent with inclusion of all exons between the selected primers and illustrates we can amplify at least 10kb by RT-PCR. The intermediately sized bands most likely represent different alternatively spliced products with various exons included in the transcripts.
Figure 6. Gel showing RT-PCR products from the dumpy gene in 3rd instar larvae and S2 cells.
Primers flanking the ol and lv regions and the ZP domain that were used to generate these products are shown in Table S2. Bands marked with asterisks were excised and sequenced.
Figure 7. Diagram of RT-PCR products exhibiting alternative splicing in the dumpy gene.
The middle line shows the intron-exon structure of the wild type dumpy gene. The ol region is shown above the line and the RT-PCR product which is missing exons 6 and 7. Below the line depicting the wild type gene is the intron-exon structure of the 3′ end of the gene showing the lv and l regions and the ZP domain. The RT-PCR products are missing a number of exons from each region. In each case, arrowheads mark the positions of primers used to obtain the RT-PCR products. These primer sequences are shown in Table S2.
The Flybase Drosophila EST database (http://flybase.org) for dumpy is highly enriched for clones with sequences from the 3′ end of the gene and essentially is non informative with regard to alternative splicing patterns. There is one EST from the database, however, that excludes exons 53 to 69 (see Figure 7). Recall that the dp lDG82 nonsense mutation is in exon 58, and according to our hypothesis this exon should be alternatively spliced.
Discussion
Our attempts to generate new dumpy mutants in an isoallelic 2nd chromosome with ethyl methanesulfonate produced a distribution of dumpy alleles similar to that of Jenkins [13] Table 5. Thus the majority of our mutants generated in crosses 1 and 2 (see Table S1) were dp olv, dp ol, and dp lv in decreasing order. Other chemical mutagens – see Table 6 in Jenkins [13]- produce similar distributions. All of these cross schemes involved screening F1s heterozygous for dp ov1 for oblique wing phenotypes and/or thoracic vortices. It's possible that the greater ease with which oblique versus vortex phenotypes are detected could bias such screens utilizing dp ov1 toward the recovery of dp olv, dp ol, dp ov, and dp o mutants, yet the latter two types are recovered very infrequently. Hence we feel the distribution of mutants accurately reflects the sizes of “targets” within the dumpy gene which, when mutant, affect one or more combinations of phenotypes. We are collaborating with the laboratory of Olga Grushko and Alexey Kondrashov at the University of Michigan where spontaneous dumpy mutants are being isolated as non-fliers at 28°C. To date, eight such mutations have been analyzed molecularly (see Table 3). As with the EMS induced mutants, lethal classes predominate and in this case they are either dp ol (1), dp olv (2) or dp lv (5) mutants. Except for two mutants, dp L2311B and dp lvD1191A, all of the others are deletions or small inversions which create frame shifts and/or stop codons. There is also a deletion in an intron with an unknown effect.
Due to our failure to recover dp v mutants in the screens utilizing crosses 1 and 2 in Table 2 and because only two such alleles currently exist in stock centers, dp v1 and dp v2, we set up crosses 3 and 4 to enrich for new vortex mutants in the isoallelic 2nd chromosome from the Australia line. Following Jenkins [13], we estimated that we would obtain one transmitted vortex mutant in 17,500 F1 flies screened (frequency of F1 dumpy mutants, ca 0.8% in his Table 4, times1/20 vortex mosaics or completes in his Table 5 times 15% of transmitted vortex mutations in his Table 6).
The number of F1s screened in cross 3 was not estimated, but in cross 4, in two separate mutageneses, we examined an estimated 24,000 flies. Five F1 flies with mosaic or complete vortices transmitted the mutation but none was a dp v mutant. Of the five mutants, four were complementing dp olv alleles and one was a homozygous viable dp ov allele. As mentioned above, there are no stocks of the dp v mutants obtained by Jenkins [13], or Grace [17]. Our failure to obtain such mutants with EMS makes it more likely, in our view, that the roo element in the 5′ end of the dumpy gene in mutants dp v1 and dp v2 is responsible for the mutant phenotype. It may also be that these alleles are, in fact, the same—the roo element is inserted between the same two base pairs in each case and were perhaps inadvertently isolated at different times in the Morgan laboratory and named as separate alleles. Clearly the vortex sublocus, so elegantly mapped by Grace [17] at the 5′ end of the dumpy gene itself, needs to be better defined mutationally. The dumpy lethal sublocus, currently defined by only two alleles dp lDG82 and dp lDG83, also needs to be further analyzed. It is not clear, however, how additional dp l alleles can be obtained, since F1 screens cannot be used. F2 screens would be tedious, although it might be possible to screen the progeny of F2 individuals crossed with dp olv flies for homozygous lethality.
The approach we have taken to define the mutations generated with EMS in isoallelic dumpy wild type alleles, i.e. producing overlapping amplicons from the entire locus and screening for base pair mismatches in reannealed duplexes with Surveyor nuclease and dHPLC as shown in Figure 3, has been very effective. The data are remarkable in that most of the mutations, including dp ol and dp lv mutants, result in stop codons either at the site of the mutations or are generated from a deletion, or cause the removal of a splice site. It is also clear that all eleven missense mutations change cysteines in EGF or DPY motifs, presumably altering or destabilizing their tertiary structures. Given the repetitive nature of the Dumpy protein, i.e. all the EGF-DPY-EGF motifs, perhaps most other kinds of missense mutations don't produce a visible phenotype. As shown in Table 3, eight of the eleven missense mutations are found in exon 11 which defines the ov sublocus in Grace's map. This is in stark contrast to the mutational spectrum in probably all other dumpy exons where the mutations are almost exclusively deletions or nonsense mutations. Grace mapped three kinds of oblique mutations viz. dp o, dp ov, and dp olv at the ov sublocus, and we also find both dpov and dp olv mutations in exon 11. The exon is unremarkable in that it encodes 4 simple EGF-DPY diads, which, although most of the protein consists of EGF-DPY-EGF triads (see Figure 2), are also found at other positions in the protein. Six of the missense mutants are in EGF modules and two are in the DPY members of the diads. Interestingly, in the EGF module in the second diad, there are four mutations affecting four of the six cysteines. Two of the mutations exhibit a dp ov phenotype and two are dp olv mutants, one of which, dp 104A, is a complementing mutant (see below).
At this point we don't know if the dumpy exon 11 is alternative or constitutive since our RT-PCR experiments did not utilize primers flanking this exon. The extreme conservation found at its 5′ end (see Figure 5) indicates it is alternatively spliced, but the presence of a dp olv nonsense mutation (dp olv105A) according to our hypothesis, would make exon 11 constitutive. We are currently analyzing the splice variants in dumpy RNA by RNA-seq. [33], [34], the results of which should clarify the status of exon 11.
As mentioned above, we believe the long standing and hitherto unexplained complexity of the dumpy gene can best be explained by extensive alternative splicing where dp olv nonsense mutations tag constitutive exons presumably located at several different places in the gene. Nonsense mutations with partial dumpy function, e.g. dp ol and dp lv mutations, will be found in alternatively spliced exons and should be more localized in the gene.
In this regard, typical characteristics of alternatively spliced exons are small size and divisibility by 3 so as not to affect the reading frame depending on their inclusion or exclusion. The dumpy gene contains 78 coding exons, many are very small, i.e. under 80bp, and the number of nucleotides in all but 1 internal exon is divisible by 3.
We predict and, indeed, have found that alternative splicing produces tissue specific isoforms of Dumpy encoded by at least several kinds of mRNAs. The dp ol, dp lv, and dp l nonsense mutations either could result in truncated isoforms only in the affected tissues or, if they are located in splicing protein binding sites in the exon, prevent the formation of the alternatively spliced transcript (see [35]). These two possibilities make different predictions about whether alternatively spliced transcripts would be found in dp ol, dp lv, and dp l mutants, viz. if the tissue specific mutation results in truncated isoforms, the alternatively spliced transcripts would be present in mutant flies but if the mutations interfere with the splicing process, the transcripts would be aberrant or absent. Thus, a comparison of RT-PCR products in tissues from wild type and mutant flies should distinguish between the two mechanisms.
How else can we explain the observation that most of the lethal classes of dumpy alleles, viz. dp ol, dp lv, dp l, and dp olv, are due to nonsense mutations? One explanation might be that some premature stop codons in the dp olv mutants result in nonsense mediated decay of the mRNA [36] and this completely removes all functions in the olv class. However, a paradox in the results is that many of the less severe dp ol, dp lv, and dp l mutations that retain some dumpy function also introduce premature stop codons that truncate the protein within its extracellular domain. Hence, these data do not appear compatible with the hypothesis that the dumpy locus generates only one molecular product. Indeed, the RT-PCR results reported here (see Figures 6 and 7) clearly show that certain exons are excluded from some dumpy mRNAs by alternative splicing. Hence, we propose that dumpy generates multiple products by alternative splicing which are specialized to particular biological functions.
As mentioned above the mutant screens outlined in cross 4, Table 2, failed to generate new dp v (vortex) mutants but did, however, produce four mutant alleles, dp olv13v, dp olvA4, dp olvB11 and dp olvB16. These came through the screen because they complemented the lethal phenotype associated with a dp lv allele. Earlier, crosses 1 and 2 produced additional complementing dp olv mutants, e.g. dp olv105A, dp olv104A, dp olv27B, dp olv48a, and dp olv6. These were also recovered as complementers of the dp lvI allele carried on the In (2LR) CyO balancer. Also several inter se crosses have revealed additional cases of complementation between these individual dp olv mutant alleles. For example, dp olv104A and dp olv6, mutants which flank the PIGSFEAST exon (number 12) fully complement for viability (i.e. 1/3rd of the F1s from a cross between flies from balanced lethal parents survive to adulthood, but still show oblique wings and vortices. Such interallelic or intragenic complementation is generally explained by association and functional complementation between mutant protein subunits [37], but when one (e.g. in dp olv6/dp olv104A heterozygotes or both alleles (e.g. in dp olv6/dp lDG82 heterozygotes) are nonsense mutations, a different explanation for the complementation must be found.
Complementing dp olv nonsense mutants, are also difficult to explain by cis alternative splicing. Alternative trans-splicing, however, could be operating in the processing of dumpy messages, perhaps only in certain tissues, and provide an explanation for the viability of some dp olv heteroallelic heterozygotes. Paradigms for putative trans-splicing events have been documented [38]–[42].
Our current RNA-seq approach to detect exon-exon junctions in the dumpy “transcriptome,” when coupled with the identification of the mutant vs. wild type codons or SNP associations in individual cDNAs should allow us to detect trans-splicing events. In this regard, we find a SNP, on the average, every 140 bases in the exons of the dumpy gene. If trans-splicing turns out to be responsible for the interallelic complementation between lethal dumpy nonsense mutants, we can begin to identify the mechanism and the gene products that are responsible for the splicing events. This can be accomplished with screens for suppressors and enhancers of the complementation patterns of different dumpy mutants.
In summary, the molecular analysis of 45 preexisting, spontaneous, and/or EMS induced dumpy mutations revealed most missense mutations were found in exon 11. All other mutations except two transposon insertions generated stop codons, were deletions, an inversion, or frameshift generated nonsense codons, even those which exhibited only one or two of the three dumpy mutant phenotypes. We present evolutionary and experimental evidence for cis alternative splicing of dumpy transcripts and argue that these observations, along with the distribution within the gene of nonsense mutations with different dumpy mutant phenotypes, makes it likely that alternative splicing underlies the genetic and phenotypic complexity of this long studied, paradigmatic Drosophila complex gene. In addition, complementation between certain dumpy nonsense mutant alleles can be explained by trans-splicing.
Materials and Methods
Drosophila strains
Dumpy mutant alleles with undefined genetic backgrounds were ordered from Bloomington or Kyoto stock centers. These are identified in Table 3 as SC. Mutants 12 and 16 were provided by Jim Fristrom and Mary Prout, and identified as MP in Table 3. They were generated by gamma ray mutagenesis and are dp olv or dp lv alleles which, when homozygous in somatic clones, give rise to wing blisters (see Prout et al. [23]). Mutants designated with an OG in Table 3 were recovered by Olga Grushko and Alexey Kondrashov in a screen for spontaneous dumpy mutants. They are present in two 2nd chromosomes designated as A and B, extracted from natural populations near Ann Arbor, MI, and made homozygous for chromosome 2. Each has a different pattern of single nucleotide polymorphisms (SNPs) in the dumpy gene. The screen selects for non fliers and, hence, the mutant alleles have an oblique wing or vortex phenotype. Most of the dumpy mutants we analyzed were generated by EMS following Jenkins [13] in stocks isoallelic for dumpy. Those labeled with a BM in Table 3 are in an isoallelic 2nd chromosome carrying cn and bw mutant alleles and generated by Brad Marshall, then an undergraduate researcher in the laboratory. Crosses 2A and 2B were performed and mutants were recovered by Michael Guertin are labeled MG in Table 3. Ross MacIntyre produced and recovered dumpy mutants from crosses 3 and 4 and are labeled RM in Table 3. In crosses 2, 3, and 4, the 2nd chromosome was marked with either net (II, O.O) or clot (II, 16.O). These chromosomes were recovered as recombinants from net dp + clot /net+ dp+ cl+ females where the net + dp + cl + chromosome had been isolated from a wild population from Australia, provided by Chip Aquadro, and made isoallelic with crosses to appropriate 2nd chromosome balancer stocks. We confirmed the identity of the dp+ alleles in the net and clot stocks by a Southern blot which allowed us to analyze the PIGSFEAST repeat number, thus confirming the dp allele came from the chromosome from the Australia line (see Carmon et al. [21]). Second, as described below, when amplicons spanning the entire dumpy gene from the two strains, net dp + and dp + clot, were denatured, reannealed, treated with the Surveyor nuclease, no base pair mismatches were detected, confirming the sequence identity of the wild type dumpy alleles.
Mutagenesis and mutant allele recovery
For the dumpy mutant alleles induced with EMS in the cn bw, net or clot chromosomes, we fed males 0.024M EMS for 24 hours following Lewis & Bacher [43] and Jenkins [13]. F1 males were then scored for mosaic or complete dumpy mutant phenotypes. Several different crosses were carried out as shown in Table 2.
In crosses 2A and 2B, mutant F1 males were crossed back to dp ov1 cl; e or net dp ov1 cl females respectively to determine which mutants transmitted a mutant allele, i.e. had a partially or completely mutant gonad. The mating of the phenotypically dumpy F1 flies indicated that only 29% transmitted the new mutation. Previous studies [13] found that 35% of the F1 dumpy mutants transmitted the mutant allele to their offspring. Mutant F2 males were then mated to In(2LR) CyO, dp lvI cl-4 [44]/ In(2LR) Gla females, and the curly winged progeny assessed for their eye color, i.e. either clot or wild type. In cross 2A, when net cl+ males were mutagenized, surviving curly sibs with wild type eyes were mated to establish a stock of the new dumpy mutant. In most cases, the newly induced mutant was lethal over the CyO, dp lvI cl-4 chromosome. In these instances, 5–10 single Gla/dumpy? males were then backcrossed to CyO, dp lvI cl-4/Gla females. Glazed eyed flies from vials with no curly winged clot eyed progeny were then mated to establish the new mutant strain. In cross 2B, when dp+ clot males were mutagenized, F1 males were crossed to dp ov1 cn bw females. If the new mutant transmitted, mutant dumpy F2 males were crossed to CyO, dp lvI cl-4/Gla females and, as in cross 2A, any curly winged clot eyed sibs were mated to recover the new mutant allele in a stock. As was usually the case, if no curly winged clot eyed flies survived, 5–10 single Glazed eyed males were backcrossed to CyO, dp lvI cl-4/Gla females and Glazed eyed flies with straight wings from vials with only Glazed or Curly/Glazed progeny were used to set up the mutant stock. In crosses 3 and 4, we hoped to enrich for dumpy vortex mutants by incorporating the e(dp v) mutant [4] on chromosome 3 in the male and female parental stocks. We also crossed F1 males showing complete or mosaic expression of the vortex phenotype to CyO, dp lvI cl-4/Gla females. If the curly winged, clot eyed (cross 3) or wild type eyed (cross 4) progeny showed a vortex mutant phenotype, males were backcrossed to CyO, dp lvI cl-4/Gla females and either curly winged, dumpy vortex or glazed eyed sibs with straight wings were used to establish a stock of the new “vortex” mutant. Each new dumpy mutant we obtained was phenotypically characterized by crossing it to dp ov1 flies and to flies carrying Df(2L)ED250, which deletes the entire dumpy gene. Thus, for example, a new dp ol mutant would show oblique wings in the F1s from a cross to dp ov1, but not mutant vortices, and the flies heterozygous for the new dumpy mutation and Df(2L)ED250 would not survive to adulthood.
Primer design
Primers were designed using Primer3 or a modification of the Primer3 algorithm available at http://flypush.imgen.bcm.tmc.edu/primer/ [45]. An initial set of 85 overlapping primer pairs, each generating an approximately 1kb product, to cover virtually the entire gene, was developed. A second set to overlap these in the region of the gene from PF to PR, as well as additional primers surrounding exon 11 and throughout intron 11, were later developed. Primers were also designed spanning the upstream region of dumpy for approximately 25kb and for exon 1. The primer sequences are listed in Table S2.
Molecular analysis of dumpy mutations
We extracted DNA from individuals heterozygous for a mutation and a progenitor second chromosome that was either cn bw, net cl + or net + cl. For mutants provided by Mary Prout and Jim Fristrom, we used heterozygotes for the mutant and their 2L progenitor chromosome (see Prout et al. [23]) as the source of DNA. PCR on this DNA provided the sequences necessary for the formation of heteroduplexes following denaturation and renaturation of the PCR products. To find the sites of the lesions in the sets of mutants, we pooled several genomic DNAs and amplified aliquots using 1.25U Optimase Polymerase (Transgenomic, Inc.) with 1.25U GoTaq polymerase (Promega) in Optimase Buffer with the primer pairs. Cycling conditions were 94°C for 2 min and 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 2 min, followed by 74°C for 5 min. Products were denatured and reannealed, treated with the Surveyor Mutation Detection Kit for WAVE Systems (Transgenomic, Inc.), and injected on the dHPLC WAVE System (Transgenomic, Inc.) using a standard sizing gradient. DNA from the original unmutated isoallelic stocks did not give any mismatched base pairs in any of the initial 85 amplicons but if one of the reannealed DNAs in the mutant pool contains a base pair mismatch, it is cleaved into two fragments which were detected as smaller, discrete peaks following dHPLC. An example of a dHPLC analysis of a cleaved amplicon is shown in Figure 3. Once an amplicon containing a mutation was detected, it was cloned using a TOPO Zero Blunt Cloning Kit (Invitrogen) and at least eight colonies were sequenced to insure the mutant site was identified.
v2 and ov1
Since the progenitor chromosome for these mutants was not known, DNA from homozygous individuals was extracted. Primer pairs not giving a product were then used for amplification with iProof Polymerase (Bio-Rad) and GC buffer. Cycling conditions were 98°C for 2 min and 35 cycles of 98°C for 5 sec, 63°C for 15 sec, 72°C for 7.5 min, followed by 72°C for 10 min. After visualization on a gel showing the presence of a long insertion, its ends were sequenced to identify the transposon.
o2 and ovDG2
DNA from homozygous individuals for dp o2 and dp ovDG2/ Df(2L)ED250 individuals was extracted. Fragments in regions suspected to contain the mutation were amplified by PCR and sequenced.
lDG82
Balanced lethal flies were crossed to dp ovDG2 and DNA was extracted from dp lDG82/dp ovDG2 F1s since both mutants were made by Dale Grace [15], [17]. Preliminary data indicated the two mutants were induced in the same progenitor chromosome and dp lDG82 was subsequently analyzed as above using Surveyor and WAVE analysis.
RT-PCR
Total RNA was extracted from 3rd instar larvae and S2 cells. RT-PCR was performed using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen). Cycling conditions were 55°C for 30 min, 94°C for 2 min, 40 cycles of 94°C for 15 sec, 55°C for 30 sec, 72°C for 8 min, followed by 72°C for 10 min. Primer pairs spanning putative alternatively spliced exons in various dumpy subloci were used as well as a pair spanning the ZP domain. Amplified products were separated in gels and bands excised and sequenced.
Computer based analyses
Evolutionary comparisons of the 30 nucleotides at the 5′ ends of dumpy exons were made in the dumpy genes from 7 Drosophila species – D. melanogaster, D. ananassae, D. pseudoobscura, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi whose genomes have been sequenced. Analysis of 30 nucleotides was chosen due to the extremely variable length of dumpy exons from 54bp to over 13kb. To do this, we aligned the sequences in MEGA 4 and used the nucleotide model to calculate the average pairwise distance with the Kimura 2-parameter correction [46].
Supporting Information
Properties of dumpy mutants derived from crosses 1, 2A, 2B, 3, and 4 shown in Table 2 - *indicates complementation with at least one other lethal allele. **oblique score according to Grace [17] in parentheses. The oblique phenotype was tested over dp ov1. ***strong = vortices on most of the dorsal thorax, intermed = 2–4 vortices, mild = 1–2 vortices when present.
(0.20 MB DOC)
Primers used in this study - *Primers used for RT-PCR products shown in Figures 6 and 7
(0.26 MB DOC)
Acknowledgments
We thank Chip Aquadro, Jim Fristrom, and Mary Prout for fly stocks. We thank Alexey Kondrashov for designing the flightless mutant screen. We also thank former undergraduate student Alex Helkin for help with defining the vortex sublocus. Lauren MacIntyre Ampel provided excellent assistance on the preparation of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by the United States Department of Agriculture Hatch Project award NYC-165426 (http://www.usda.gov/) and National Institutes of Health grant 1R21HD059073-01A1 (http://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Morgan TH. The origin of nine wing mutations in drosophila. Science. 1911;33:496–499. doi: 10.1126/science.33.848.496. [DOI] [PubMed] [Google Scholar]
- 2.Morgan TH, Sturtevant AH, Muller HJ, Bridges CB. The Mechanism of Mendelian Heredity. New York: Henry Holt and Company; 1915. 262 [Google Scholar]
- 3.Altenburg E. The genetic basis of truncate wing, - An inconstant and modifiable character in Drosophila. Genetics. 1920;5:1–59. doi: 10.1093/genetics/5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridges CB, Mohr OL. The inheritance of the mutant character “vortex”. Genetics. 1919;4:283–306. doi: 10.1093/genetics/4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan TH. Data relating to six mutants of Drosophila. In: Sturtevant AH, Bridges CB, Morgan TH, Li JC, editors. Contributions to the genetics of Drosophila simulans and Drosophila melanogaster. Washington: Carnegie Institution of Washington; 1929. pp. 171–199. [Google Scholar]
- 6.Muller HJ. Variation due to change in the individual gene. Am Nat. 1922;56:32–50. [Google Scholar]
- 7.Mohr OL. Modifications of the sex-ratio through a sex-linked semi-lethal in Drosophila melanogaster.(Besides notes on an autosomal section deficiency). 1923. pp. 266–287. Studia Mendeliana, ad centesimum diem natalem Gregorii Mendelii a grata patria celebrandum, adiuvante ministerio Pragensi edita Brunae.
- 8.Mohr OL. Exaggeration and Inhibition Phenomena encountered in the analysis of an autosomal dominant Zeitschrift fur induktive Abstammungs- und. Vererbungslehre. 1929;50:113–200. [Google Scholar]
- 9.Carlson EA. Allelism, Complementation, and Pseudoallelism at the dumpy locus in Drosophila melanogaster. Genetics. 1959;44:347–373. doi: 10.1093/genetics/44.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson EA. Comparative Genetics of Complex Loci. Q Rev Biol. 1959;34:33–67. doi: 10.1086/402574. [DOI] [PubMed] [Google Scholar]
- 11.Carlson EA, Southin JL. Comparative Mutagenesis of Dumpy Locus in Drosophila melanogaster .1. X-Ray Treatment of Mature Sperm - Frequency and Distribution. Genetics. 1962;47:321–336. doi: 10.1093/genetics/47.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Southin JL. An analysis of eight classes of somatic and gonadal mutation at dumpy locus in Drosophila melanogaster. Mutat Res. 1966;3:54–65. doi: 10.1016/0027-5107(66)90007-8. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins JB. Mutagenesis at a complex locus in Drosophila with the monofunctional alkylating agent, ethyl methanesulfonate. Genetics. 1967;57:783–793. doi: 10.1093/genetics/57.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sederoff R. Rare Pseudoallelic Crossover between 2 Phenotypically Identical Alleles at a Restricted Sublocus of Dumpy in Drosophila Melanogaster. Nature. 1967;216:1348–1349. doi: 10.1038/2161348b0. [DOI] [PubMed] [Google Scholar]
- 15.Grace D. Genetic analysis of dumpy Region in Drosophila - Its multigenic composition. Mutat Res. 1970;10:489–496. doi: 10.1016/0027-5107(70)90008-4. [DOI] [PubMed] [Google Scholar]
- 16.Montgomerie DW. Recombination and mutation analysis of lethals at the dumpy locus in Drosophila melanogaster. Montreal, Canada: McGill University; 1974. 168 [Google Scholar]
- 17.Grace D. Genetic analysis of the dumpy complex locus in Drosophila melanogaster - Complementation, Fine-Structure and Function. Genetics. 1980;94:647–662. doi: 10.1093/genetics/94.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segraves WA, Louis C, Tsubota S, Schedl P, Rawls JM, et al. The rudimentary locus of Drosophila melanogaster. J Mol Biol. 1984;175:1–17. doi: 10.1016/0022-2836(84)90441-8. [DOI] [PubMed] [Google Scholar]
- 19.Jack J, Delotto Y. Structure and regulation of a complex locus - the cut gene of Drosophila. Genetics. 1995;139:1689–1700. doi: 10.1093/genetics/139.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkin MB, Becker MN, Mulvey D, Phan I, Chao A, et al. Drosophila Dumpy is a gigantic extracellular protein required to maintain tension at epidermal-cuticle attachment sites. Curr Biol. 2000;10:559–567. doi: 10.1016/s0960-9822(00)00482-6. [DOI] [PubMed] [Google Scholar]
- 21.Carmon A, Wilkin M, Hassan J, Baron M, MacIntyre R. Concerted evolution within the Drosophila dumpy gene. Genetics. 2007;176:309–325. doi: 10.1534/genetics.106.060897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carmon A, Larson M, Wayne M, MacIntyre R. The rate of unequal crossing over in the dumpy gene from Drosophila melanogaster. J Mol Evol. 2010;70:260–265. doi: 10.1007/s00239-010-9327-1. [DOI] [PubMed] [Google Scholar]
- 23.Prout M, Damania Z, Soong J, Fristrom D, Fristrom JW. Autosomal mutations affecting adhesion between wing surfaces in Drosophila melanogaster. Genetics. 1997;146:275–285. doi: 10.1093/genetics/146.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokel C, Prokop A, Brown NH. Papillote and Piopio: Drosophila ZP-domain proteins required for cell adhesion to the apical extracellular matrix and microtubule organization. J Cell Sci. 2005;118:633–642. doi: 10.1242/jcs.01619. [DOI] [PubMed] [Google Scholar]
- 25.Jazwinska A, Ribeiro C, Affolter M. Epithelial tube morphogenesis during Drosophila tracheal development requires Piopio, a luminal ZP protein. Nat Cell Biol. 2003;5:895–901. doi: 10.1038/ncb1049. [DOI] [PubMed] [Google Scholar]
- 26.Mahoney MB, Parks AL, Ruddy DA, Tiong SYK, Esengil H, et al. Presenilin-based genetic screens in Drosophila melanogaster identify novel Notch pathway modifiers. Genetics. 2006;172:2309–2324. doi: 10.1534/genetics.104.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmon A, Topbas F, Baron M, MacIntyre R. dumpy interacts with a large number of genes in the developing wing of Drosophila melanogaster. Fly. 2010;4:117–127. doi: 10.4161/fly.4.2.11953. [DOI] [PubMed] [Google Scholar]
- 28.Xing Y, Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. P Natl Acad Sci USA. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing Y, Lee C. Alternative splicing and RNA selection pressure - evolutionary consequences for eukaryotic genomes. Nat Rev Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 30.Lu HC, Lin L, Sato S, Xing Y, Lee CJ. Predicting Functional Alternative Splicing by Measuring RNA Selection Pressure from Multigenome Alignments. Plos Comput Biol. 2009;5:-. doi: 10.1371/journal.pcbi.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graveley BR. Mutually exclusive splicing of the insect Dscam Pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson S, Blanchette M, Park J, Savva Y, Yeo GW, et al. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 36.Behm-Ansmant I, Izaurralde E. Quality control of gene expression: a stepwise assembly pathway for the surveillance comp ex that triggers nonsense-mediated mRNA decay. Gene Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- 37.Fincham JRS. Complementation maps and their interpretation. Genetic Complementation. New York: W. A. Benjamin, Inc; 1966. pp. 90–112. [Google Scholar]
- 38.Horiuchi T, Giniger E, Aigaki T. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Gene Dev. 2003;17:2496–2501. doi: 10.1101/gad.1137303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauss V, Dorn R. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene. 2004;331:165–176. doi: 10.1016/j.gene.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 41.Rowley JD, Blumenthal T. Medicine - The cart before the horse. Science. 2008;321:1302–1304. doi: 10.1126/science.1163791. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Wang JL, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 43.Lewis E, Bacher F. Method of feeding ethylmethanesulfonate (EMS) to Drosophila males. DIS. 1968;43:193. [Google Scholar]
- 44.Davis MB, Macintyre RJ. A genetic analysis of the alpha-Glycerophosphate Oxidase locus in Drosophila melanogaster. Genetics. 1988;120:755–766. doi: 10.1093/genetics/120.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: S K, S M, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ, USA: Humana Press; 2000. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Properties of dumpy mutants derived from crosses 1, 2A, 2B, 3, and 4 shown in Table 2 - *indicates complementation with at least one other lethal allele. **oblique score according to Grace [17] in parentheses. The oblique phenotype was tested over dp ov1. ***strong = vortices on most of the dorsal thorax, intermed = 2–4 vortices, mild = 1–2 vortices when present.
(0.20 MB DOC)
Primers used in this study - *Primers used for RT-PCR products shown in Figures 6 and 7
(0.26 MB DOC)