Abstract
Cardiac complications and heart failure are the leading cause of death in type 2 diabetic patients. Mitochondrial dysfunction is central in the pathogenesis of the type 2 diabetic heart. However, it is unclear whether this dysfunction is specific for a particular subcellular region. The purpose of this study was to determine whether mitochondrial dysfunction in the type 2 diabetic heart is specific to a spatially distinct subset of mitochondria. We investigated mitochondrial morphology, function, and proteomic composition of subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM) in 18-wk-old db/db mice. Oxidative damage was assessed in subpopulations through the measurement of lipid peroxidation byproducts and nitrotyrosine residues. Proteomic profiles and posttranslational modifications were assessed in mitochondrial subpopulations using iTRAQ and multi-dimensional protein identification technologies, respectively. SSM from db/db hearts had altered morphology, including a decrease in size and internal complexity, whereas db/db IFM were increased in internal complexity. Db/db SSM displayed decreased state 3 respiration rates, electron transport chain activities, ATP synthase activities, and mitochondrial membrane potential and increased oxidative damage, with no change in IFM. Proteomic assessment revealed a greater impact on db/db SSM compared with db/db IFM. Inner mitochondrial membrane proteins, including electron transport chain, ATP synthesis, and mitochondrial protein import machinery, were predominantly decreased. We provide evidence that mitochondrial dysfunction in the type 2 diabetic heart is associated with a specific subcellular locale. Furthermore, mitochondrial morphological and functional indexes are impacted differently during type 2 diabetic insult and may result from the modulation of spatially distinct mitochondrial proteomes.
Keywords: mitochondria, diabetes, proteomics
type 2 diabetes mellitus is linked to a reduced lifespan in humans due in part to cardiovascular complications and heart failure (19). Mitochondrial dysfunction and altered myocardial metabolism contribute to cardiovascular complications during type 2 diabetes mellitus (3, 4). A previous study (42) observed a decrease in cardiac function and decreased cardiac efficiency in db/db mice, a model of type 2 diabetes mellitus. An increase in circulating free fatty acids present with type 2 diabetes mellitus leads to a pooling of fatty acids in the mitochondrion, facilitating an enhanced oxidative milieu. An examination of total mitochondria from db/db mouse hearts revealed respiration and oxidative phosphorylation deficits, due in part to an increased oxidative environment in the mitochondrion (3).
The cardiomyocyte consists of two biochemically and spatially distinct mitochondrial subpopulations: subsarcolemmal mitochondria (SSM), which are located beneath the plasma membrane, and interfibrillar mitochondria (IFM), which are situated between the myofibrils (33). These two mitochondrial subpopulations respond differently to physiological stimuli, including type 1 diabetes mellitus (25, 27, 30, 37). Previously, we (12) reported differential effects on spatially distinct mitochondrial subpopulations in terms of morphology, function, and oxidative parameters after streptozotocin-induced type 1 diabetic insult, with the IFM subpopulation being the most affected. However, a previous study (37) observed decreased complex II activity and mitochondrial DNA copy number in SSM from the skeletal muscle of type 2 diabetic patients, with no significant effects on IFM. The examination of mitochondrial cardiac proteomic profiles has revealed changes in specific mitochondrial constituents suggesting that the alteration of key proteins involved in substrate utilization, electron transport chain (ETC) function, antioxidant status, and other important mitochondrial processes may be associated with the pathogenesis of diabetes mellitus (6, 20, 47). A recent study (23) examining the impact of endurance exercise revealed distinct subpopulation-specific mitochondrial proteome alterations. Nevertheless, to date, no study has examined the cardiac mitochondrial subpopulation response in a type 2 diabetic model.
The goal of the present study was to determine how spatially distinct mitochondrial subpopulations in the heart of db/db mice are impacted and to discern the effects on subpopulation-specific mitochondrial proteomes. Our results suggest that the SSM subpopulation displays greater dysfunction in the db/db heart, which may be due to specific alterations in the SSM proteome. These data highlight the importance and relevance of taking into account subcellular location when examining mitochondria during diabetic insult.
MATERIALS AND METHODS
Experimental Animals
The animal experiments in this study conformed with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the West Virginia University Animal Care and Use Committee. Male db/db mice (strain BKS.Cg-m +/+ Leprdb/J) and wild-type (WT) littermate controls (Jackson Laboratories, Bar Harbor, ME) were housed in the West Virginia University Health Sciences Center animal facility. Mice were given unlimited access to a rodent diet and water. All mouse groups were aged to 18 wk and then killed for the following experiments.
Preparation of Individual Mitochondrial Subpopulations
Db/db mice and their littermate controls were killed, and their hearts were excised. Hearts were rinsed in PBS (pH 7.4), blotted dry, and then weighed. SSM and IFM were isolated on ice following the methods of Palmer et al. (33) with minor modifications (12, 13). Briefly, the ventricles were minced and homogenized 1:10 (wt/vol) in cold Chappel-Perry buffer [containing (in mmol/l) 100 KCl, 50 MOPS, 5 MgSO4·7H2O, 1 EGTA, and 1 ATP (pH 7.4)] at 4°C. Homogenates were then centrifuged at 700 g for 10 min. The supernatant containing SSM was extracted and centrifuged again at 10,000 g to isolate SSM. The SSM pellet was washed and centrifuged two more times at 10,000 g and then once more at 10,000 g to obtain a clean SSM fraction. The remaining pellet from the 700-g spin was resuspended in KCl-MOPS-EGTA buffer [containing (in mmol/l) 100 KCl, 50 MOPS, and 0.5 EGTA (pH 7.4)] and exposed to 5 mg/g trypsin for 10 min. After 10 min, the IFM pellet was diluted twofold with buffer plus protease inhibitor cocktail (Biovision, Mountain View, CA) to inhibit trypsin and spun down at 700 g for 10 min. The IFM-containing supernatant was saved, and the pellet was resuspended and spun down again at 700 g for 10 min to maximize the IFM yield. Next, supernatants were combined and spun down at 10,000 g to yield IFM. IFM were washed several times and spun down at a final spin of 10,000 g for 10 min. Pellets were resuspended in a sucrose buffer containing (in mmol/l) 220 sucrose, 70 mannitol, 10 Tris·HCl, and 1 EDTA (pH 7.4), and protein concentrations were determined using the Bradford method with BSA as the standard (5).
Mitochondria Size and Internal Complexity
To index mitochondrial subpopulation size and complexity, we performed flow cytometry analyses using a FACS Calibur equipped with a 15-MW 488-nm argon laser and 633-nm red diode laser (Becton Dickinson, San Jose, CA) as previously described (12, 13). Briefly, Mitotracker deep red 633 (Invitrogen, Carlsbad, CA), which moves into intact mitochondria due to membrane potential (ΔΨm), was used to selectively stain intact mitochondria (emission wavelength: 633 nm, fluorescent 633-nm red diode laser) and debris excluded by gating for only mitotracker deep red 633-positive events (intact mitochondria). Forward scatter (FSC) and side scatter (SSC) detectors were used to examine size (FSC) and complexity (SSC) in isolated mitochondrial subpopulations, and data were represented as histograms plotted against the number of gated events as previously described (12, 13). To confirm the absolute mitochondria size, we used a flow cytometry size calibration kit (Invitrogen), which uses a set of microsphere suspensions (0.5–6 μm) to serve as reliable size references for flow cytometric analyses as previously described (12, 13). All flow cytometric measurements were performed under the supervision of the West Virginia University Flow Cytometry Core Facility.
ETC Respiration
State 3 and state 4 respiration rates were assessed in isolated mitochondrial subpopulations as previously described (7, 8, 21) with slight modifications (13). After the mitochondrial subpopulation isolation, samples were resuspended in respiration buffer containing 80 in mmol/l KCl, 50 in mmol/l MOPS, 1 in mmol/l EGTA, 5 in mmol/l KH2PO4, and 1 mg/ml BSA, and equal volumes were loaded into a Gilson chamber (Gilson, Middleton, WI) attached to a Yellow Springs Instruments 5300 biological oxygen monitor (Yellow Springs Instruments, Yellow Springs, OH). The substrates glutamate (5 mM) + malate (5 mM) as well as palmitoylcarnitine (40 μM) were used to initiate maximal respiration, and measurements of state 3 (1 mM ADP) and state 4 (ADP-limited) respiration were made as previously described (7).
ETC Complex Activities
ETC activities of complexes I, III, and IV were measured spectrophotometrically as previously described (12, 46). Briefly, complex I activity was determined by measuring the oxidation of NADH at 340 nm as previously described (46). The assay mixture for complex I contained 25 mM potassium phosphate buffer (pH 7.2), 5 mM MgCl2, 2 mM KCN, 2.5 mg/ml BSA, 50 μM NADH, 10 μM decylubiquinone, and 2 μg/ml antimycin A. The reaction was initiated by the addition of purified mitochondria, and enzyme activity was measured for 3 min, with values recorded every 10 s after the initiation of the reaction. Complex I-specific activity was inhibited by 2 μg/ml rotenone. Complex III activity was determined as previously described (12, 46) by following the reduction of cytochrome c at 550 nm in the presence of reduced decylubiquinone (50 μM). Briefly, the assay buffer for complex III consisted of 500 mM sucrose, 2 mM EDTA, 100 mM Tris·HCl, (pH 7.4), 1 mM cytochrome c, 200 mM KCN, 1 mg/ml antimycin A, and reduced decylubiquinone. Finally, complex IV activity was determined by measuring the oxidation of cytochrome c at 550 nm. Briefly, the assay mixture for complex IV consisted of 10 mM phosphate buffer (pH 7.4) and 20 μM reduced cytochrome c. Protein content was determined as described above (5), and values are expressed as activities (in nmol substrate consumed·min−1·mg protein−1).
ATP Synthase Activity
ATP synthase activity was measured in frozen-thawed mitochondria as oligomycin-sensitive ATPase activity using an assay coupled with pyruvate kinase, which converts ADP to ATP and produces pyruvate from phosphoenolpyruvate (16, 34, 39). Briefly, isolated mitochondria subpopulations were incubated in a buffer containing (in mmol/l) 20 HEPES, 5 MgCl2, 100 KCl, 5 KCN, 2.5 phosphoenolpyruvate, and 0.2 NADH with 0.1 mg/ml pyruvate kinase and 0.1 mg/ml lactate dehydrogenase (pH 7.5–8.0). The reaction was initiated by the addition of ATP to a desired final concentration (1 mM) and followed by the decrease in NADH absorption at 340-nm wavelength (16). Absorbance was measured on a Biotek Synergy HT plate reader (Biotek, Winooski, VT), and protein content was assessed as described above (5) with final values expressed as micromoles consumed per minute per milligram of protein, which was equal to nanomoles of NADH oxidized per minute per milligram of protein.
Mitochondrial ΔΨm
Mitochondrial ΔΨm was measured by flow cytometry using the ratiometric dye 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol carbocyanine iodide (JC-1; Molecular Probes, Carlsbad, CA), which is a lipophilic cation that selectively enters into mitochondria (36). Isolated mitochondrial subpopulations were incubated with JC-1 for 15 min at 37°C, and 20,000 gated events were analyzed per sample. Changes in ΔΨm were reflected in the degree of color change from green to orange as ΔΨm increased. The shift to orange is due to the dye forming aggregates upon membrane polarization, causing shifts in emitted light from 530 nm (green) to 590 nm (orange). The addition of 200 μM of dinitrophenol, which collapses ΔΨm, was used as an assay control. Measurements were performed on freshly isolated mitochondria subpopulations.
Western Blot Analyses
SDS-PAGE was run on 4–12% gradient gels as previously described (26) with equal amounts of protein loaded for each study treatment. Relative amounts of subpopulation-specific mitochondrial adenine nucleotide translocase (ANT), cytochrome c oxidase IV, and ubiquinol cytochrome c reductase core protein 2 (UQCRC2) were assessed through the use of specific antibodies, including the following: anti-ANT goat antibody (product no. sc-9300, Santa Cruz Biotechnology, Santa Cruz, CA), anti-cytochrome oxidase IV rabbit antibody (product no. 4866, Cell Signaling, Danvers, MA), uncoupling protein 3 (UCP3; product no. PA1-028, Affinity Bioreagents, Golden, CO), and anti-UQCRC2 mouse antibody (product no. MS304, Mitosciences, Eugene, OR). Secondary antibodies were goat anti-rabbit IgG horseradish peroxidase conjugate (product no. 10004301, Cayman Chemical, Ann Arbor, MI) for cytochrome oxidase IV, donkey anti-goat IgG horseradish peroxidase conjugate (product no. sc-2020, Santa Cruz Biotechnology) for ANT, and goat-anti mouse conjugate (product no. 31430, Pierce Biotech, Rockford, IL) for UQCRC2. Detection of signals was performed according to the manufacturer's instructions using Pierce ECL Western Blotting Substrate (Pierce, Rockford, IL). Autoradiographic signals were assessed using a G:Box Bioimaging System (Syngene, Frederick, MD), and data were captured and analyzed using GeneSnap/GeneTools software (Syngene, Frederick, MD). Control for protein loading was confirmed by Ponceau staining.
Lipid Peroxidation Products
Peroxidation of lipids was assessed by the measurement of malondialdehyde (MDA) and 4-hydroxyalkenal (4-HAE), stable end products formed from the oxidation of polyunsaturated fatty acids and esters, as previously described (12, 13). Equal volumes of freshly isolated mitochondrial subpopulations were analyzed for MDA and 4-HAE using a colorimetric assay kit (Oxford Biomedical Research, Oxford, MI). This assay is based on the reaction of a chromogenic reagent, N-methyl-2-phenylindole, with MDA and 4-HAE at 45°C. One molecule of either MDA or 4-HAE reacts with two molecules of the reagent to yield a stable chromophore with a maximal absorbance at 586 nm. Absorbance was measured on a Biotek Synergy HT plate reader (Biotek, Winooski, VT), and protein content was assessed as described above (5) with final values expressed per milligram of protein.
Protein Nitrotyrosine Content
Oxidatively modified proteins were examined by measuring nitrosylated groups introduced into protein side chains using a commercially available kit (Cell Sciences, Canton, MA) as previously described (12). Nitrotyrosine-containing proteins were measured using solid-phase ELISA based on the sandwich principle. Samples were incubated in microtiter wells coated with antibodies recognizing nitrotyrosine residues. After an incubation period and a wash, a biotinylated secondary antibody (tracer) was added, followed by a wash and the addition of a streptavidin-peroxidase conjugate. Color development was measured spectrophotometrically at 450 nm after the addition of tetramethylbenzidine using a Biotek Synergy HT plate reader (Biotek), and values were compared against known nitrotyrosine standards.
iTRAQ Labeling
Pooled SSM and IFM subpopulations (n = 4) from db/db and WT hearts were lysed and precipitated overnight in acetone at −20°C, and pellets were resuspended in 20 μl of 0.5 M triethylammonium bicarbonate (pH 8.5). Protein contents were determined using a two-dimensional Quant Kit (Amersham, Piscataway, NJ), and 100 μg of each pooled sample were then denatured with 0.1% SDS and reduced with 5 mM tris-(2-carboxyethyl)phosphine. After an incubation for 1 h at 60°C, cysteines were blocked with 10 mM methylmethane thiosulfonate in isopropanol, and samples were incubated at room temperature for 10 min. Sequencing grade trypsin (10 μl; Applied Biosystems, Foster City, CA) was added in a trypsin-to-protein ratio of 1:20, and samples were incubated at 37°C overnight. Digested samples were labeled with iTRAQ reagents following the protocol provided by the vendor (Applied Biosystems).
After digestion and iTRAQ labeling, ultracomplex protein digests were combined to create a 400-μg pooled protein digest sample that contained equal fractions of each of the four labeled samples for subsequent multi-dimensional protein identification technology (MudPIT) analysis (28). After lyophilization, the digest mixture was reconstituted in strong cation exchange (SCX) loading buffer (5 mM ammonium formate in 20% acetonitrile; pH 3.0) to be fractionated with SCX SpinTips (Protea Biosciences, Morgantown, WV) per the manufacturer's protocol. Briefly, the sample solution was loaded centrifugally onto the SCX SpinTip. The nonadsorbing solution that passed through the SCX SpinTip was collected. Eight different elution solutions were used to fractionate the peptides (20, 60, 100, 150, 200, 250, 400, and 500 mM ammonium formate in 10% acetonitrile) in a step-wise manner, for a total of nine sample fractions. Collected fractions were cleaned by repeated lyophilization, reconstituted in a 0.1 M acetic acid solution, and then lyophilized to dryness. Fractions were then submitted for liquid chromotography (LC)-matrix-assisted laser desorption/ionization (MALDI)-time of flight (TOF)/TOF mass spectral analysis for protein identification, characterization, and differential expression analysis.
Mass Spectrometry Analyses with iTRAQ Labeling
The LC-MALDI mass spectrometry system used was an ABI Tempo LC MALDI spotter with Tempo LC MALDI version 2.00.09 data acquisition and processing software. Lyophilized SCX sample fractions were reconstituted in LC aqueous run buffer (0.1% trifluoroacetic acid and 2% acetonitrile) and were injected onto a Zorbax C18 chromatographic column (150 × 0.3 mm, Agilent Technologies, Wilmington, DE). Peptides were eluted from the column using an acetonitrile-trifluoroacetic acid gradient (2–72% acetonitrile in 35 min) and spotted directly onto a MALDI plate in 6-s spot fractions. MALDI spots were analyzed using an ABI 4800 MALDI-TOF/TOF analyzer operated with 4000 Series Explorer software. Mass spectroscopy (MS) acquisition was in positive ion reflector mode with 400 laser shots/spectrum performed. The 15 strongest precursors per spot were chosen for MS/MS, and the MALDI spot was interrogated until at least four peaks in the MS/MS spectra achieved a signal-to-noise ratio of ≥70.
The resulting MS/MS spectra were analyzed using ABI Protein ProteinPilot software 2.0 (Applied Biosystems). Spectral data were searched against the mouse protein database (National Center for Biotechnology Information nr.fasta database customized to select for all mouse proteins) for the identification of the peptides and corresponding proteins. In ProteinPilot, the sample type was selected as iTRAQ 4Plex for the retrieval of the isotopic tag information from the mass spectra. After database correlation analysis, the proteins were grouped, scored, and normalized against one to four isotope correction factors. The Pro Group algorithm of ProteinPilot generated a ProtScore, which is a cumulative score from each of the peptides used by the algorithm in the protein identification. Protein scores (ProtScore) above 2.0, 1.0, and 0.47 reflect percent confidence levels of >99%, >90%, and >66%, respectively. Each peptide match showed the iTRAQ isotopic labels, carbamidomethylated cysteines, and other posttranslational modifications present as mass spectral shifts identified during the database correlation analysis. Each protein identified also showed the differential protein expression compared against the other iTRAQ-labeled samples for relative quantitation.
Statistics
Means and SEs were calculated for all data sets. Data were analyzed with one-way ANOVA methods to evaluate the main treatment effect: diabetes induction (GraphPad Software, La Jolla, CA). Fisher's least-significant-difference post hoc tests were performed to determine significant differences among means. When appropriate, a Student's t-test was used. P < 0.05 was considered as significant.
RESULTS
db/db Mouse Characterization
At 18 wk, body weights were significantly increased (P < 0.05; Supplemental Material, Supplemental Table S1)1 in db/db mice compared with WT mice, which is in agreement with another report (29). Analyses of fasting glucose levels, circulating free fatty acids, and insulin levels revealed significant increases (P < 0.05 for all three; Supplemental Table S1) in db/db mice compared with WT mice, which is also in agreement with another report (29). Cardiac contractile function was assessed in isolated perfused hearts. Rates of contraction (+dP/dt), relaxation (−dP/dt), and developed pressures were all significantly decreased in db/db hearts compared with WT hearts (P < 0.05 for all three; Supplemental Table S2), demonstrating decreased contractility in the db/db heart. These data are in agreement with others examining the db/db model of type 2 diabetes mellitus (14).
Mitochondrial Subpopulation Morphology
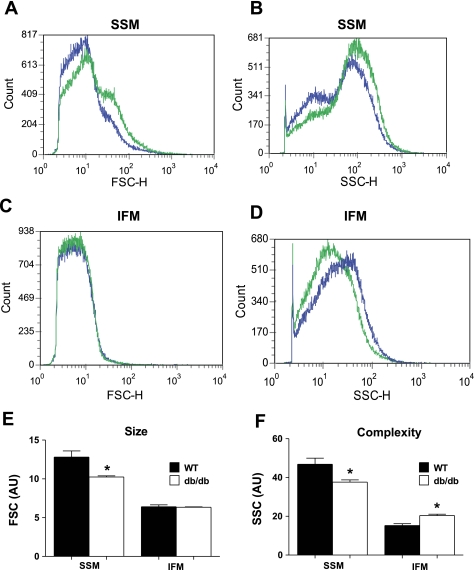
FSC, indicative of size, was significantly decreased in db/db SSM compared with WT SSM (P < 0.05; Fig. 1, A and E), yet no significant change in size in db/db IFM compared with WT IFM was observed (Fig. 1, C and E). Mitochondrial internal complexity (SSC) was significantly decreased in db/db SSM compared with WT SSM (P < 0.05; Fig. 1, B and F), whereas db/db IFM internal complexity was increased compared with WT IFM (P < 0.05; Fig. 1, D and F).
Fig. 1.
Mitochondrial subpopulation size [forward scatter (FSC)] and internal complexity [side scatter (SSC)] in wild-type (WT) and db/db hearts. The relative size and internal complexity of distinct mitochondrial subpopulations were determined using flow cytometric analyses. Mitochondrial subpopulations were stained with mitotracker deep red 633, a membrane potential (ΔΨm)-dependent dye, and gated based on incorporation of the dye. Analysis of FSC and SSC were calculated per 20,000 gated events for all mitochondrial subpopulations. A–D: representative histograms of FSC (A and C) and SSC (B and D) in WT subsarcolemmal mitochondrial (SSM) and interfibrillar mitochondria (IFM) (green) versus db/db SSM and IFM (blue). E and F: FSC (E) and SSC (F) are expressed in arbitrary units (AU). Values are ± SE; n = 4 for each group. *P < 0.05 for WT vs. db/db.
Mitochondrial Subpopulation Functional Assessment
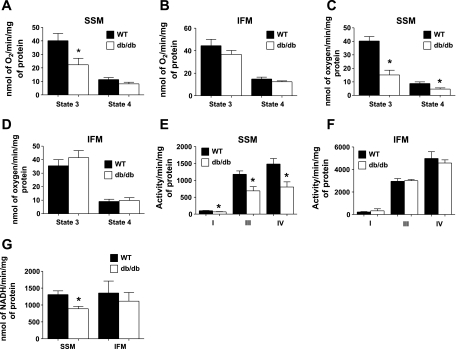
Assessment of state 3 and state 4 respiration using glutamate + malate revealed a significant decrease in state 3 respiration in db/db SSM compared with WT SSM (P < 0.05; Fig. 2A), with no significant changes between WT and db/db IFM (Fig. 2B). Furthermore, fatty acid-stimulated state 3 and state 4 respiration with palmitoylcarnitine displayed significantly decreased respiration in only db/db SSM (P < 0.05; Fig. 2C). ETC complex I, III, and IV activities were significantly decreased in db/db SSM compared with WT SSM (P < 0.05 for all three; Fig. 2E), with no significant differences between db/db IFM and WT IFM (Fig. 2F). Finally, ATP synthase activity was significantly decreased in db/db SSM compared with WT SSM (P < 0.05; Fig. 2G), with no significant differences between db/db and WT IFM (Fig. 2G).
Fig. 2.
Respiratory capacity of mitochondrial subpopulations in WT and db/db hearts. A–G: state 3 and state 4 respiration rates with glutamate + malate (A and B), palmitoylcarnitine (C and D), electron transport chain (ETC) complex activities (E and F), and ATP synthase activity (G) were assessed in isolated mitochondrial subpopulations [SSM (A, C, and E) and IFM (B, D, and F)] from hearts of 18-wk-old WT and db/db mice. State 3 and state 4 respiration rates were determined in the presence of the substrates glutamate + malate as well as palmitoylcarnitine, and state 3 respiration was examined upon the addition of ADP. ETC complex I, III, and IV activities were assessed spectrophotometrically by measuring the oxidation of NADH (complex I), reduction of cytochrome c (complex III), and oxidation of cytochrome c (complex IV). Respiration rates are expressed in nmol·min−1·mg protein−1, whereas enzymatic activities are expressed in activity·min−1·mg protein−1. Values are means ± SE; n = 6 for each group. *P < 0.05 for WT vs. db/db.
Mitochondrial Subpopulation ΔΨm
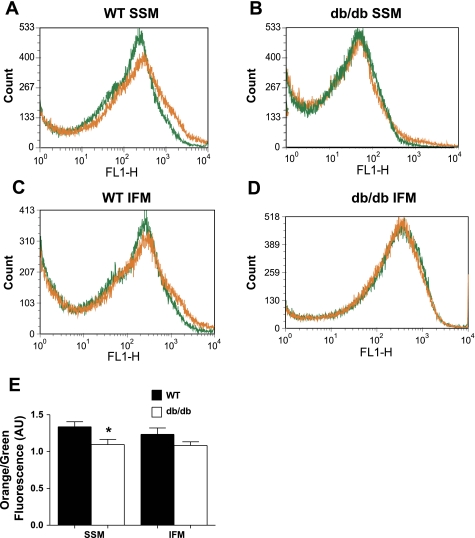
Inner mitochondrial membrane (IMM) potential (orange-to-green fluorescence ratio) was significantly decreased in db/db SSM compared with WT SSM (P < 0.05; Fig. 3, A, B, and E) due to a decrease in overall orange fluorescence. No significant differences between db/db and WT IFM were observed (Fig. 3, C–E).
Fig. 3.
Flow cytometric analysis of mitochondrial subpopulation ΔΨm in WT and db/db hearts. ΔΨm was assessed by staining mitochondrial subpopulations with JC-1 dye and assessing the shift from green to orange fluorescence with flow cytometry. A–D: representative histograms showing green and orange fluorescence in WT SSM (A) versus db/db SSM (B) and WT IFM (C) versus db/db IFM (D). E: ΔΨm was calculated based on orange-to-green fluorescence ratios in WT versus db/db cardiac mitochondrial subpopulations. Orange-to-green fluorescence ratios are expressed in AU. Values are means ± SE; n = 4 for each group. *P < 0.05 for WT vs. db/db.
Mitochondrial Subpopulation Protein Expression of ANT and UCP3
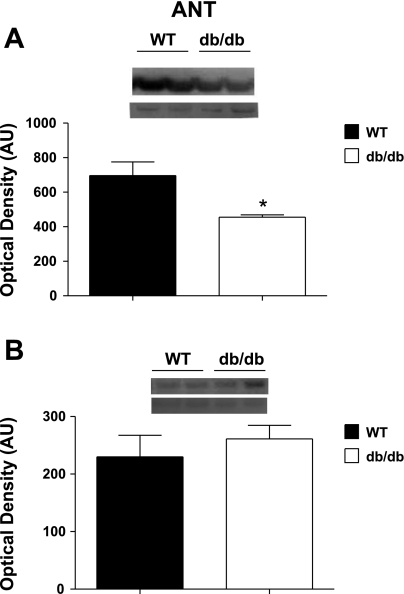
Western blot analysis of total ANT protein expression was significantly down in db/db SSM compared with WT SSM (P < 0.05; Fig. 4A), with no significant differences between db/db and WT IFM (Fig. 4B). In contrast, UCP3 expression was significantly increased in db/db SSM compared with WT (P < 0.05; Supplemental Fig. S1A), with no significant differences between db/db and WT IFM (Supplemental Fig. S1A).
Fig. 4.
Adenine nucleotide translocase (ANT) protein expression in mitochondrial subpopulations from WT and db/db hearts. A and B: representative Western blots (top) and densitometric analyses (bottom) for total ANT protein in SSM (A) and IFM (B) from WT and db/db hearts. Values are means ± SE; n = 5 for each group. *P < 0.05 for WT vs. db/db.
Mitochondrial Subpopulation Oxidative Damage
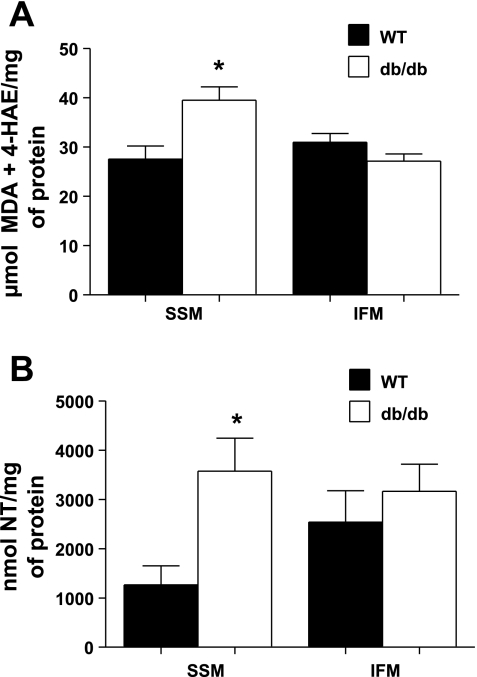
MDA and 4-HAE lipid peroxidation byproducts were significantly increased in db/db SSM compared with WT SSM (P < 0.05; Fig. 5A), with no significant changes in db/db IFM compared with WT IFM (Fig. 5A). Similarly, there was a significant increase in 3-nitrotyrosine formation in db/db SSM compared with WT SSM (P < 0.05; Fig. 5B), with no changes in db/db IFM compared with WT IFM (Fig. 5B).
Fig. 5.
Oxidative damage in mitochondrial subpopulations from WT and db/db hearts. Oxidative damage to lipids was assessed in mitochondrial subpopulations by measuring lipid peroxidation byproducts [malondialdehyde (MDA) and 4-hydroxyalkenal (4-HAE)] using a colorimetric assay and compared against a standard curve of known 4-HAE and MDA concentrations. Data are expressed in μmol MDA + 4-HAE/mg mitochondrial protein (A). Oxidative damage to proteins was assessed by quantifying 3-nitrotyrosine (NT) residues through a sandwich ELISA, and concentrations were determined by comparing results against a known protein NT standard curve. Results are expressed in nmol NT/mg protein (B). Values for both lipid peroxidation and protein nitrosylation are means ± SE; n = 5 for each group. *P < 0.05 for WT vs. db/db.
Mitochondrial Subpopulation Proteome Analyses
To determine whether the db/db phenotype differentially impacts spatially distinct mitochondrial subpopulation proteomes, we used an iTRAQ approach, which enabled the simultaneous identification and relative quantification of mitochondrial proteins through isobaric peptide tagging. Figure 6A shows an example of an MS/MS fragment ion (spectra) for a peptide that was used to identify a protein, cytochrome c oxidase subunit VIIa1. Figure 6B shows an example of an iTRAQ tag mass spectral signature (mass-to-charge ratios: 114, 115, 116, and 117) used to relatively quantitate cytochrome c oxidase subunit VIIa1 within the four labeled sample groups (see Fig. 6 for groups). A second example of the associated iTRAQ data is included for the protein UQCRC2 (Supplemental Fig. S2, A and B).
Fig. 6.
Representative iTRAQ spectra and Western blot analysis. Isolated mitochondrial subpopulations from WT and db/db hearts were labeled with iTRAQ reagents [114 (db/db SSM), 115 (db/db IFM), 116 (WT SSM), and 117 (WT IFM)] and combined for analysis with mass spectrometry. A: representative spectra of the simultaneous quantitation of cytochrome c oxidase subunit VIIa1 peptide in WT and db/db mitochondrial subpopulations. B: spectra for the reporter groups of the iTRAQ reagents (114, 115, 116, and 117) from cytochrome c oxidase subunit VIIa1 peptide mass spectrometry (MS)/MS spectra. These spectra were used along with other peptides to simultaneously quantify cytochrome c oxidase subunit VIIa1 in WT and db/db mitochondrial subpopulations. C and D: representative Western blots (top) and densitometric analyses (bottom) of cytochrome c oxidase subunit VIIa1 in SSM (C) and IFM (D) from WT and db/db hearts. Values are means ± SE; n = 5 for each group. *P < 0.05 for WT vs. db/db.
Proteins of fatty acid oxidation.
Fatty acid oxidation (FAO) proteins were significantly altered in both db/db SSM and IFM compared with WT (P < 0.05; Table 1). However, five of eight FAO proteins identified were significantly elevated in db/db SSM compared with WT SSM (P < 0.05; Table 1). In contrast, three of eight proteins identified were upregulated and three of eight proteins identified were downregulated in db/db IFM.
Table 1.
Proteomic analysis of mitochondrial subpopulations from WT and db/db hearts
| SSMD/SSMC | IFMD/IFMC | SSMC/IFMC | |
|---|---|---|---|
| Mitochondrial fatty acid β-oxidation | |||
| Hydroxyacyl-CoA dehydrogenase | 1.08 | 1.21 | 1.26 |
| Glutamate oxaloacetate transaminase 2 | 1.13 | 0.74* | 0.75 |
| Mitochondrial trifunctional protein, β-subunit | 1.20 | 1.19 | NS |
| Acyl-CoA dehydrogenase, medium chain | 1.16 | NS | 0.80 |
| 3-Ketoacyl-CoA thiolase, mitochondrial | 1.24 | NS | NS |
| Acetyl-CoA acetyltransferase 1 | NS | 0.30* | 0.54 |
| Dihyrolipoamide dehydrogenase | NS | 0.75* | 0.85 |
| Dodecenoyl-CoA δ-isomerase | NS | 1.29 | NS |
| Mitochondrial respiratory chain | |||
| ATP synthase, H+-transporting mitochondrial F0 complex, subunit d | 0.90* | NS | 0.88 |
| ATP synthase H+-transporting F0 subunit F isoform 2 | 0.90* | 1.25 | 1.35 |
| Ubquinol cytochrome c reductase core protein 2 | 0.76* | NS | NS |
| Cytochrome b-c1 complex subunit 1 | NS | 1.44 | NS |
| Cytochrome c1, heme protein, mitochondrial | 0.85* | NS | 1.21 |
| Electron-transferring flavoprotein, β-polypeptide | 0.88* | 1.44 | 1.79 |
| Succinate dehydrogenase Fp subunit | NS | 0.68 | 0.66 |
| NADH dehydogenase (ubiquinone)-1α subcomplex 9 | NS | 1.16 | NS |
| NADH dehydrogenase (ubiquinone)-1β subcomplex 10 | 0.81* | NS | NS |
| NADH dehydrogenase (ubiquinone) Fe-S protein 6 | 0.70* | NS | 0.79 |
| NADH dehydrogenase ubiquinone FE-S protein 7 | 0.76* | NS | NS |
| Cytochrome c oxidase subunit VIc | 0.88* | 1.28 | 1.41 |
| Cytochrome c oxidase subunit Va | 0.75* | 1.20 | 1.75 |
| Cytochrome c oxidase subunit VIIa1 | 0.64* | NS | 0.82 |
| Cytochrome c oxidase subunit IV isoform 1 | NS | 1.16 | NS |
| Cytochrome c oxidase subunit VIb polypeptide 1 | NS | 0.81* | NS |
| Cytochrome c oxidase subunit II | NS | 1.32 | NS |
| Tricarboxylic acid cycle | |||
| Isocitrate dehydrogenase 2 (NADP+) | 1.12 | 1.23 | NS |
| Isocitrate dehydrogenase 3 (NAD+), α | NS | 1.28 | 1.35 |
| Malate dehydrogenase 2, NAD | NS | 0.79* | NS |
| Succinyl-CoA ligase [ADP-forming] subunit-β, mitochondrial | NS | 1.30 | |
| Amino acid metabolism | |||
| Dihydrolipamide S-succinyltransferase | NS | 0.61* | NS |
| Oxidative stress related | |||
| Peroxiredoxin V | 0.80* | NS | NS |
| Transport proteins | |||
| Carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 | 1.15 | 0.87* | 1.17 |
| Glucose-related protein 75, mitochondrial heat shock protein70 | 0.80* | NS | 1.26 |
| Solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | NS | 1.28 | 0.64 |
| Voltage-dependent anion-selective channel 2 | 1.24 | NS | 0.67 |
| Solute carrier family 25 (adenine nucleotide translocator), member 4 | 0.80* | 1.28 | NS |
| Carnitine O-palmitoyltransferase 1, muscle isoform | NS | 1.37 | 0.647 |
| Voltage-dependent anion-selective channel 1 | NS | 0.70* | 0.77 |
| Voltage-dependent anion-selective channel 3 | NS | 0.50* | NS |
| Miscellaneous | |||
| NipSnap homolog 2 | NS | 1.46 | 1.36 |
| Es 1 protein | 1.15 | 0.67* | 0.85 |
| Elongation factor tu, mitochondrial | NS | 0.46* | 0.73 |
| Aldehyde dehydrogenase | 0.69* | NS | NS |
| Mirq-like protein | NS | 1.21 | NS |
| Desmin | 0.66* | NS | NS |
n = 4 for each group. Shown is the iTRAQ analysis of identified and significantly changed proteins, which were categorized into groups consisting of fatty acid oxidation, tricarboxylic acid cycle, mitochondrial respiratory chain, amino acid-related, oxidative stress, transport, and miscellaneous proteins in isolated subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM) from wild-type [WT; control (C)] and db/db [diabetic (D)] hearts. Treatment differences are indicated ratiometrically, with positive changes unmarked and negative changes marked with an asterisk (*). NS, no significant differences between any group; all other values were significant (P < 0.05 for WT vs. db/db).
Proteins of the mitochondrial respiratory chain.
A number of proteins of the mitochondrial respiratory chain were significantly altered in both db/db SSM and db/db IFM compared with WT controls. Of the 18 respiratory chain proteins identified, 11 proteins were significantly decreased in db/db SSM compared with WT SSM, whereas 8 of 18 respiratory chain proteins identified were increased in expression in db/db IFM compared with WT IFM (P < 0.05 for all; Table 1). In db/db SSM, 3 of 12 proteins were from complex I, 2 of 12 proteins were from complex III, 3 of 12 proteins were from complex IV, and 2 of 12 proteins were from complex V. Western blot analyses of UQCRC2 (Supplemental Fig. S2, C and D), a protein of complex III, and cytochrome c oxidase subunit VIIa1 (Fig. 6, C and D), a protein of complex IV, provided confirmation of the observed iTRAQ results.
Proteins of the tricarboxylic acid cycle.
Proteins of the tricarboxylic acid cycle (TCA) were significantly altered in both db/db SSM and IFM compared with WT (P < 0.05; Table 1). However, only one of four TCA proteins identified was increased in db/db SSM, whereas three of four proteins identified were upregulated in db/db IFM compared with WT IFM.
Antioxidant defense proteins.
Peroxiredoxin V (PRDX5) was the only antioxidant identified in our iTRAQ analyses that was significantly decreased in db/db SSM compared with WT SSM, whereas there were no significant changes observed in IFM antioxidant defense components (P < 0.05; Table 1).
Mitochondrial transport proteins.
ANT1 was significantly decreased in db/db SSM compared with WT SSM but upregulated in db/db IFM (P < 0.05; Table 1). Western blot analysis confirmed the overall decrease in total ANT expression in db/db SSM, whereas total ANT was unchanged in db/db IFM (Fig. 4, C and D). Glucose-regulated protein 75 was significantly decreased in db/db SSM compared with WT SSM (P < 0.05; Table 1), whereas there were no significant changes in db/db IFM compared with WT IFM.
Mitochondrial Subpopulation Posttranslational Modifications
Fifteen proteins presented with some type of posttranslational modifications in db/db SSM that were nonexistent in WT SSM, whereas ten proteins were present in db/db IFM (Supplemental Table S4). Seven of fifteen proteins identified were of the ETC, and they included specific posttranslational modifications such as acetylations, deamidations, and oxidations. Interestingly, 3 of 10 proteins identified in db/db IFM were of the ETC, and they included modifications such as acetylations, deamidations, and methylations. Of greatest interest was that the majority of the modified proteins were IMM proteins, suggesting that this locus may play a key role in the pathogenesis of cardiac dysfunction associated with type 2 diabetes mellitus.
DISCUSSION
Mitochondrial dysfunction is a major contributor to the development of various pathologies, including type 1 and type 2 diabetes mellitus (18, 24, 25, 35, 37, 38). Nevertheless, mitochondrial analyses are complicated by the fact that two spatially and functionally distinct populations of mitochondria exist in the cardiomyocyte: SSM and IFM. We and others (12, 25, 27, 30, 37) have shown that these two mitochondrial populations respond differently to pathological insults. The goal of this study was to determine whether the subcellular location was associated with a differential impact on cardiac mitochondria in a type 2 diabetes mellitus model, the db/db mouse. Our results indicate that SSM, located below the sarcolemma, display greater dysfunctional profiles compared with IFM, which are situated between the myofibrils. These dysfunctional profiles are the result of a differential impact on a number of mitochondrial indexes including structure, function, oxidative milieu, and proteome make up.
Morphological alterations have been associated with mitochondrial dysfunction (17, 24, 37, 44). Kelley et al. (24) reported a decrease in mitochondrial size and a decrease in the abundance of SSM from skeletal muscle of type 2 diabetic patients, whereas we showed a decrease in the size and complexity of cardiac SSM. Taken together, these studies suggest that type 2 diabetes mellitus imparts a specific influence on SSM structure. Interestingly, we observed a significant increase in IFM complexity (Fig. 1). It is unclear as to why such a phenomenon occurred, and it may be the result of an increased abundance of ETC components in db/db IFM, which we observed in our proteome analyses (Table 1). Regardless, the result is interesting and warrants further investigation.
Studies have suggested that mitochondrial respiration is significantly altered in the db/db heart. However, these analyses have focused on total mitochondria (3, 4). In the present study, we observed decreases in state 3 respiration and ETC complex I, III, and IV activities exclusively in db/db SSM (Fig. 2). Our results are in agreement with others reporting decreases in the succinate-driven respiration of SSM from the vastus lateralis muscle of type 2 diabetic patients (37). Additionally, we observed a significant decrease in ATP synthase activity solely in db/db SSM. These findings indicate that the ability of db/db SSM to generate ATP may be compromised; thus, those processes, as suggested by others, that rely upon ATP from this source, such as signal transduction pathways, as well as substrate transport across the plasma membrane, may be specifically impacted (37). Furthermore, we observed a significant decrease in ΔΨm in db/db SSM with no change in IFM (Fig. 3). Alterations to ΔΨm have been indicated to impact F1F0-ATP synthase function (2) and, as a result, may have contributed to the diminished function observed in db/db SSM. ANT, which catalyzes the exchange of ADP and ATP across the IMM, was significantly decreased only in db/db SSM (Fig. 4A). This observation was confirmed by the iTRAQ analyses in which ANT1 was significantly decreased in db/db SSM but increased in db/db IFM. The increase in ANT in IFM was not observed in the Western blot analyses, which may be due to the fact that total ANT was assessed and not ANT1. Inhibition of mitochondrial ANT has been proposed to contribute to cellular dysfunction in obesity and type 2 diabetes mellitus by increasing ROS and adenosine (10). Our finding of decreased ANT expression is in agreement with another report (3) and supports the hypothesis that decreases in ANT expression may contribute to a reduction in the ability to generate ATP, which was also observed (Fig. 3).
Mitochondrial oxidative stress has been reported in models of both type 1 (12, 18, 38, 44) and type 2 (3, 37) diabetes mellitus and may play a central role in mitochondrial dysfunction. In the present study, lipid peroxidation and protein nitration products were increased only in db/db SSM (Fig. 5), suggesting that type 2 diabetes mellitus presents an enhanced oxidative environment solely to SSM. Increased ROS production and oxidative stress byproducts in a type 2 diabetic model have been previously reported (3) and suggested to impact submitochondrial components, particularly in the IMM. Enhanced ROS production and lipid peroxidation byproducts can disrupt the fluidity and integrity of the IMM and further modify protein and lipid constituents within the membrane, altering overall mitochondrial function (9). Furthermore, db/db SSM displayed decreases in PRDX5, an antioxidant that reduces H2O2 and alkyl hydroperoxides. Loss of PRDX5 may have contributed to the enhanced oxidative environment observed in this particular population of mitochondria. Interestingly, PRDX5 was the only major antioxidant significantly decreased in db/db SSM. The result suggests that loss of PRDX5 may contribute to the enhanced oxidative milieu observed in db/db SSM. It is important to point out that other mechanisms may have also been involved in the increased oxidative stress observed in db/db SSM, such as the accumulation of neutral fatty acids, which are susceptible to oxidation (3, 40).
Several groups (6, 11, 20, 44, 47) have identified mitochondrial proteomic alterations with various type 1 diabetic models. However, no examination of spatially distinct mitochondrial subpopulation profiles has been performed in any pathological model, including diabetes mellitus. Kavazis et al. (23) examined the impact of exercise training on cardiac SSM and IFM proteomes and observed subpopulation-specific changes. We examined the impact of type 2 diabetic insult on spatially distinct cardiac mitochondrial proteomes. The use of iTRAQ isobaric labeling technology coupled with MudPIT-based mass spectral analyses of the labeled samples provided significant advantages over previously used methods (two-dimensional gel electrophoresis, two-dimensional differential gel electrophoresis, and label-free ultraperformance LC-MS). These advantages include global protein identification and characterization of the complex samples, simple and robust isotopic labeling that does not bias analytical separations, and simultaneous identification and relative quantification of the proteomes of the four samples. Using the iTRAQ approach, we observed an increase in FAO proteins in both SSM and IFM of db/db hearts (Table 1), which was probably due, in part, to the enhanced lipid environment present with type 2 diabetes mellitus (1). However, although the effect occurred in both subpopulations, the result was more pronounced in SSM, which saw a greater number of FAO proteins increased in abundance, suggesting a more enhanced fatty acid environment presented to this subpopulation. We also observed an increase in circulating free fatty acid levels (Supplemental Table S1). Because SSM are located at the periphery of the cell, they may be impacted to a greater extent by an enhanced lipid environment, compared with IFM, which reside in the interior of the cell. Nielson et al. (31) observed lipid accumulation specifically surrounding SSM in type 2 diabetic skeletal muscle, suggesting that SSM may be subject to a more enhanced lipid environment in the type 2 diabetic state based on its proximity. In addition, we observed increases in UCP3 protein expression in db/db SSM (Supplemental Fig. S1), which is in agreement with other authors (41). Because UCP3 has been suggested to be upregulated in the presence of enhanced free fatty acids, its expression may be indicative of a greater fatty acid environment in db/db SSM. However, although we observed an upregulation of FAO proteins in SSM, the ability to use fatty acid substrates (palmitoylcarnitine) was diminished in db/db SSM (Fig. 2), suggesting that the mitochondrial dysfunction displayed by db/db SSM at 18 wk may be due to limitations to the ETC as opposed to FAO.
In general, ETC complex proteins were primarily decreased in db/db SSM, whereas ETC and TCA proteins were increased in IFM. The changes observed in db/db SSM were not specific to one ETC complex, indicating that both proximal and distal portions of the respiratory chain were impacted. These results indicate that protein loss in SSM may be contributing to the decrease in oxidative capacity observed in SSM. Furthermore, the majority of the proteins lost in db/db SSM reside in the IMM, including ETC proteins, which may have contributed to the decrease in oxidative capacity observed in SSM. Taken together, these results suggest that the formation and/or organization of ETC constituents may have been impacted to a greater extent in db/db SSM, which may have contributed to the respiratory dysfunction observed. In contrast, the preservation of proteins involved in mitochondrial bioenergetics in db/db IFM may have been sufficient to prevent the loss of respiratory capacity.
Our proteomic analyses revealed changes to a number of other key mitochondrial proteins (Table 1). Several proteins involved in protein/substrate import necessary for proper mitochondrial function were significantly altered by type 2 diabetes mellitus. Glucose-regulated protein 75, a mitochondrial chaperone and the active motor component of the mitochondrial import process, was significantly decreased only in db/db SSM, which may have implications for the nuclear-encoded mitochondrial protein import process. Since the vast majority of mitochondrial proteins are encoded by nuclear sources, such a finding may have repercussions for a number of essential mitochondrial functions, including structure, function, and antioxidant capacity.
Because the assessment of abundance does not entirely account for protein functionality, we examined posttranslational modifications in the db/db heart of mitochondrial subpopulations. Although both mitochondrial subpopulations saw increased posttranslational modifications as a result of type 2 diabetic insult, the result was more pronounced in SSM, which saw a greater number of posttranslational modifications (Supplemental Table S4). Of particular interest was the observation that ETC proteins displayed oxidations at specific amino acid residues, and this phenomenon occurred only in db/db SSM. Oxidation to these proteins found within the ETC may alter the structure and/or function of each protein and subsequently contribute to the diminished respiratory capacity reported. As a result, this may compromise the ability of SSM to provide adequate ATP for cellular processes (Fig. 2). Additionally, our findings suggest that IFM show less protein modifications and protein loss compared with SSM, suggesting a potential adaptive response to prevent diabetes-induced mitochondrial dysfunction. It should be noted that although the MudPIT-based mass spectral method provides a wealth of global protein identification and characterization information, the larger number of peptides analyzed from an ultracomplex sample can mask those species containing posttranslational modifications, due to the greater natural abundance and higher ionization efficiencies of the nonmodified forms. Other methods exist for assessing posttranslational modifications that may use selective enrichment strategies or enhanced sensitivity; thus, additional studies may provide complementary information (15).
The most interesting finding in the present study was that in a type 2 diabetic model, cardiac SSM are affected to a greater extent by the pathology compared with cardiac IFM. These results are in contrast with our previous findings indicating that in a type 1 diabetic model (streptozotocin), IFM are most impacted by the pathology. These findings are probably due to differences in the phenotypes elicited by the two conditions, resulting from the unique pathological etiologies specific for each model. For example, higher free fatty acids and lipid deposition associated with the subsarcolemmal region during type 2 diabetes mellitus may put SSM at greater risk due to their location at the cell periphery compared with IFM (31). It is plausible that an increased lipid environment in and surrounding SSM may enhance the oxidative environment within SSM, contributing to their dysfunction. Such a phenomenon would manifest during type 2 diabetes mellitus to a greater extent with an enhanced lipid environment compared with type 1 diabetes mellitus. Regardless, it is important to point out that in multiple models of both type 1 diabetes mellitus (12, 22, 43) and type 2 diabetes mellitus (4), oxidative stress appears to be an underlying contributor to mitochondrial dysfunction. However, the mechanism in which enhanced oxidative stress manifests may be different between the type 1 and type 2 diabetic heart. Furthermore, it is important to point out that diminished function in either subpopulation may have negative implications on cardiac contraction regardless of the subpopulation. Although it has been speculated that each subpopulation provides ATP for specific cellular functions, inability to provide ATP for any of these functions may translate into contractile abnormalities in the heart.
In conclusion, mitochondrial dysfunction in the type 2 diabetic heart is confined primarily to those mitochondria residing below the plasma membrane, SSM, with a minimal effect on those mitochondria situated between the myofibrils, IFM. Of particular interest is the observation that proteins of the IMM, an important locus containing the ETC, ATP synthesis machinery, and protein trafficking constituents, were specifically impacted in SSM as a result of type 2 diabetes mellitus. Functional aspects of the IMM, such as ΔΨm, respiration, ETC activities, and ATP synthesis, were concomitantly diminished in SSM. Taken together, the combination of findings provided by our study offer evidence that not all cardiac mitochondria are impacted equally by type 2 diabetic insult but rather mitochondrial dysfunction is specific to a particular subcellular locale. Our analyses of mitochondrial subpopulation proteomic compositional changes resulting from type 2 diabetic insult reveal submitochondrial loci at risk and identify specific proteomic targets that may be of interest for therapeutic and diagnostic interventions.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant DP2-DK-083095. This work was also supported by American Heart Association (AHA) Grant-In-Aid 0855484D. E. Dabkowski is a recipient of AHA Predoctoral Fellowship 0815406D. Flow cytometry experiments were supported in part by NIH Grants RR-020866 and RR-16440.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
The authors thank Dr. Christopher Cuff as well as contributions from the West Virginia University Flow Cytometry Core Facility.
Footnotes
Supplemental Material for this article is available online at the American Journal of Physiology-Heart and Circulatory Physiology website.
REFERENCES
- 1.Abdul-Ghani MA, Muller FL, Liu Y, Chavez AO, Balas B, Zuo P, Chang Z, Tripathy D, Jani R, Molina-Carrion M, Monroy A, Folli F, Van Remmen H, DeFronzo RA. Deleterious action of FA metabolites on ATP synthesis: possible link between lipotoxicity, mitochondrial dysfunction, and insulin resistance. Am J Physiol Endocrinol Metab 295: E678–E685, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Bornhovd C, Vogel F, Neupert W, Reichert AS. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J Biol Chem 281: 13990–13998, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Boudina S, Abel ED. Mitochondrial uncoupling: a key contributor to reduced cardiac efficiency in diabetes. Physiology (Bethesda) 21: 250–258, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Boudina S, Sena S, Theobald H, Sheng X, Wright JJ, Hu XX, Aziz S, Johnson JI, Bugger H, Zaha VG, Abel ED. Mitochondrial energetics in the heart in obesity-related diabetes: direct evidence for increased uncoupled respiration and activation of uncoupling proteins. Diabetes 56: 2457–2466, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254, 1976 [DOI] [PubMed] [Google Scholar]
- 6.Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, Ganesan B, Weimer BC, Abel ED. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58: 1986–1997, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J Biol Chem 217: 383–393, 1955 [PubMed] [Google Scholar]
- 8.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. VI. The effects of adenosine diphosphate on azide-treated mitochondria. J Biol Chem 221: 477–489, 1956 [PubMed] [Google Scholar]
- 9.Chen JJ, Yu BP. Alterations in mitochondrial membrane fluidity by lipid peroxidation products. Free Radic Biol Med 17: 411–418, 1994 [DOI] [PubMed] [Google Scholar]
- 10.Ciapaite J, Bakker SJ, Diamant M, van Eikenhorst G, Heine RJ, Westerhoff HV, Krab K. Metabolic control of mitochondrial properties by adenine nucleotide translocator determines palmitoyl-CoA effects. Implications for a mechanism linking obesity and type 2 diabetes. FEBS J 273: 5288–5302, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Dabkowski E. Quantitative proteomic analysis of distinct mitochondrial subpopulations in diabetic myocardium. FASEB J 1226.36: 2008 [Google Scholar]
- 12.Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, Peer CJ, Callery PS, Hollander JM. Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol 296: H359–H369, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med 45: 855–865, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Daniels A, van Bilsen M, Janssen BJ, Brouns AE, Cleutjens JP, Roemen TH, Schaart G, van der Velden J, van der Vusse GJ, van Nieuwenhoven FA. Impaired cardiac functional reserve in type 2 diabetic db/db mice is associated with metabolic, but not structural, remodelling. Acta Physiol (Oxf). In press [DOI] [PubMed] [Google Scholar]
- 15.Desiderio DM, Nibbering NM. Redox Proteomics: From Protein Modifications to Cellular Dysfunction and Diseases, edited by Dalle-Donne I, Scaloni A, Butterfield DA. Hoboken, NJ: Wiley, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Feniouk BA, Suzuki T, Yoshida M. Regulatory interplay between proton motive force, ADP, phosphate, and subunit epsilon in bacterial ATP synthase. J Biol Chem 282: 764–772, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Figueiredo PA, Ferreira RM, Appell HJ, Duarte JA. Age-induced morphological, biochemical, and functional alterations in isolated mitochondria from murine skeletal muscle. J Gerontol A Biol Sci Med Sci 63: 350–359, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Flarsheim CE, Grupp IL, Matlib MA. Mitochondrial dysfunction accompanies diastolic dysfunction in diabetic rat heart. Am J Physiol Heart Circ Physiol 271: H192–H202, 1996 [DOI] [PubMed] [Google Scholar]
- 19.Garcia MJ, McNamara PM, Gordon T, Kannel WB. Morbidity and mortality in diabetics in the Framingham population. Sixteen year follow-up study. Diabetes 23: 105–111, 1974 [DOI] [PubMed] [Google Scholar]
- 20.Hamblin M, Friedman DB, Hill S, Caprioli RM, Smith HM, Hill MF. Alterations in the diabetic myocardial proteome coupled with increased myocardial oxidative stress underlies diabetic cardiomyopathy. J Mol Cell Cardiol 42: 884–895, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofhaus G, Shakeley RM, Attardi G. Use of polarography to detect respiration defects in cell cultures. Meth Enzymol 264: 476–483, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Kaul N, Siveski-Iliskovic N, Thomas TP, Hill M, Khaper N, Singal PK. Probucol improves antioxidant activity and modulates development of diabetic cardiomyopathy. Nutrition 11: 551–554, 1995 [PubMed] [Google Scholar]
- 23.Kavazis AN, Alvarez S, Talbert E, Lee Y, Powers SK. Exercise training induces a cardioprotective phenotype and alterations in cardiac subsarcolemmal and intermyofibrillar mitochondrial proteins. Am J Physiol Heart Circ Physiol 297: H144–H152, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950, 2002 [DOI] [PubMed] [Google Scholar]
- 25.King KL, Young ME, Kerner J, Huang H, O'Shea KM, Alexson SE, Hoppel CL, Stanley WC. Diabetes or peroxisome proliferator-activated receptor alpha agonist increases mitochondrial thioesterase I activity in heart. J Lipid Res 48: 1511–1517, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685, 1970 [DOI] [PubMed] [Google Scholar]
- 27.Lesnefsky EJ, Slabe TJ, Stoll MS, Minkler PE, Hoppel CL. Myocardial ischemia selectively depletes cardiolipin in rabbit heart subsarcolemmal mitochondria. Am J Physiol Heart Circ Physiol 280: H2770–H2778, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Lin D. Multidimensional protein identification technology as an effective tool for proteomics. Am Genomic/Proteomic Technol 1: 38–46, 2001 [Google Scholar]
- 29.Masaki Y, Kato A, Kato C, Fujii E, Adachi K, Miyoshi A, Suzuki M. Segmentation of the pathophysiological stages of diabetic changes in the db/db mouse. J Toxicol Pathol 22: 133–137, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mollica MP, Lionetti L, Crescenzo R, D'Andrea E, Ferraro M, Liverini G, Iossa S. Heterogeneous bioenergetic behaviour of subsarcolemmal and intermyofibrillar mitochondria in fed and fasted rats. Cell Mol Life Sci 63: 358–366, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen J, Mogensen M, Vind BF, Sahlin K, Hojlund K, Schroder HD, Ortenblad N. Increased subsarcolemmal lipids in type 2 diabetes: effect of training on localization of lipids, mitochondria, and glycogen in sedentary human skeletal muscle. Am J Physiol Endocrinol Metab 298: E706–E713, 2010 [DOI] [PubMed] [Google Scholar]
- 33.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977 [PubMed] [Google Scholar]
- 34.Pullman ME, Penefsky HS, Datta A, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J Biol Chem 235: 3322–3329, 1960 [PubMed] [Google Scholar]
- 35.Rabol R, Boushel R, Dela F. Mitochondrial oxidative function and type 2 diabetes. Appl Physiol Nutr Metab 31: 675–683, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Reers M, Smith TW, Chen LB. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry 30: 4480–4486, 1991 [DOI] [PubMed] [Google Scholar]
- 37.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54: 8–14, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Rolo AP, Palmeira CM. Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol 212: 167–178, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Rosca MG, Okere IA, Sharma N, Stanley WC, Recchia FA, Hoppel CL. Altered expression of the adenine nucleotide translocase isoforms and decreased ATP synthase activity in skeletal muscle mitochondria in heart failure. J Mol Cell Cardiol 46: 927–935, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Russell AP, Gastaldi G, Bobbioni-Harsch E, Arboit P, Gobelet C, Deriaz O, Golay A, Witztum JL, Giacobino JP. Lipid peroxidation in skeletal muscle of obese compared with endurance-trained humans: a case of good vs. bad lipids? FEBS Lett 551: 104–106, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Schrauwen P, Hesselink MK. Oxidative capacity, lipotoxicity, and mitochondrial damage in type 2 diabetes. Diabetes 53: 1412–1417, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Semeniuk LM, Kryski AJ, Severson DL. Echocardiographic assessment of cardiac function in diabetic db/db and transgenic db/db-hGLUT4 mice. Am J Physiol Heart Circ Physiol 283: H976–H982, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Shen X, Ye G, Metreveli NS, Epstein PN. Cardiomyocyte defects in diabetic models and protection with cardiac-targeted transgenes. Methods Mol Med 112: 379–388, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab 287: E896–E905, 2004 [DOI] [PubMed] [Google Scholar]
- 45.Taegtmeyer H, McNulty P, Young ME. Adaptation and maladaptation of the heart in diabetes: part I: general concepts. Circulation 105: 1727–1733, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Meth Enzymol 264: 484–509, 1996 [DOI] [PubMed] [Google Scholar]
- 47.Turko IV, Murad F. Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem 278: 35844–35849, 2003 [DOI] [PubMed] [Google Scholar]