Aminoacyl-tRNA synthetases (aaRSs) are the enzymes normally responsible for the attachment of amino acids (aa) to tRNAs. Numerous paralogous proteins of aaRSs have been identified in a wide range of organisms, but the functions of most of these aaRS-like proteins are yet to be determined. In PNAS, the study by Mocibob et al. (1) identifies a paralog of seryl-tRNA synthetase that does not aminoacylate a tRNA, but instead, aminoacylates an aa carrier protein. This exciting discovery provides an unforeseen function for the aaRS architecture and also uncovers a possible evolutionary link between ribosome-catalyzed translation and nonribosomal peptide synthesis.
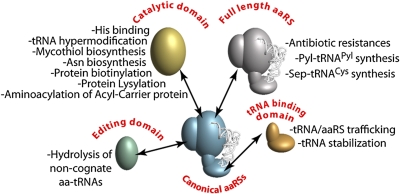
aaRSs are an ancient and ubiquitous family of enzymes responsible for the specific pairing of each of the 20 aa used in protein synthesis with their cognate tRNAs, thereby providing the 20 different aminoacyl-tRNAs (aa-tRNAs) necessary for ribosomes to translate mRNAs. Consistent with this role in defining the genetic code, aaRSs display a high specificity for their aa and tRNA, a property that helps to ensure fidelity during protein synthesis. The aaRSs are distributed between two equally populated, evolutionary unrelated classes of enzymes. aaRSs within the same class display common structural features and evolutionary origin. During the course of evolution, duplication and divergence of aaRS genes yielded enzymes with different specificities, allowing various aa to infiltrate the genetic code. Although all extant genomes possess a set of tRNA aminoacylation systems adequate for protein biosynthesis, many organisms also encode additional paralogs of aaRSs. Recent efforts to the functional characterization of these paralogs have shown that these enzymes are sometimes not only involved in tRNA aminoacylation but have also evolved to accomplish a wide range of other functions (Fig. 1).
Fig. 1.
Functions of free-standing aaRS paralogs.
Several aaRS paralogs have been identified as isoenzymes (i.e., bona fide duplicates with the same aa and tRNA specificity as the canonical enzyme) (2). The most commonly known function for such duplications, which probably occurred recently in the evolution of aaRSs, is that one isoenzyme displays sequence variations in its active site that provide resistance to natural inhibitors, which block the activity of the second isoenzyme (3, 4). One of the more notable examples is the duplication of isoleucyl-tRNA synthetase, which provides resistance to the antibiotic mupirocin (reviewed in ref. 5). In other cases, it has been shown that what at first appeared to be isoenzymes are instead aaRSs, which synthesize aa-tRNAs outside of the canonical 20 normally used in protein synthesis. In some archaeons, for instance, pyrrolysyl-tRNA synthetase (PylRS) is responsible for the synthesis of Pyl-tRNAPyl, which is used to translate certain UAG stop codons as pyrrolysine (6). Phosphoseryl-tRNA synthetase is another example of a noncanonical aaRSs that is responsible for the synthesis of phosphoseryl-tRNACys, an intermediate for the formation of Cys-tRNACys in methanogenic archaea (7). Despite their narrow distribution within some archaeons, phylogenetic analyses showed that these enzymes arose very early in the evolution of aaRSs and probably existed in the last universal common ancestor but have not been retained in most organisms (8).
aaRSs are modular enzymes with a core catalytic domain responsible for ATP-dependent acyl-adenylate synthesis, which allows transfer of activated aa on to the 3′ end of tRNA. It is believed that additional modules appeared in aaRSs to maintain the accuracy of aminoacylation and protein synthesis as the number of aa increased in the genetic code. Most of the additional domains present in aaRSs assist RNA recognition (RNA binding domains) or possess a proofreading activity hydrolyzing incorrect aminoacylation products (editing domains). Interestingly, paralogs of both the core and appended domains of aaRSs are encoded in a wide range of genomes as stand-alone proteins. Although many of these paralogs play roles similar to those observed in aaRSs (9, 10), they are also involved in various other processes, such as tRNA and aaRS trafficking between the cytoplasm and nucleus (11) and the stabilization of tRNA structures (12). Beyond these various activities, all of which involve normal tRNA recognition, other aaRS paralogs either bind tRNA in an unusual fashion (13) or sometimes have roles that do not require tRNA at all. HisZ, a paralog of the catalytic domain of HisRS, has no tRNA binding domain and does not have any ligase activity, but instead, acts as an aa-sensing subunit, regulating the first step of His biosynthesis (14). Most paralogs of aaRS catalytic domains characterized to date still function as ligases but often use an amino group, not hydroxyl, as an aa acceptor. For example, the CysRS paralog MshC transfers Cys on the amino group of a precursor of mycothiol (15), and BirA, a distant paralog of SerRS, activates and transfers biotin onto Lys of biotin-carrier proteins (16). In bacteria and archaeons, AsnRS paralogs (AsnA) synthesize Asn using ammonia as the acceptor group of Asp, which is activated by its β-carboxylic group (17). In proteobacteria, PoxA, a paralog of LysRS, transfers Lys on the ε-amino group of a conserved Lys residue of elongation factor P, resulting in a posttranslationally modified protein required for virulence of Salmonella typhimurium (18). One of the keys to identifying the role of PoxA came from analyzing the general conservation of the genomic context in which it is encoded (19), a strategy that also proved important in the recent work of Mocibob et al. (1).
Mocibob et al. (1) notice that several bacteria encode proteins that resemble archaeal SerRSs but are unable to aminoacylate tRNA. Structural investigations showed that these atypical SerRS paralogs are deprived of a tRNA binding domain, whereas the other structural features crucial for SerRS activity are still intact. Furthermore, biochemical studies also showed that Ser is not the main substrate of the SerRS paralogs, but rather, Gly and Ala are both efficiently activated by these enzymes. The SerRS paralogs are encoded in a region that also contains a gene for a putative acyl-carrier protein predicted to use phosphopantetheine as a prosthetic group and acyl-acceptor. The proximity of these genes prompted Mocibob et al. (1) to investigate if the SerRS paralogs possess aminoacyl-[carrier protein] ligase (aaCP ligase) activity, which they verified by showing the transfer of Gly or Ala to the thiol function of the prosthetic group bound to the putative carrier protein. The exact role of the acylated carrier protein is currently unknown,
Several bacteria encode proteins that resemble archaeal SerRSs but are unable to aminoacylate tRNA.
but proteins with a similar prosthetic group are used for diverse biosynthetic processes involving acyl condensation (e.g., synthesis of peptides, oligosaccharides, and fatty acids). The unearthing of an aaCP ligase activity supported by an aaRS paralog is an exciting discovery, because it could represent a missing link between thioester- and ribosome-catalyzed peptide synthesis. Such a connection was proposed several years ago in a series of investigations by Jakubowski (20), who observed that aaRSs of both classes weakly aminoacylate several thiolated compounds, among which CoA and pantetheine are known to be used in nonribosomal peptide synthesis. This activity was thought to be a relic from ancestral aaRSs, which may have acted as aaCP ligases in nonribosomal peptide synthesis before acquiring their aa-tRNA synthetase (ligase) activity. Before the discovery by Mocibob et al. (1) , no evidence from among contemporary metabolic functions had been found to support this scenario. Furthermore, apart from the functional resemblance between aaRS and aaCP ligases, there is not any structural similarity between these two types of enzymes, an observation that pleads in favor of a separate evolutionary history for these proteins. Further phylogenetic and structural investigations will now be necessary to provide more clues to determine whether these SerRS paralogs are a molecular fossil and whether acyl-adenylate synthesis in the ribosomal and nonribosomal protein synthesis worlds has a common evolutionary origin.
Acknowledgments
Work on tRNA and aaRS paralogs in the authors’ laboratory is supported by National Institutes of Health Research Grant R01GM065183.
Footnotes
The authors declare no conflict of interest.
See companion article on page 14585.
References
- 1.Mocibob M, et al. Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis. Proc Natl Acad Sci USA. 2010;107:14585–14590. doi: 10.1073/pnas.1007470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lévêque F, Plateau P, Dessen P, Blanquet S. Homology of lysS and lysU, the two Escherichia coli genes encoding distinct lysyl-tRNA synthetase species. Nucleic Acids Res. 1990;18:305–312. doi: 10.1093/nar/18.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brevet A, Chen J, Lévêque F, Blanquet S, Plateau P. Comparison of the enzymatic properties of the two Escherichia coli lysyl-tRNA synthetase species. J Biol Chem. 1995;270:14439–14444. doi: 10.1074/jbc.270.24.14439. [DOI] [PubMed] [Google Scholar]
- 4.Zeng Y, Roy H, Patil PB, Ibba M, Chen S. Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974. Antimicrob Agents Chemother. 2009;53:4619–4627. doi: 10.1128/AAC.00782-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JR, et al. Horizontal transfer of drug-resistant aminoacyl-transfer-RNA synthetases of anthrax and Gram-positive pathogens. EMBO Rep. 2003;4:692–698. doi: 10.1038/sj.embor.embor881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight SK, et al. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 7.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 8.Kavran JM, et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. An isolated class II aminoacyl-tRNA synthetase insertion domain is functional in amino acid editing. J Biol Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- 11.Frechin M, et al. Yeast mitochondrial Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009;23:1119–1130. doi: 10.1101/gad.518109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushiro T, Schimmel P. Trbp111 selectively binds a noncovalently assembled tRNA-like structure. Proc Natl Acad Sci USA. 2002;99:16631–16635. doi: 10.1073/pnas.262667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaise M, et al. Glu-Q-tRNA(Asp) synthetase coded by the yadB gene, a new paralog of aminoacyl-tRNA synthetase that glutamylates tRNA(Asp) anticodon. Biochimie. 2005;87:847–861. doi: 10.1016/j.biochi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Champagne KS, Piscitelli E, Francklyn CS. Substrate recognition by the hetero-octameric ATP phosphoribosyltransferase from Lactococcus lactis. Biochemistry. 2006;45:14933–14943. doi: 10.1021/bi061802v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sareen D, Steffek M, Newton GL, Fahey RC. ATP-dependent L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry. 2002;41:6885–6890. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- 16.Artymiuk PJ, Rice DW, Poirrette AR, Willet P. A tale of two synthetases. Nat Struct Biol. 1994;1:758–760. doi: 10.1038/nsb1194-758. [DOI] [PubMed] [Google Scholar]
- 17.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc Natl Acad Sci USA. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarre WW, et al. PoxA, YjeK and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailly M, de Crécy-Lagard V. Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol Direct. 2010;5:3. doi: 10.1186/1745-6150-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakubowski H. Aminoacylation of coenzyme A and pantetheine by aminoacyl-tRNA synthetases: Possible link between noncoded and coded peptide synthesis. Biochemistry. 1998;37:5147–5153. doi: 10.1021/bi972528v. [DOI] [PubMed] [Google Scholar]