Abstract
Tumors with mutant BRAF and some with mutant RAS are dependent upon ERK signaling for proliferation, and their growth is suppressed by MAPK/ERK kinase (MEK) inhibitors. In contrast, tumor cells with human EGF receptor (HER) kinase activation proliferate in a MEK-independent manner. These findings have led to the development of RAF and MEK inhibitors as anticancer agents. Like MEK inhibitors, the RAF inhibitor PLX4032 inhibits the proliferation of BRAFV600E tumor cells but not that of HER kinase-dependent tumors. However, tumors with RAS mutation that are sensitive to MEK inhibition are insensitive to PLX4032. MEK inhibitors inhibit ERK phosphorylation in all normal and tumor cells, whereas PLX4032 inhibits ERK signaling only in tumor cells expressing BRAFV600E. In contrast, the drug activates MEK and ERK phosphorylation in cells with wild-type BRAF. In BRAFV600E tumor cells, MEK and RAF inhibitors affect the expression of a common set of genes. PLX4032 inhibits ERK signaling output in mutant BRAF cells, whereas it transiently activates the expression of these genes in tumor cells with wild-type RAF. Thus, PLX4032 inhibits ERK signaling output in a mutant BRAF-selective manner. These data explain why the drug selectively inhibits the growth of mutant BRAF tumors and suggest that it will not cause toxicity resulting from the inhibition of ERK signaling in normal cells. This selectivity may lead to a broader therapeutic index and help explain the greater antitumor activity observed with this drug than with MEK inhibitors.
The prevalence of deregulation of ERK signaling in human tumors (1–3) suggests that drugs that inhibit the pathway might have significant therapeutic activity (4–7). Selective, allosteric MAPK/ERK kinase (MEK) inhibitors (MEKi) have been useful for determining the utility and feasibility of therapeutic targeting of the ERK pathway. In such studies, the consequences of ERK activation in tumors have been shown to vary as a function of the mechanism of pathway activation (8, 9). Tumors with BRAF mutation require ERK signaling for G1 progression and sometimes survival and almost always are sensitive to MEK inhibition. In contrast, tumors in which ERK activation is caused by receptor tyrosine kinase (RTK) deregulation grow in a MEK/ERK-independent manner (8, 9). The role of ERK signaling in tumors with activated mutant RAS is more complex, and both MEK-dependent and -independent subsets of mutant RAS tumor cell lines have been identified (10, 11).
Receptor activation of ERK signaling is regulated by negative feedback (12), but mutant BRAF is unresponsive to inhibition of MEK-dependent feedback (13, 14). Thus, the V600E mutation both elevates the catalytic activity of BRAF (15) and renders it insensitive to negative feedback, leading to hyperactivation of ERK signaling and dependence of the tumor cell on the pathway. These findings prompted clinical trials of MEKi in patients with advanced cancer, including those with BRAF mutation (16–18). In these trials, significant inhibition of ERK has been achieved in normal and tumor tissue. The predominant toxicity has been skin rash (17). Despite promising preclinical data, however, the antitumor effects of MEKi have been modest. For example, in a recent phase II trial only 12% of patients with melanoma whose tumors harbored BRAFV600E had a partial response to the MEKi AZD6244 according to the response evaluation criteria in solid tumors (RECIST) (18). Many factors may be responsible for the modest clinical activity of MEKi. Other substrates of RAF have been identified (19–21), and thus MEKi may suppress only some of the consequences of activated RAF kinase. To test the possibility that inhibition of MEK and RAF have different consequences, we compared the effects of an MEKi with those elicited by an ATP-competitive RAF inhibitor (RAFi) PLX4032 that binds to BRAFV600E, wild-type CRAF, and wild-type BRAF (BRAFWT) with Ki50 values of 35, 48, and 110 nM, respectively (22).
Results
MEK Inhibition and RAF Inhibition Affect Expression of the Same Genes in BRAFV600E Melanomas.
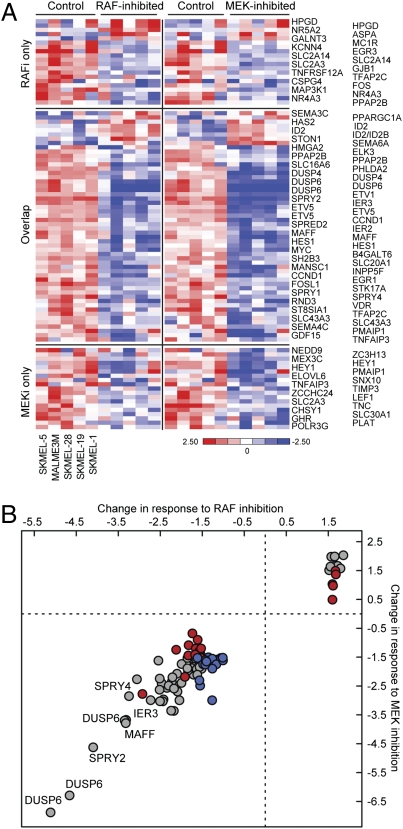
We previously used the MEKi PD0325901 to identify a set of 52 genes that comprises the transcriptional output of the ERK pathway in BRAFV600E tumors. To investigate whether non-MEK substrates of BRAFV600E play a role in mediating its effects, we compared the effects of PLX4032 and PD0325901 on the transcriptome of these cells. We determined the effects of 8 h of exposure to either drug on gene expression in five melanoma cell lines harboring BRAFV600E mutation (Malme3M, SkMel-1, SkMel-5, SkMel-19, and SkMel-28). Fifty-nine MEKi-dependent genes and 58 RAFi-dependent genes were identified (Methods), represented by 73 and 74 probe sets, respectively. Forty-four of these genes overlapped statistically in their response to both inhibitors (Fig. 1A). However, most of the genes whose expression changed significantly upon MEK inhibition changed similarly and to roughly the same extent with RAF inhibition, and vice versa (Fig. 1B and Table S1). We found, therefore, that the identification of genes selectively affected by only one drug was a statistical artifact, because the change in expression of these genes was just above or below the statistically defined cutoff (of fold change and/or q value). We conclude that the ERK-dependent transcriptional output is characterized by the differential expression of 73 genes (Fig. 1A, “RAFi-only” plus “MEKi-only” plus “overlap”) and found no genes that were differentially regulated by the RAFi. A 4-fold higher concentration of PLX4032 yielded the same results (Fig. S1). In summary, there was no evidence for MEK-independent effects of PLX4032 on the transcriptome of BRAFV600E tumor cells.
Fig. 1.
Identification of RAF- and MEK-dependent gene expression in BRAFV600E melanoma cells. (A) Heat map representation of 93 probe sets (73 genes), in each of five BRAFV600E cell lines in the presence of DMSO (control), 250 nM PLX4032 (RAFi), or 50 nM PD0325901 (MEKi). Genes were categorized by their change in expression in response to PLX4032 (“RAFi only”), to PD0325901 (“MEKi only”), or to both (“overlap”). (B) The correlation between the average change in expression of each of the 73 ERK-dependent genes across BRAFV600E melanoma cells in response to either RAFi or MEKi. The plot distinguishes genes responding to “RAFi only” (red), to “MEKi only” (blue), or to both (gray).
Only Mutant BRAF Tumors Are Sensitive to PLX4032.
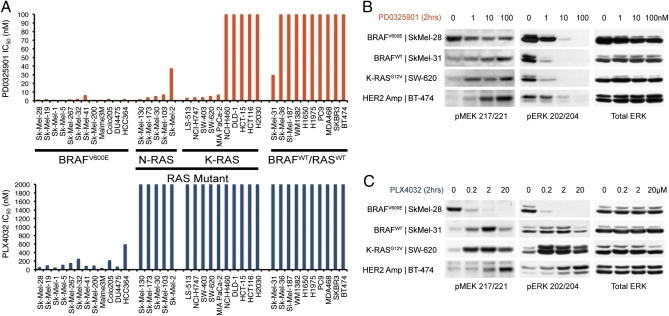
To compare the effects of PLX4032 with those of PD0325901 on the proliferation of cells with ERK activation, we screened 12 BRAFV600E, 15 RAS-mutant, and 10 BRAFWT/RASWT cancer cell lines (Fig. 2A). All cell lines with BRAFV600E mutation were sensitive to both the MEKi and RAFi, whereas both MEKi and RAFi were ineffective in inhibiting BRAFWT/RASWT tumor cells, including those with activated human EGF receptor (HER)-family RTKs. Only some mutant RAS tumor cells were sensitive to MEK inhibition (Fig. 2A). The mutant RAS tumor cells that were MEKi-resistant also were insensitive to PLX4032. In contrast, however, MEKi-sensitive, mutant RAS tumor cells were uniformly insensitive to PLX4032 (Fig. 2A). Thus, sensitivity to PLX4032 was limited exclusively to cell lines harboring BRAFV600E.
Fig. 2.
PLX4032 selectively inhibits the growth of BRAFV600E-mutant cell lines. (A) Day five IC50 values for PD0325901 (orange) and PLX4032 (blue) in a panel of 37 cell lines with BRAFV600E mutation, with RAS mutation, and with BRAFWT/RASWT, determined using the Alamar Blue assay (Methods). (B and C) MAPK activity as measured by immunoblot detection of pMEK1/2 (Ser217/221) and pERK1/2 (Thr202/Tyr204) in select BRAFV600E, RAS mutant, and BRAFWT/RASWT cell lines from A, after 2-h exposure to PD0325901 (B) or PLX4032 (C) over a range of concentrations.
PLX4032 Inhibits MEK and ERK Phosphorylation Only in Tumors with BRAFV600E Mutation and Activates Phosphorylation in BRAFWT Cells.
The insensitivity of MEK-dependent RAS-mutant tumors to PLX4032 was puzzling initially. PD0325901 (1–10 nM) inhibits phosphorylated ERK (pERK) in all cell lines examined (Fig. 2B) (8). ERK causes feedback inhibition of pathways that activate RAF signaling (12, 23, 24), and thus levels of phosphorylated MEK (pMEK) are low in tumor cells in which the pathway is activated by HER2 (BT-474), mutant RAS (SW-620), and other cell lines with BRAFWT (SkMel-31) but are elevated in those containing BRAFV600E (SkMel-28). Exposure of BRAFWT cells to the MEKi relieves this feedback and induces MEK phosphorylation, whereas in mutant BRAF tumors MEK inhibition does not further induce pMEK (Fig. 2B) (13).
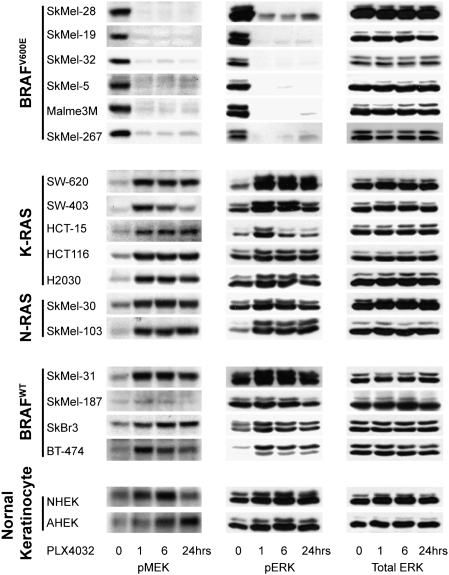
PLX4032 rapidly suppressed pMEK and pERK in all cell lines with BRAFV600E (Figs. 2C and 3). In marked contrast, however, PLX4032 induced MEK and ERK phosphorylation in BRAFWT cells (Fig. 3), including those with mutant N- and K-RAS, those with RTK activation, and in two normal keratinocyte cultures. Induction of MEK/ERK phosphorylation was rapid and persisted for at least 24 h. Induction occurred over a range of concentrations, including those that inhibit signaling in cells with mutant BRAF, but began to diminish at 20 μM PLX4032 (Fig. 2C). Thus, the sensitivity of ERK signaling to PLX4032 was confined to tumor cells with BRAFV600E and correlates with inhibition of proliferation. Paradoxical activation of RAF by other RAFi has been noted previously and attributed to relief of negative feedback (25). We and others have tested six other ATP-competitive inhibitors of RAF kinase, including an irreversible inhibitor, and found that all induce phosphorylation of MEK and ERK in cells with BRAFWT, so this property probably is common to this class of compounds (26).
Fig. 3.
PLX4032 inhibits ERK phosphorylation only in BRAFV600E-mutant cell lines. Immunoblots of pMEK, pERK, and total ERK for a panel of tumor and primary human keratinocyte cell lines. Cells were treated with 2 μM PLX4032 for 1, 6, and 24 h.
PLX4032 Inhibits ERK Signaling Output in Cells with BRAFV600E Mutation.
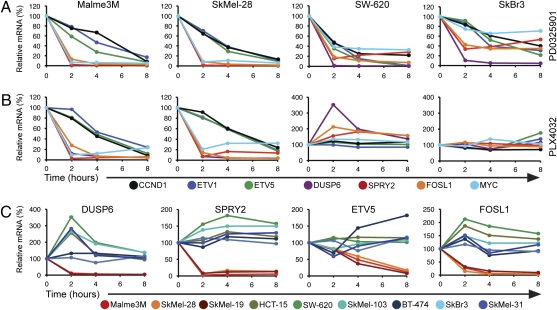
The induction of pERK by PLX4032 suggests that RAF inhibition may activate ERK signaling and perhaps accelerate the proliferation of BRAFWT cells. However, activation of the ERK signaling pathway is regulated by negative feedback at multiple sites. Phosphorylation and activation of ERK induces the expression of dual-specificity phosphatases (DUSPs) (27). Because increasing ERK signaling output increases DUSP expression and thereby enhances ERK dephosphorylation, pERK is a poor measure of ERK signaling output. Although pERK levels are similar in tumors with BRAF mutation and those with RTK activation, only the former have increased ERK output (13). Thus, induction of pERK in BRAFWT tumors may not reflect an increase in ERK signaling output. We therefore evaluated the effects of PLX4032 on ERK-dependent transcriptional output in tumor cells with mutant and wild-type BRAF. The set of 73 ERK-dependent genes that comprise ERK transcriptional output includes transcription factors important in mediating transformation (ETV1, ETV5, FOSL1, and MYC), feedback regulators (SPRY2 and DUSP6) (27, 28), and downstream effectors of ERK-signaling, including cyclin D1 (CCND1). We used quantitative RT-PCR to measure changes in the expression of some of these genes in cells exposed to PLX4032 or to PD0325901.
The MEKi reduced the expression of these genes in all cell lines tested (Fig. 4A), including MEKi-sensitive BRAFV600E cell lines (Malme3M and SkMel-28), mutant RAS (SW-620) tumor cells with elevated ERK output, and the MEKi-insensitive, HER2-dependent, SkBr3 cell line in which ERK output is low. In SkBr3, although the steady-state ERK output is low and the change in expression modest, the expression of each gene fell with MEK inhibition. In each of the cell lines, DUSP6, SPRY2, FOSL1, and MYC were down-regulated rapidly by 80–95% after 2 h of drug exposure. Down-regulation of the other genes (ETV1, ETV5, CCND1) occurred with slower kinetics. In BRAFV600E cell lines, the effects of PLX4032 were essentially identical to those of PD0325901: down-regulation of all seven genes (Fig. 4B and Fig. S2A). Thus, in agreement with the microarray data (Fig. 1), the consequences of RAF and MEK inhibition were nearly identical in BRAFV600E cells.
Fig. 4.
PLX4032 inhibited the ERK-dependent transcriptional output of BRAFV600E cell lines. The expression levels of ERK output genes were determined by RT-PCR in the designated cell lines. Cells were treated with 50 nM PD0325901 (A) or 250 nM PLX4032 (B) for 0, 2, 4, and 8 h. Values are expressed as the relative mRNA level in drug-treated samples compared with time 0. (C) Relative mRNA levels of DUSP6, SPRY2, ETV5, and FOSL1 in PLX4032-treated BRAFV600E and BRAFWT cell lines over time. Untreated RNA expression is set at 100% for each individual cell line.
Very different results were obtained in BRAFWT tumor cells. Some of these BRAFWT cells are sensitive to MEK inhibition, including a subset of tumors with mutant N- or K-RAS. In such cells [SW-620 (K-RAS), SkMel-103 (N-RAS), and SkMel-31 (BRAFWT/RASWT)], MEKi inhibited ERK output (Fig. 4A), but PLX4032 did not (Fig. 4 B and C). Instead, in these cell lines, PLX4032 caused a rapid induction of DUSP6, SPRY2, and, in some cells, FOSL1 and MYC expression. Induction of DUSP6 (2.5- to 3.5-fold), SPRY2 (1.4- to 1.8-fold), and FOSL1 (1.4- to 2.1-fold) was most marked, whereas other genes were not significantly induced. However, the induction of DUSP6 and SPRY2 in cells with RAS mutation was transient compared with the persistence of pMEK and pERK induction (Fig. 3), and their expression re-mained only slightly elevated (1.5-to 1.7-fold) 8 h after treatment. Tumor cells with activated HER kinases (e.g., BT-474 and SkBr3) have low ERK output and are insensitive to MEK inhibition. PLX4032 treatment caused pMEK and pERK induction in these cells as well (Fig. 3). However, despite durable induction of pERK at 250 nM, expression of ERK-dependent genes was affected marginally or not at all in these and in KIT-driven (WM1382; ref. 29) tumor cells.
We further quantified ERK transcriptional output by taking the mean of Z-scores of the 73 genes in the set as a measure of aggregate ERK-dependent gene expression (Methods). Aggregate expression was elevated in BRAFV600E tumors, less so in tumors with mutant RAS, and not at all in tumors with HER2 amplification (Fig. S2B). The MEKi caused a reduction in ERK transcriptional output in all these cells, but the degree of change varied as a function of the mode of pathway activation, with the greatest change in BRAFV600E cells. PLX4032 caused a similar reduction in BRAFV600E tumor cells. In contrast, the aggregate expression value changed only minimally in HER2-activated cells in response to either inhibitor. Moreover, in RTK cells and in mutant RAS cells, the aggregate gene expression in response to PLX4032 changed in the direction opposite that observed with the MEKi and was induced rather than inhibited. The small absolute change in aggregate expression in PLX4032-treated BRAFWT cell lines is consistent with the conclusion that ERK signaling is activated only transiently by the RAFi in this context.
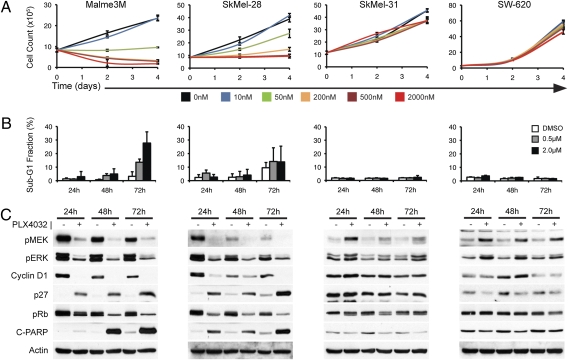
PLX4032 Induces RB-Hypophosphorylation, G1 Arrest, and Apoptosis in Cells with Mutant BRAF and Not in Those with BRAFWT.
Cyclin D1 expression, RB phosphorylation, G1/S progression, and proliferation are dependent on MEK/ERK signaling in tumors with mutant BRAF and on some tumorswith mutant RAS (8). Because PLX4032 inhibits pERK and ERK signaling to a degree indistinguishable from that of MEKi in mutant BRAF cells, RAF inhibition would be expected to have the same consequences. Indeed, PLX4032 caused the growth arrest of the BRAFV600E melanoma cells (SkMel-28 and Malme3M; Fig. 5A and Fig. S3). In both cell lines, inhibition of signaling by PLX4032 was followed by a reduction in cyclin D1, induction of p27 expression, hypophosphorylation of RB, and G1 arrest of cell growth (Fig. 5C and Fig. S3). The response of SkMel-28 cells to PLX4032 was primarily cytostatic, with marginal induction of poly(ADP-ribose) polymerase (PARP) cleavage and sub-G1 DNA after 72 h, whereas significant cell death occurred in Malme3M cells (Fig. 5 B and C). These results are identical to those obtained with MEK inhibition in BRAFV600E tumors (8).
Fig. 5.
Biologic effects of RAF inhibition occur only in BRAFV600E-mutant cell lines. (A) Growth kinetics of BRAFV600E cell lines (Malme3M and SkMel-28) compared with a MEK/ERK-dependent K-RAS–mutant cell line (SW-620) and a BRAFWT/RASWT cell line (SkMel-31) in the presence of PLX4032 (10 nM–2 μM) or vehicle alone (0 nM), as determined by cell count. (B) Percentage of cells in the sub-G1 population, as determined by FACS, following 24-, 48-, and 72-h exposures to 0.5 μM and 2.0 μM PLX4032 or control, for each of the cell lines shown in A. (C) Immunoblot detection of pMEK and pERK in lysate from the same cell lines, following treatment with vehicle (−) or 0.5 μM PLX4032 (+) over a 72-h time course. Cell-cycle regulation and apoptosis induction in the same samples was assessed by immunoblot detection of cyclin D1, p27, RB phosphorylation, and PARP cleavage.
PLX4032 induced pMEK and pERK in mutant RAS or RTK-driven tumor cells. However, we did not observe evidence for striking growth acceleration upon treatment with PLX4032 in detailed studies of eight BRAFWT cell lines (three mutant RAS, four with RTK activation, and one with no known RAS or RTK mutation) (Fig. 5 and Fig. S4). As shown in Fig. 5 for the SkMel-31 (BRAFWT/RASWT melanoma) and SW-620 (mutant K-RAS colon carcinoma) cell lines, PLX4032 potently induced pMEK and pERK but had no effect on cyclin D1 expression, RB phosphorylation, G1 progression, or proliferation. These results are consistent with the findings in Fig. 4, showing only a partial and transient induction of ERK transcriptional output by PLX4032 in these models.
Discussion
We previously used selective MEK inhibitors to show that the output of the ERK pathway and its functional consequences in tumor cells vary as a function of the mechanism of pathway deregulation and are not linearly related to levels of pERK (8, 9, 13). BRAFV600E is refractory to negative feedback and causes increased steady-state output of ERK signaling. Tumor cells with BRAFV600E mutation are dependent on ERK activity for cell-cycle progression and proliferation and almost always are sensitive to MEK inhibition. A subset of tumors with RAS mutation also is dependent on ERK signaling, although in these tumors ERK signaling output is elevated to a lesser degree than in V600E tumors (13). In contrast, tumors with HER kinase activation have low ERK signaling output and are MEKi insensitive.
Despite promising preclinical data, MEK inhibitors have been only modestly effective in clinical trials, with the MEKi AZD6244 demonstrating a 12% RECIST partial response rate in BRAFV600E melanoma patients (17, 18). There are many potential reasons for the insensitivity of most tumors to these compounds. It is possible that they have suboptimal potency or pharmacokinetics. Melanomas harbor other genetic changes, including PTEN mutations that might decrease their dependency on ERK signaling (30, 31). Furthermore, as expected, the dose of MEK inhibitors is limited by rash and other side effects (16, 17). In patients, MEK inhibitors significantly inhibit ERK phosphorylation at their maximally tolerated dose, but a greater degree of inhibition may be required to inhibit growth effectively or induce tumor cell death. It also is possible that important biologic effects of mutant BRAF are not mediated by MEK, because RAF has other putative substrates, including those that regulate cell survival (19–21).
Here, we used an ATP-competitive RAF inhibitor to address this question. We found no evidence that RAF inhibition with PLX4032 elicited different effects than MEK inhibitors in tumors with BRAFV600E. If PLX4032 causes additional biologic effects through non-MEK substrates, one might expect a different pattern of changes in gene expression than that caused by MEK inhibitors. This was not the case. Specifically, microarray analysis of changes in transcription induced by the inhibitors did not identify genes affected by PLX4032 and not PD0325901. Thus, when exposed to a RAFi, BRAFV600E melanomas growing in tissue culture showed no novel, transcriptionally mediated biologic effects that could not be attributed to inhibition of ERK signaling.
In fact, we found that the RAF inhibitor was less active than the MEK inhibitor in one notable context. BRAFV600E tumors were uniformly sensitive to both RAF and MEK inhibitors, whereas MEKi-insensitive tumors with wild-type or mutant RAS also were insensitive to PLX4032. Mutant RAS tumors that are MEKi sensitive and thus dependent upon ERK signaling were, however, resistant to the effects of PLX4032. This finding was explained when the effects of PLX4032 on signaling were examined in detail. MEK inhibition suppresses ERK signaling in all cells, whether or not proliferation was ERK dependent. In contrast, although PLX4032 inhibited MEK and ERK phosphorylation in tumors with BRAFV600E, it rapidly induced MEK and ERK phosphorylation in all other tumor cells tested and in normal keratinocytes. Induction of ERK phosphorylation by other putative RAF inhibitors has been described recently (7, 25, 32) and suggests that paradoxical induction of ERK phosphorylation is common property of ATP-competitive RAF-kinase inhibitors in cells with BRAFWT, but not BRAFV600E (26, 33). The mechanism of ERK induction in cells with BRAFWT has been attributed to negative feedback (23) or to selective inhibition of BRAF causing activation of CRAF (32). We recently demonstrated that pan-RAF inhibitors bind to and transactivate RAF dimers in a manner dependent upon activated RAS (26). In BRAFV600E cells, levels of RAS activity are too low to support activation, so the drug inhibits BRAF activity and ERK phosphorylation.
The results reported here have profound implications for the therapeutic use of RAF inhibitors. They imply that these drugs can be used only to treat tumors with mutant BRAF and will increase the expression of phosphorylated ERK in other tumors, including ERK-dependent tumors with mutant RAS. Furthermore, induction of ERK phosphorylation in tumors or normal cells could have deleterious effects, including acceleration of the growth of BRAFWT tumors. It therefore will be necessary to genotype prospectively the tumors of all patients treated with this agent to ascertain whether they contain BRAFV600E to avoid exposing cancer patients to an agent that will have no efficacy and potentially may accelerate cancer progression.
What, then, is the potential utility of such an agent, in comparison with MEK inhibitors? RAF inhibitors would be predicted to activate rather than inhibit ERK signaling in normal cells and thus should not cause the toxicity that is incurred with MEK inhibitors. Therefore administration of higher doses of the RAF inhibitors that result in more complete inhibition of ERK signaling in tumors than is feasible with MEK inhibitors may be possible, resulting in a wider therapeutic index and greater clinical efficacy. In a recent phase I clinical trial of PLX4032 in melanoma patients, these predictions have been borne out strikingly (34). High doses of drug administered daily result in serum concentrations of 40–60 μM with only modest toxicity. The most common toxicity of PLX4032 is a skin rash that is distinct from that induced by EGFR and MEK inhibitors, suggesting that the rash induced by the RAF inhibitor is not caused by ERK inhibition. No tumor responses were observed in patients with BRAFWT tumors. In contrast, in patients with BRAFV600E tumors, the efficacy of the drug has been extraordinary. At the recommended phase II dose, almost all tumors exhibited regression with ~80% achieving a RECIST partial response. Because the effects of PD0325901 and PLX4032 in melanoma models are so similar, it is likely that the superior effectiveness of the latter is caused in large part by its selective and more complete inhibition of ERK signaling in BRAFV600E-expressing tumors. The prolonged half-life of PLX4032 also may allow a more constant inhibition of the target that is tolerated because it is tumor specific.
An unexpected toxicity of treatment with PLX4032 has been the development of squamous cell carcinomas (keratoacanthoma-type) in about one third of patients treated at the maximum tolerated dose (34). Whether this development results from the induction of ERK signaling by PLX4032 is unknown. Others also have noted ERK activation in tumors with BRAFWT that have been exposed to RAF inhibitors, and it has been suggested that this may lead to the acceleration of the growth of such tumors (7, 33). We found that induction of ERK phosphorylation in mutant RAS tumor cells induced only transient expression of some ERK-dependent genes. The expression of these genes had returned to baseline 8 h after the addition of PLX4032 to BRAFWT cells, whereas levels of pMEK and pERK remained elevated. The mechanism through which ERK output returns to baseline is unknown but probably reflects an increase in negative feedback pathways. The lack of induction of steady-state ERK output is consistent with the absence of PLX4032-induced changes in growth rate in these BRAFWT tumors. These data suggest that patients with BRAFWT tumors treated with RAF inhibitors may not be at risk for tumor acceleration. Of course, this may not be true for tumors of all lineages. The mechanism of induction of keratoacanthomas in PLX4032-treated patients and its relationship to induction of ERK activation will require further investigation and may provide insight as to why an increase in tumor cell growth has been noted so prominently in this cell lineage.
Methods
Materials.
PLX4032 was obtained from Plexxikon, Inc. PD0325901 was synthesized according to its published structure (35). Drugs for in vitro studies were dissolved in DMSO stock solutions and stored at −20 °C.
Cell Culture.
Cancer cell lines were obtained from the American Type Culture Collection or were provided by the Houghton laboratory, Memorial Sloan-Kettering Cancer Center, and maintained as described (8). For proliferation assays, cells were plated in 96-well plates, at 2,000–3,000 cells/well, and after 24 h were treated with inhibitors. IC50 values were calculated using Alamar Blue (Trek Diagnostics) and Softmax Pro (Molecular Devices), as described (8). For Western blots, cell extracts were prepared and immunoblotting was performed as described (13).
Cell Cycle and Apoptosis.
Cells were seeded in 6-cm dishes at a density of 0.5–1.0 × 106 cells per dish 1 d before treatment with PLX4032 (0.5 or 2 μM) or DMSO. Both adherent and floating cells were harvested and stained with ethidium bromide using the method of Nusse et al. (36). Flow cytometric analysis was used for quantitation of cell-cycle distribution and apoptosis (subG1) as described (8).
Microarray Analysis.
Cells were treated with drug or vehicle for 8 h. RNA was extracted, labeled, and hybridized to HG-U133A2.0 expression arrays (Affymetrix) as previously reported (13). Chips then were processed, scanned, and quantified, and the data were normalized as described (13). Differential expression was identified with an empirical Bayes approach using the Linear Models for Microarray Analysis (37), and genes were selected based on an absolute log2 fold change ≥1.5 and false discovery rate <1%. Among the selected genes, the expression of multiple probe sets per gene was averaged. Average-linkage hierarchical clustering of an uncentered Pearson correlation similarity matrix was executed with Cluster and visualized with TreeView (38). We referenced http://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE10086) for gene expression changes in response to PD0325901 (13). An aggregate measure of expression of the 73-gene signature was calculated by standardizing normalized expression per array by converting expression levels across all probes to a Z-score. The arithmetic mean of Z-scores across the 73 genes in each sample served as aggregate expression value.
RT-PCR Analysis.
RNA was extracted using the RNeasy Mini Kit (Qiagen), reverse-transcribed, and used for quantitative RT-PCR as described (13). Relative expression of target genes was calculated using the ΔΔCt method and normalized to the mRNA content of three housekeeping genes. Values are reported as percentages relative to the untreated control for each cell line (2−ΔΔCt).
Supplementary Material
Acknowledgments
We acknowledge D. You, Y. Zhao, and M. Hassimi for processing Affymetrix arrays and A. Houghton, C. Liu, and H. Gallardo (Memorial Sloan-Kettering Cancer Center) for providing cell lines. This study was supported by grants from the National Institutes of Health (to C.A.P., D.B.S., and N.R.), the Melanoma Research Alliance (to N.R. and D.B.S.), and the STARR Foundation (to N.R. and D.B.S.).
Footnotes
Conflict of interest statement: N.R. has major consulting/advisory roles with Roche (>$10,000/y). D.B.S. and P.B.C have minor consulting/advisory roles with Roche (>$10,000/y). J.T. and G.B. are full-time employees of Plexxikon.
Data deposition: The sequence reported in this paper has been deposited in Microarray Gene Expression Omnibus: http://www.ncbi.nlm.nih.gov/geo/ (accession no.GSE20051).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008990107/-/DCSupplemental.
References
- 1.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 2.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 4.Sebolt-Leopold JS, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 5.Yeh TC, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 6.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeflich KP, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69:3042–3051. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 8.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pratilas CA, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wee S, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 11.Halilovic E, et al. PIK3CA mutation uncouples tumor growth and Cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-0409. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougherty MK, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 13.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friday BB, et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 2008;68:6145–6153. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 15.Wan PT, et al. Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 16.Adjei AA, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LoRusso PM, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral MAPK/ERK kinase inhibitor PD-0325901 in patients with advanced cancers. Clin Cancer Res. 2010;16:1924–1937. doi: 10.1158/1078-0432.CCR-09-1883. [DOI] [PubMed] [Google Scholar]
- 18.Dummer R, et al. AZD6244 (ARRY-142886) vs temozolomide (TMZ) in patients (pts) with advanced melanoma: An open-label, randomized, multicenter, phase II study. J Clin Oncol. 2008;26(suppl) Abstract no. 9033. [Google Scholar]
- 19.Ichijo H, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Fujii K, Zhang L, Roberts T, Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci USA. 2001;98:7783–7788. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neill E, Rushworth L, Baccarini M, Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science. 2004;306:2267–2270. doi: 10.1126/science.1103233. [DOI] [PubMed] [Google Scholar]
- 22.Tsai J, et al. Development of a novel inhibitor of cogenic b-Raf. AACR Meeting Abstracts. 2006;2006(1):571. [Google Scholar]
- 23.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 24.Courtois-Cox S, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall-Jackson CA, et al. Paradoxical activation of Raf by a novel Raf inhibitor. Chem Biol. 1999;6:559–568. doi: 10.1016/s1074-5521(99)80088-x. [DOI] [PubMed] [Google Scholar]
- 26.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 28.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: A developing story. Nat Rev Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 29.Smalley KS, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008;68:5743–5752. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies MA, et al. Integrated molecular and clinical analysis of AKT activation in metastatic melanoma. Clin Cancer Res. 2009;15:7538–7546. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 34.Flaherty K, et al. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009;27(suppl) Abstract no. 9000. [Google Scholar]
- 35.Barrett SD, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18:6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 36.Nüsse M, Beisker W, Hoffmann C, Tarnok A. Flow cytometric analysis of G1- and G2/M-phase subpopulations in mammalian cell nuclei using side scatter and DNA content measurements. Cytometry. 1990;11:813–821. doi: 10.1002/cyto.990110707. [DOI] [PubMed] [Google Scholar]
- 37.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 38.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.