Abstract
Histone deacetylase inhibitors (HDACi) developed as anti-cancer agents have a high degree of selectivity for killing cancer cells. HDACi induce acetylation of histones and nonhistone proteins, which affect gene expression, cell cycle progression, cell migration, and cell death. The mechanism of the tumor selective action of HDACi is unclear. Here, we show that the HDACi, vorinostat (Suberoylanilide hydroxamic acid, SAHA), induces DNA double-strand breaks (DSBs) in normal (HFS) and cancer (LNCaP, A549) cells. Normal cells in contrast to cancer cells repair the DSBs despite continued culture with vorinostat. In transformed cells, phosphorylated H2AX (γH2AX), a marker of DNA DSBs, levels increased with continued culture with vorinostat, whereas in normal cells, this marker decreased with time. Vorinostat induced the accumulation of acetylated histones within 30 min, which could alter chromatin structure-exposing DNA to damage. After a 24-h culture of cells with vorinostat, and reculture without the HDACi, γH2AX was undetectable by 2 h in normal cells, while persisting in transformed cells for the duration of culture. Further, we found that vorinostat suppressed DNA DSB repair proteins, e.g., RAD50, MRE11, in cancer but not normal cells. Thus, the HDACi, vorinostat, induces DNA damage which normal but not cancer cells can repair. This DNA damage is associated with cancer cell death. These findings can explain, in part, the selectivity of vorinostat in causing cancer cell death at concentrations that cause little or no normal cell death.
Keywords: γH2AX, DNA repair proteins, vorinostat, histones
Histone deacetylase inhibitors (HDACi) are being developed as a promising new class of drugs for cancers (1–3) and a potential therapy for nononcologic disorders, including neurodegenerative diseases (2, 4, 5). A number of HDACi are in clinical trials for various hematologic and solid neoplastic diseases (1–3). Vorinostat (suberoylanilide hydroxamic acid, SAHA) was the first HDACi approved by the US Food and Drug Administration for clinical use in treating cutaneous T cell lymphoma (6, 7). Vorinostat has anti-cancer activity in studies with transformed cells in culture, tumor-bearing animal models and in clinical trials (1, 6–8). In studies with cells in culture, HDACi induced transformed cell death at concentrations that did not affect normal cell viability (9–12). It has been found that vorinostat at doses that inhibited almost 100% of the growth of human prostate cancer xenographs in mice caused no detectable toxicity as evaluated by weight loss and extensive necropsy studies (13). In clinical trials with vorinostat, it was shown that the drug was well tolerated at doses that had significant anti-tumor activity (14, 15). The mechanisms of this selective anti-tumor cell activity of vorinostat are not well understood.
This study has investigated the effect of vorinostat on normal and transformed cell genomic integrity. Vorinostat inhibits class I HDACs, HDACs 1, 2, 3, and 8, and class IIb HDAC, HDAC 6 (1–3, 16). Analysis of lysine transacetylase targets of transformed cells found 3,600 acetylated lysine sites in 1,750 proteins (17). The inhibition of HDACs with vorinostat altered only ≈10% of these lysine acetylation sites. The lysine sites affected by vorinostat inhibition of HDACs were in proteins that included core histones, H3 and H4, and the variant histone, H2AX (18). Acetylation of lysines of histones neutralizes their positive charge, altering DNA–protein structure in chromatin (19).
HDACi can induce transformed cell growth arrest and cell death by one or more pathways (1, 20–22). HDAC inhibition leads to genomic instability (23). There is no evidence that HDACi directly cause DNA mutations. The histone hyperacetylation induced by HDACi causes structural alterations in chromatin, which may expose portions of DNA that are normally protected by heterochromatin to DNA-damaging agents such as: UV, x-ray, cytotoxic drugs, or reactive oxygen species (ROS). In previous studies, our laboratory found that vorinostat caused an accumulation of ROS and caspase activation in certain transformed cells, but not in normal cells (9). HDACi can also down-regulate the levels of DNA repair proteins (24–26).
In this study, we show that vorinostat induced the accumulation of the phosphorylated histone H2AX, γH2AX, an early marker of DNA DSBs, in both normal and transformed cells. It was found that in both normal and transformed cells, vorinostat induced rapid accumulation of acetylated histones H3 and H4, suggesting that altered chromatin structures may be a factor in exposing DNA to damage. Normal cells, but not transformed cells, repair the DNA DSBs, as evidenced by the disappearance of γH2AX when the cultures are washed free of the HDACi. Vorinostat caused the down-regulation of certain DNA repair proteins in transformed but not normal cells, which may contribute to the failure of the cancer cells to repair the DNA DSBs. There are several reports that the combination of vorinostat or other HDACi with DNA-damaging agents are synergistic in inducing transformed cell death (1–3, 22–28). These findings provide an understanding, in part, of vorinostat selectivity in causing transformed cell death at concentrations that cause little or no normal cell death.
Results
Vorinostat Induces Death of Transformed but Not Normal Cells.
Normal human foreskin fibroblast (HFS) cells and transformed cells, human prostate cancer cells (LNCaP), and human lung adenocarcinoma cells (A549) were cultured with 1, 2.5, 5, or 10 μM vorinostat for up to 96 h. Vorinostat concentrations of 2.5–10 μM inhibited cell growth of normal and transformed cells (Fig. S1) (29). After 72 h of culture with 5 μM vorinostat, there was >80% loss of cell viability of LNCaP and 30% of A549 cells, but no detectable loss of cell viability of the normal cells, HFS. These results are consistent with reported findings that normal cells are relatively resistant to vorinostat induced cell death (9). LNCaP cells are more sensitive to vorinostat induced cell death than are A549 cells.
Vorinostat Induces the Accumulation of Acetylated Proteins and of γH2AX in Normal and Transformed Cells.
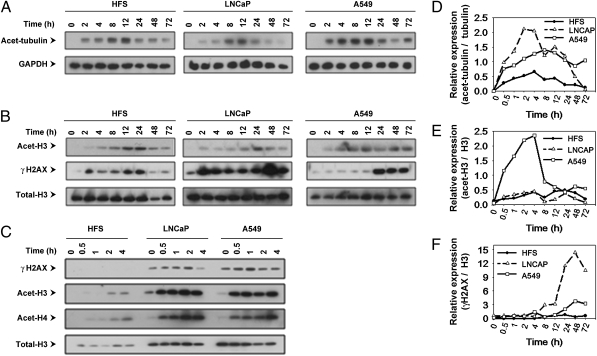
Because HDACs are inhibited by vorinostat (1, 3, 16, 20–22), we assayed the acetylation patterns of protein targets of the deacetylases. In HFS, LNCaP, and A549 cells in culture with 5 μM vorinostat, accumulation of acetylated α-tubulin, which is a substrate of HDAC6, was detected at 2 h with peak levels at ≈12 h and declining toward control levels by 72 h (Fig. 1 A and D). The pattern of vorinostat-induced accumulation of acetylated α-tubulin was similar in the normal and transformed cells.
Fig. 1.
Vorinostat induces accumulation of acetylated α-tubulin, histone H3, and γH2AX in normal (HFS) and transformed (LNCaP, A549) cells. Cells were cultured with 5 μM vorinostat for indicated times. Immunoblots are of acetylated α-tubulin (A), acetylated histone H3 and γH2AX (B), immunoblots of γH2AX and acetylated histones H3 and H4 at early time points (C), GAPDH, and total histone H3 are indicated as the loading controls. Graphs, expressed as a ratio to the loading control, of the accumulation of acetylated α-tubulin (D), acetylated histone H3 (E), and γH2AX (F). Graphs were prepared by quantifying immunoblot bands by using TINA 2.0 software (Ray Tests).
Vorinostat induced accumulation of acetylated histone H3, which is a substrate of class I HDACs. In HFS cells, the accumulation of acetylated histone H3 was detectable at 2 h, with peak levels between 12 and 24 h and declining by 48 h (Fig. 1 B, C, and E). In LNCaP and A549, the accumulation of acetylated histone H3 is detectable within 30 min and persists for the duration of the culture up to 72 h (Fig. 1 B, C, and E).
We found a striking difference in the pattern of accumulation of γH2AX when comparing normal and transformed cells (Fig. 1 B, C, and F). In HFS cells cultured with 5 μM vorinostat, accumulation of γH2AX was detectable at 2 h, reached a peak between 12 and 24 h, and decreased thereafter in culture with the HDACi for up to 72 h. In both LNCaP and A549 cells, there is a detectable level of γH2AX accumulation in culture without the HDACi. Vorinostat increased the accumulation of γH2AX in LNCaP and in A549, which persists for the duration of the culture up to 72 h (Fig. 1 B, C, and F). These findings suggest that vorinostat induced DNA DSBs in both normal and transformed cells, but normal cells, in contrast to transformed cells, can repair the DNA DSBs as evidenced by the decline in γH2AX accumulation in HFS cells despite continued exposure to vorinostat. The vorinostat-induced accumulation of acetylated histone H3 and γH2AX is greater in LNCaP than A549 (Fig. 1 B–D). These findings are associated with the greater sensitivity of LNCaP compared with A549 to vorinostat-induced cell death (Fig. S1).
Vorinostat-Induced γH2AX Foci in Transformed and in Normal Cells.
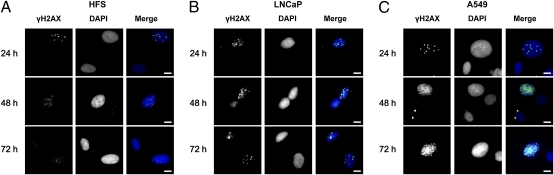
We further evaluated vorinostat-induced γH2AX foci in HFS, LNCaP, and A549 cells by immunofluorescence microscopy visualization (Fig. 2). In HFS nuclei, γH2AX foci were present at 24 h and were not detectable at 48 and 72 h (Fig. 2A). γH2AX foci were present in LNCaP and A549 nuclei and persisted for the duration of the culture (Fig. 2 B and C). In these studies, at least 100 cells were scored (range 100–428 cells) per time point. At 48 h of culture with 5 μM vorinostat, the foci per nucleus in HFS averaged 0.9% without HDACi, 4.8% with HDACi; in LNCaP, averaged 0.02% without inhibitor and 15.3% with HDACi; and in A549, averaged 2.7% without HDACi and 21.7% with HDACi (Table 1). These findings are consistent with the biochemical evidence of the pattern of vorinostat-induced foci of γH2AX and indicate that DNA DSBs persist in transformed cells but not in normal cells.
Fig. 2.
Vorinostat induced accumulation of γH2AX foci in normal (HFS) and transformed (LNCaP, A549) cells. Cells were cultured with 5 μM vorinostat for indicated times and probed with γH2AX antibody and DAPI (nuclei). Representative images were observed by immunofluorescence, γH2AX, DAPI, and merged images of γH2AX and DAPI fluorescence staining. (A) HFS cells. (B) LNCaP cells. (C) A549 cells. (Scale bars: 10 μm.)
Table 1.
Vorinostat-induced yH2AX foci in normal (HFS) and transformed (LNCaP, A549) cells
| Measurement | HFS | LNCaP | A549 | |||
| No Tx | HDACi | No Tx | HDACi | No Tx | HDACi | |
| Total cells, n | 423 | 143 | 132 | 286 | 208 | 100 |
| Foci per nucleus,* % | 0.9 | 4.8 | 0.02 | 15.3 | 2.7 | 21.7 |
*Ratio of phospho-H2AX area over DAPI area was measured by MetaMorph 7.6.5 (Molecular Devices). Each value in table was obtained by examining at least 100 cells.
Normal but Not Transformed Cells Repair DNA DSBs Upon Removal of HDACi.
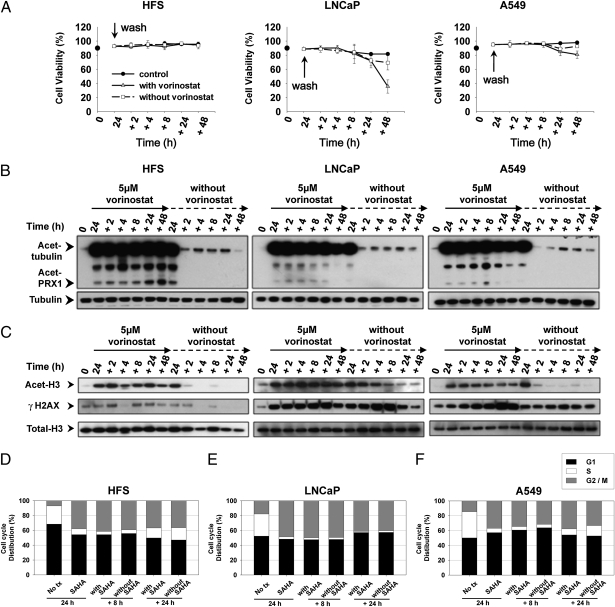
We next evaluated the capacity of normal and transformed cells to repair vorinostat-induced DNA DSBs when cells were washed free of the HDACi. After a 24-h culture with vorinostat, HFS, LNCaP, and A549 cells were washed free of medium and divided into two separate cultures, one with vorinostat and one without the HDACi. In cultures with vorinostat, there was a decrease in viability of LNCaP and A549 cells of 60% and 20%, respectively, at 48 h (Fig. 3A) but no loss of viability of HFS cells (Fig. 3A). Pretreatment with vorinostat for 24 h induced a reduction of cells in S phase both in normal and transformed cells (Fig. 3 D–F). In the following culture with or without vorinostat, HFS cells in S phase increased over time (Fig. 3D), but not in LNCaP (Fig. 3E) and, to a lesser extent, in A549 cells (Fig. 3F).
Fig. 3.
Normal cells recover from vorinostat induced DNA DSBs after removal of HDACi but not transformed cells. Washout experiments were performed as described in Materials and Methods. (A) Graphs represent cell viability after replacing medium with or without vorinostat, after 24-h culture with vorinostat. Each time point is the mean of three independent experiments; each bar is ±SD. (B) Immunoblots of acetylated tubulin and acetylated peroxiredoxins (prx1). Total α-tubulin is the loading control. (C) Immunoblots of acetylated histone H3 and γH2AX. Total histone H3 is the loading control. Cell cycle distribution of HFS (D), LNCaP (E), and A549 cells (F) at times indicated.
In normal (HFS) and transformed cells (LNCaP and A549), there was the accumulation of acetylated α-tubulin and acetylated peroxiredoxin, substrates of HDAC6 (30), during the 24-h culture with the inhibitor (Fig. 3B). In cultures washed free of medium with vorinostat and replaced with fresh media without the HDACi, within 2 h, there was a return toward control levels of acetylated α-tubulin and acetylated peroxiredoxin in both the normal and transformed cell lines (Fig. 3B). The acetylated histone H3, which accumulated during the 24-h culture with vorinostat, returned to undetectable levels in the culture with fresh medium without vorinostat within 2 h in HFS and A549 and by 8–24 h in LNCaP (Fig. 3C). These findings are consistent with vorinostat being a reversible inhibitor of HDACs, class I HDACs, and HDAC6 (6) and that these enzymes were active on removal of the inhibitor.
In all three cell lines, culture with vorinostat caused the accumulation of γH2AX, which is more marked in transformed than normal cells (Fig. 3C). Upon removal of medium with vorinostat and culture in fresh medium without the HDACi, within 2 h, there was a marked decrease in the accumulation of γH2AX in HFS cells. In transformed cells, there was persistence of the accumulation of γH2AX for the duration of the culture without vorinostat (Fig. 3C). LNCaP cells could not recover from either vorinostat-induced S phase reduction or DNA DSBs. A549 cells recovered a limited capacity to enter S phase, but γH2AX accumulation persisted, indicating these transformed cells could not recover from vorinostat-induced DNA DSBs (Fig. 3 E and F). These findings are consistent with the normal but not transformed cells having the capacity to repair DNA DSBs as evidenced by the disappearance of γH2AX.
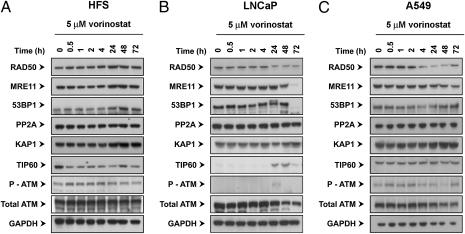
Vorinostat Can Suppress Expression of DNA DSB Repair Proteins in Transformed but Not Normal Cells.
We next assayed the levels of expression of certain DNA DSB repair proteins in normal and transformed cells cultured with vorinostat (Fig. 4). During a 72-h culture in HFS cells, vorinostat did not cause detectable changes in the levels of: RAD50, MRE11, 53BP1, PP2A, KAP1, TIP60, or phosphorylated ATM (p-ATM) (Fig. 4A). Vorinostat did cause a decrease in RAD50 and MRE11 levels in LNCaP and A549 by 24–48 h (Fig. 4 B and C). Accumulation of p-ATM occurred in LNCaP but not in A549 cells. There was no detectable TIP60 in LNCaP during the initial 4 h of culture with HDACi. The detectable decreases in levels of repair proteins in transformed cells cultured with vorinostat occur later than the increases in acetylated histones and of γH2AX, an early marker of DNA DSB sites.
Fig. 4.
Vorinostat can suppress expression of DNA DSB repair proteins in transformed, but not normal, cells. Cells were cultured with 5 μM vorinostat for indicated times. Immunoblots were probed with antibodies as described in Materials and Methods. DNA damage repair proteins assayed: RAD50, MRE11, 53BP1, KAP1, TIP60, p-ATM, and ATM.
Discussion
This study addresses the question why an HDACi, vorinostat, is selective in causing cancer cell death at doses that cause little or no death of normal cells (6–9, 13–15). We show that vorinostat induces accumulation of γH2AX, a marker of DNA DSBs, in both normal and transformed cells (18). The accumulation of γH2AX is one of the earliest events after formation of DNA DSBs. γH2AX interacts with the MRN complex, which is a multiprotein complex including RAD50, MRE11, and other proteins involved in DNA DSB repair (18, 31–33). This complex recruits ATM and several other proteins involved in DNA damage repair. DNA DSBs are associated with the spreading of γH2AX to larger domains on either side of a DSB and is evidenced in our findings of increasing accumulation of γH2AX in transformed but not normal cells exposed to vorinostat over time. The normal cells (HFS), but not the transformed cells (LNCaP and A549) have the capacity to repair the DNA DSBs upon removal of the inhibitor or even during continued exposure to the inhibitor, as evidenced by the decreased accumulation of γH2AX. This finding reflects, in part, the fact that transformed cells have many genomic and other molecular defects that contribute to failure in the DNA damage response (23, 34). Among DNA lesions, the most harmful is the breakage of DNA double strands (31, 35). DNA DSB damage can lead to altered gene expression and induction of apoptosis.
This study found that vorinostat suppressed expression of DNA damage repair proteins, e.g., RAD50 and MRE11, in transformed but not the normal cells. It has also been reported the HDACi can cause accumulation of acetylated Ku70 associated with DNA DSBs (26). Previous reports have found that HDACi can suppress DNA damage repair proteins (24–26). We found that vorinostat induced a decrease of cells in S phase of the cell cycle associated with impaired DNA repair mechanisms in transformed cells.
Vorinostat induced the accumulation of acetylated histones within 30 min of culture in transformed cells. Acetylation of histones causes a change in chromatin structure, exposing DNA to intrinsic damaging agents, e.g., ROS, and to extrinsic damage including radiation and cytotoxic agents (1, 2, 8, 9, 22–24, 36, 37). Vorinostat in combination therapy with DNA damaging agents has shown greater anti-cancer activity than HDACi alone (27, 37–43).
The failure of transformed cells to repair vorinostat-induced DNA DSBs does not reflect an irreversible inhibition of HDACs, because high levels of acetylated α-tubulin, acetylated peroxiredoxin and acetylated histones disappear, upon removal of the inhibitor in continued culture of both normal and transformed cells. Thus, intermittent dosing of vorinostat could minimize toxic effects on normal cells. The present findings are consistent with the in vivo observation that vorinostat induces accumulation of acetylated histones in normal peripheral mononuclear cells within 1–2 h after a single oral dose, which returns to control levels within 10–12 h (14).
In sum, this finding that normal, but not transformed cells, can repair vorinostat-induced DNA DSBs can explain, in part, the selectivity of this HDACi in causing cancer cell death.
Materials and Methods
Cell Lines, Reagents, and Antibodies.
HFS (human normal foreskin fibroblast) was purchased from Yale Skin Diseases Research Center Core. A549 (non-small cell lung cancer cell line) and LNCaP (prostate cancer cell line) were purchased from American Type Culture Collection and cultured per directions of the supplier. Vorinostat was synthesized as reported (29) and was dissolved in DMSO. Antibodies used were as follows: anti-phosphorylated histone H2AX (Abcam), anti-53BP1 and KAP1 (Bethyl Laboratories), anti-MRE11 (Novus), anti-RAD50 (R&D Systems), anti-acetylated histone H3 and total histone H3 (Active Motif), anti-p-ATM (Rockland), anti-total ATM (Genetex), anti-acetylated α-tubulin antibody (Sigma), anti-total α-tubulin and TIP60 (Santa Cruz Biotechnology), anti-PP2A (Millipore), and anti-GAPDH (Thermo Scientific).
Cell Growth and Viability.
Each cell culture was performed in triplicate and cell growth and viability performed as described (29). Graphs were constructed by using Sigma Plot software (Systat Software). Data were expressed as mean ± SD.
Immunoblotting Analysis.
Cells were collected, washed with ice-cold PBS, and lysed with cell lysis buffer (50 mM Tris·Cl at pH 8.0, 120 mM NaCl, 0.5% Nonidet P-40, 5 mM EDTA) containing protease inhibitors mixture (Boehringer Mannheim) on ice for 30 min. The lysates were cleared by centrifugation at 18,000 × g for 20 min at 4 °C. For washout experiment, after treatment with 5 μM vorinostat for 24 h, the culture medium was replaced with fresh medium with or without inhibitor. At each time point, cell viability analyses were performed and cell pellets were collected. Equal amounts of protein were separated on 4–12% Bis-Tris gels (Invitrogen) and transferred by semidry transfer iBlot system (Invitrogen). The membranes were blocked with 3% BSA in PBST for 1 h, probed with the appropriate primary antibodies at 4 °C overnight, and followed by HRP-conjugated secondary antibodies. Proteins were detected by using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). NuPAGE Large Protein Analysis System (Invitrogen) was used for detection of large proteins (e.g., ATM, 53BP1, and KAP1).
Histone Extraction.
Cells (1 × 106) were washed with PBS and lysed with 50 μL of histone lysis buffer (10 mM Tris·Cl at pH 6.5, 10 mM MgCl2, 25 mM KCl, 1% Triton X-100, 8.6% Sucrose) containing protease inhibitors mixture. After centrifuge at 1,000 × g for 5 min at 4 °C, supernatants were kept for analysis of levels of acetylated tubulin. The pellets were gently resuspended in TE buffer (10 mM Tris·Cl at pH 7.4, 13 mM EDTA), and then centrifuged for 5 min at 600 × g at 4 °C. The pellets were resuspended in ice cold 0.2 M H2SO4, incubated on ice for 1 h, and vortexed 10 s every 15 min during the incubation. Samples were centrifuged for 10 min at 10,000 × g at 4 °C. The supernatants were incubated with cold acetone for at least 1 h. The histone pellets were obtained by centrifugation for 10 min at 10,000 × g at 4 °C. After drying the pellet, histones were resolved with distilled water. Graphs were prepared by quantifying immunoblotting bands by using TINA 2.0 software (Ray Tests).
Immunofluorescence Staining.
Cells were fixed with 4% paraformaldehyde containing 2% sucrose for 10 min and then washed with PBS. Cells were permeabilized with 0.25% Triton X-100 for 10 min, blocked with 1% BSA for 1 h, and incubated with phosphorylated H2AX antibody (Abcam) in PBS containing 1% BSA for 1 h at room temperature. Cells were then washed with PBS and incubated with Alexa 488-conjugated goat anti-mouse IgG antibody (Molecular Probes) for 1 h. Cells were washed with PBST, and then stained for 5 min with DAPI. The images were captured by a fluorescent microscope, IX71 (Olympus), with Normarski optics by using ORCA CCD camera (Hamamatsu). The image analysis was done by using IP LAB (BioVision Technologies), MetaMorph 7.6.5 (Molecular Devices), Image J (National Institutes of Health), and Photoshop CS4 (Adobe) software.
Cell Cycle Analysis.
To determine cell cycle distribution in attached cells, 1 × 106 cells were fixed in 70% methanol at −20 °C. After centrifugation at 600 × g for 5 min, cell pellets were resuspended in PI staining solution (50 μg/μL), which contains RNase A (100 μg/mL) and incubated for 30 min at 37 °C. Cells were transferred to tube with top-filter and analyzed by FACSCalibur. Analysis was performed with the FlowJo software (version 8.8.4; TreeStar).
Statistical Analyses.
Data are expressed as mean ± SD derived minimally from three independent experiments. Statistical significance was calculated by using the two population Student's t test.
Supplementary Material
Acknowledgments
We thank Joann Perrone and Mabel Miranda for their assistance in the preparation of this manuscript. These studies were supported, in part, by National Institute of Cancer Grant P30CA08748-44, The David Koch Foundation, the CapCure Foundation, and Experimental Therapeutics at Memorial Sloan–Kettering Cancer Center.
Footnotes
Conflict of interest statement: Memorial Sloan–Kettering Cancer Center and Columbia University hold patents on suberoylanilide hydroxamic acid (SAHA, vorinostat) and related compounds that were exclusively licensed in 2001 to ATON Pharma, a biotechnology start-up that was wholly acquired by Merck, Inc., in April 2004.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008522107/-/DCSupplemental.
References
- 1.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: Current status and overview of recent clinical trials. Drugs. 2009;69:1911–1934. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Adcock IM. HDAC inhibitors as anti-inflammatory agents. Br J Pharmacol. 2007;150:829–831. doi: 10.1038/sj.bjp.0707166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 6.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 7.Duvic M, Vu J. Vorinostat in cutaneous T-cell lymphoma. Drugs Today (Barc) 2007;43:585–599. doi: 10.1358/dot.2007.43.9.1112980. [DOI] [PubMed] [Google Scholar]
- 8.Rosato RR, Grant S. Histone deacetylase inhibitors: Insights into mechanisms of lethality. Expert Opin Ther Targets. 2005;9:809–824. doi: 10.1517/14728222.9.4.809. [DOI] [PubMed] [Google Scholar]
- 9.Ungerstedt JS, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atadja P, et al. Selective growth inhibition of tumor cells by a novel histone deacetylase inhibitor, NVP-LAQ824. Cancer Res. 2004;64:689–695. doi: 10.1158/0008-5472.can-03-2043. [DOI] [PubMed] [Google Scholar]
- 11.Qiu L, et al. Anti-tumour activity in vitro and in vivo of selective differentiating agents containing hydroxamate. Br J Cancer. 1999;80:1252–1258. doi: 10.1038/sj.bjc.6690493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piekarz RL, Bates SE. Epigenetic modifiers: Basic understanding and clinical development. Clin Cancer Res. 2009;15:3918–3926. doi: 10.1158/1078-0432.CCR-08-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler LM, et al. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- 14.Kelly WK, Marks PA. Drug insight: Histone deacetylase inhibitors—development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat Clin Pract Oncol. 2005;2:150–157. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 15.Duvic M, Vu J. Vorinostat: A new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 16.Bradner JE, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.Pilch DR, et al. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 20.Batta K, Das C, Gadad S, Shandilya J, Kundu TK. Reversible acetylation of non histone proteins: Role in cellular function and disease. Subcell Biochem. 2007;41:193–212. [PubMed] [Google Scholar]
- 21.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 23.Eot-Houllier G, Fulcrand G, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase inhibitors and genomic instability. Cancer Lett. 2009;274:169–176. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Munshi A, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res. 2005;11:4912–4922. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 25.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280:125–133. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 26.Chen CS, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67:5318–5327. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 27.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: Current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280:201–210. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15:3970–3977. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 29.Richon VM, et al. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parmigiani RB, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci USA. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardo B, Gómez-González B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–1056. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrini JH. DNA replication reaches the breaking point. Cell. 2009;137:211–212. doi: 10.1016/j.cell.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 34.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abreu PA, et al. DNA methylation: A promising target for the twenty-first century. Expert Opin Ther Targets. 2008;12:1035–1047. doi: 10.1517/14728222.12.8.1035. [DOI] [PubMed] [Google Scholar]
- 36.Ruefli AA, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98:10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MS, et al. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 38.Abujamra AL, Dos Santos MP, Roesler R, Schwartsmann G, Brunetto AL. Histone deacetylase inhibitors: A new perspective for the treatment of leukemia. Leuk Res. 2010;34:687–695. doi: 10.1016/j.leukres.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Botrugno OA, Santoro F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280:134–144. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Ramalingam SS, et al. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13:3605–3610. doi: 10.1158/1078-0432.CCR-07-0162. [DOI] [PubMed] [Google Scholar]
- 41.Shiozawa K, et al. Preclinical studies of vorinostat (suberoylanilide hydroxamic acid) combined with cytosine arabinoside and etoposide for treatment of acute leukemias. Clin Cancer Res. 2009;15:1698–1707. doi: 10.1158/1078-0432.CCR-08-1587. [DOI] [PubMed] [Google Scholar]
- 42.Kano Y, et al. Cytotoxic effects of histone deacetylase inhibitor FK228 (depsipeptide, formally named FR901228) in combination with conventional anti-leukemia/lymphoma agents against human leukemia/lymphoma cell lines. Invest New Drugs. 2007;25:31–40. doi: 10.1007/s10637-006-9000-0. [DOI] [PubMed] [Google Scholar]
- 43.Bishton M, Kenealy M, Johnstone R, Rasheed W, Prince HM. Epigenetic targets in hematological malignancies: Combination therapies with HDACis and demethylating agents. Expert Rev Anticancer Ther. 2007;7:1439–1449. doi: 10.1586/14737140.7.10.1439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.