Abstract
Direct determination of functional biomolecular chemistry of clinically relevant tissues in vivo is a challenging task. Current approaches, based on tissue retrieval by biopsy and subsequent solubilization, are limited in terms of accurate representation of tissue constituents, reproducibility, and retention of functionality of solubilized tissue biomolecules. Using a pool of known surfactants, we designed and screened a large combinatorial library of surfactant formulations, which led to the discovery of rare synergistic formulations that greatly enhance tissue solubilization as well as preserve bioactivity of solubilized molecules, in particular proteins. By combining these formulations with a short ultrasound application, we developed a tissue sampling method—STAMP (Surfactant-based Tissue Acquisition for Molecular Profiling)—for rapid one-step determination of functional tissue chemistry in vivo. We specifically demonstrate STAMP-assisted profiling of a multitude of proteins, lipids, and genomic DNA in skin and mucosal tissues. Applications of this sampling methodology to rapid molecular diagnostics of cutaneous allergies and infectious diseases are also presented.
Keywords: tissue profiling, tissue diagnostics, dermatology
Systems biology studies focused on investigating functional interactions among biomolecules in their physiological tissue environment have paved the way to understanding disease mechanisms, discovery of drugs, and disease biomarkers for better diagnostics (1–4). Further advances in this exciting field hinge upon the development of technologies that allow accurate characterization of tissue’s functional biomolecular chemistry. A major challenge in this analysis is the ability to retrieve intact and functional biomolecules from target tissues in a reproducible and effective way (5–7). This challenge becomes especially formidable when direct collection of biomolecules from living tissues in vivo is desired.
Current approaches for molecular profiling of clinically relevant tissue samples require successful implementation of two steps: (i) harvesting a tissue sample, which is typically done by biopsy, and (ii) recovery of proteins from the harvested tissue sample, which is done through tissue solubilization (8). Biopsy has been the mainstay for diagnosis of malignancies; however, it suffers from a variety of limitations when used for molecular profiling purposes. In addition to being invasive, potential tissue degradation during procurement and handling of biopsies can lead to a significant loss and distortion of molecular information (9).
Even when tissues are harvested by biopsies, preparation of their liquefied extracts in an efficient and reproducible manner is a nontrivial hurdle (10). Several methods, including mechanical disruption, high pressure homogenization, enzymatic digestion of tissue matrix, surfactant-mediated solubilization, or their combinations, have been described for tissue solubilization (6, 8). These approaches are generally nonstandardized, involve multiple steps, and a major fraction of tissue constituents is often sacrificed in the process (8, 11). Furthermore, it is challenging to achieve sufficient solubilization without changing the native chemistry of tissue components, particularly proteins (10, 11). The process of tissue solubilization is often a compromise between the extent of solubilization and preservation of the functionality and structural integrity of liquefied biomolecules. Surfactants are often used to disband macromolecular tissue assemblies and facilitate dissolution. Nonionic and zwitterionic surfactants are preferred for functionality retention due to their mild and nondenaturing character; regrettably, these surfactants are highly ineffective in solubilizing tissues. In contrast, ionic surfactants are highly potent in solubilizing tissues; however, they denature proteins—often irreversibly—and profoundly alter the charge properties of biomolecules in solution, rendering them unusable for functional purposes. Collectively, these issues limit the applications of tissue-based molecular profiling, particularly in vivo.
In order to address these limitations, we describe a method termed Surfactant-based Tissue Acquisition for Molecular Profiling (STAMP), which combines the two key steps, tissue acquisition and solubilization, into a single-step process that can be used in vivo for profiling functionally active molecules in clinically relevant tissues. In particular, using existing surfactants as building blocks, we designed and screened over 150 binary combinations of surfactants to search for a family of surfactant formulations that are highly potent in solubilizing tissues, yet maintain the functionality of solubilized biomolecules. STAMP methodology synergistically combines these unique surfactant mixtures with a short in situ ultrasound application for rapid tissue solubilization. We demonstrate STAMP-assisted profiling of a multitude of proteins, lipids, and genomic DNA in skin and mucosal tissues. Applications of this sampling methodology to rapid molecular diagnostics of cutaneous allergies and infectious diseases are also presented.
Results
Discovery of Unique Surfactant Formulations for Enhanced Tissue Solubilization and Protein Functionality Retention.
A library of 153 binary surfactant formulations was created using 19 surfactants belonging to four distinct categories: (i) anionic surfactants, (ii) cationic surfactants, (iii) zwitterionic surfactants, and (iv) nonionic surfactants. Only a handful of surfactants from these categories (primarily, nonionic surfactants) have been traditionally utilized for solubilizing functional tissue constituents (6, 12). By combining nonionic surfactants with other types of surfactants that have been previously described for their high solubilization ability (anionic, cationic, and zwitterionic surfactants), we sought to discover families of surfactant formulations that may simultaneously possess superior solubilization as well as nondenaturing capabilities.
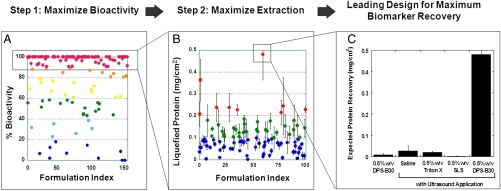
The surfactant library was first screened to identify nondenaturing formulations that retain bioactivity of a model protein, IgE antibody (Fig. 1A). The formulations spanned a wide range of IgE denaturing potentials with an increasing number of denaturing surfactants showing a synergistic gain in IgE functionality retention upon combination with gentler nonionic surfactants (more details in Fig. S1). Nondenaturing potential, averaged over all binary surfactant formulations, was significantly higher than their constituent single surfactant formulations (p < 0.01).
Fig. 1.
Discovery of synergistic surfactant formulations for enhanced recovery and functionality retention of tissue biomolecules. (A) The surfactant library was screened for identifying nondenaturing surfactant formulations that retained bioactivity of a model protein—IgE antibody. (B) Surfactant formulations exhibiting high bioactivity retention (≥90%) were further screened for their ability to solubilize proteins from porcine skin tissue. Solubilization potential of the formulations was evaluated in conjunction with a short ultrasound application. (C) Compared to conventional surfactants used in tissue solubilization studies, the leading surfactant formulation identified from screening studies—0.5% (wt/vol) DPS-B30—demonstrated a high (> 100-fold) expected protein recovery (defined as the product of fractional bioactivity retained (data reported in A) and total solubilized protein (data reported in B). Incubation of 0.5% (wt/vol) DPS-B30 without ultrasound application, and ultrasound application without incorporation of surfactant yielded significantly lower protein recovery. Errors bars indicate SD N > or = 4.
Surfactant formulations that exhibited high bioactivity retention (≥90%; Fig. 1A) were further screened for their ability to solubilize tissue proteins in conjunction with a short ultrasound application. Because highly keratinized tissues are among the toughest biological assemblies to solubilize (13), porcine skin was used as a model tissue for these studies. A majority of formulations solubilized close to 0.1 mg of protein per cm2 of skin tissue upon ultrasound application (Fig. 1B), only a handful of formulations achieved protein solubilization exceeding 0.3 mg/cm2, and only one mixture—0.5% (wt/vol) 3-(Decyl dimethyl ammonio) propane sulfonate and polyethylene glycol dodecyl ether (DPS-B30)—yielded values close to 0.5 mg/cm2. Application of 0.5% (wt/vol) DPS-B30 alone or ultrasound alone did not result in high protein solubilization (0.009 ± 0.007 mg/cm2 and 0.027 ± 0.025 mg/cm2, respectively).
Fig. 1C compares the performance of 0.5% (wt/vol) DPS-B30, with 0.5% (wt/vol) sodium lauryl sulfate (SLS), a surfactant commonly combined with ultrasound for transdermal drug delivery applications. The results are reported as expected functional protein recovery, defined as the product of fractional bioactivity retained (data from Fig. 1A) and total solubilized protein (data from Fig. 1B). In addition to possessing only a moderate solubilization ability (∼0.07 mg/cm2), SLS is highly denaturing, which results in a low expected functional protein recovery (∼0.002 mg/cm2). In contrast, DPS-B30 not only solubilized more skin proteins (0.48 mg/cm2) but also preserved protein activity, resulting in an excess of 230-fold enhancement in functional protein recovery potential over SLS. Similarly, the ability of DPS-B30 to harvest functional proteins was about 25-fold higher over commonly used nondenaturing surfactant 0.5% (wt/vol) Triton X-100.
In Vitro Validation of STAMP.
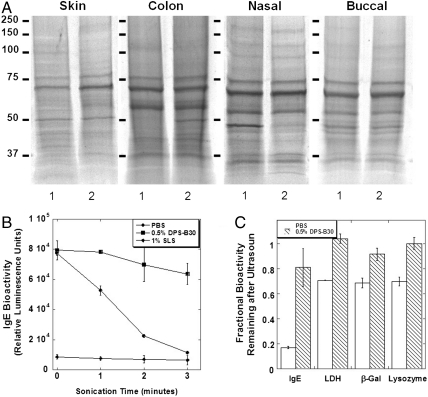
Using the leading surfactant formulation [0.5% (wt/vol) DPS-B30] and porcine skin and mucosa (colon, nasal, and buccal mucosa) as model tissues, we found that STAMP (the combination of ultrasound and surfactants) can sample a comprehensive repertoire of proteins in the same proportions as they natively exist in tissues (gel electrophoretic profiles, Fig. 2A). Gel densitometry analysis confirmed quantitative comparison between STAMP samples and homogenates (Table S1 and Fig. S2; most bands were statistically identical except for 57- to 53-kDa band for colon and 59- to 54-kDa band for nasal tissues). To assess whether STAMP-solubilized proteins remained functionally viable during sampling, we subjected four representative proteins, IgE, lactate dehydrogenase (LDH), β-galactosidase (β-Gal), and lysozyme, to the STAMP procedure and followed their bioactivity over time. A progressively sharp decrease in functionality was observed for IgE dissolved in PBS; however, DPS-B30 formulation protected IgE proteins against ultrasonic denaturing stress (Fig. 2B). As a control, irrespective of ultrasound treatment, IgE antibodies dissolved in SLS showed complete denaturation. Similar trends were also observed for LDH, β-Gal, and lysozyme (Fig. 2C).
Fig. 2.
STAMP facilitated sampling of skin and mucosal tissues, and protection of protein bioactivity from sonication stress. (A) Electrophoretic profiles of STAMP samples (lane 2) from porcine skin and various mucosal tissues (colon, nasal, and buccal mucosa) demonstrated that STAMP can sample a comprehensive repertoire of proteins in the same proportions as they natively exist in tissues (homogenized tissue; lane 1). (B and C) STAMP, when performed with 0.5% (wt/vol) DPS-B30 formulation, protected representative proteins (IgE, LDH, β-Gal, and lysozyme) against ultrasonic denaturing stress; however, a progressively sharp decrease in bioactivity was observed for proteins prepared in PBS. IgE prepared in SLS showed a complete state of denaturation. Errors bars indicate SD N = 5 (B and C).
Direct in Vivo Tissue Sampling and Molecular Diagnostics by STAMP.
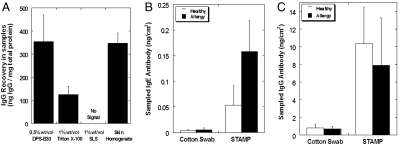
STAMP sampled functionally active proteins, in our case, IgG antibodies (Fig. 3A), from skin at a concentration that was statistically indistinguishable from the native IgG concentration in skin. IgG concentration (wt/wt) was defined as the total amount of IgG (ng) sampled by STAMP normalized by total amount of protein (mg) sampled. Sampling with conventional surfactants (TritonX-100 and SLS) was unable to represent the actual concentration of IgGs in skin. While 1% TritonX-100 recovered some IgG, it lowered its proportion in the sample by a factor of 3 compared to native skin. SLS, on the other hand, was not able to preserve the native structure of IgG.
Fig. 3.
In vivo sampling and tissue-based molecular diagnostics with STAMP methodology. (A) Sampling of IgG from mouse skin was investigated using different surfactant formulations. While sampling with conventional surfactants (TritonX-100 and SLS) was unable to represent the actual concentration of IgGs in skin, STAMP sampled IgG at a concentration that was statistically indistinguishable from the native IgG concentration in skin. (B and C) Sampling of diagnostic biomarkers localized in tissue microenvironment was investigated using a mouse model of allergic dermatitis. STAMP rapidly sampled about 3-fold higher amount of allergy biomarkers (IgE antibodies; B) from eczematic skin than healthy mouse skin (p < 0.05). No statistically significant difference was found between the amounts of IgG antibodies in the STAMP samples from allergic and healthy mice skin (C). Cotton swabbing, performed as a control procedure, sampled significantly lower amounts of IgE and IgG antibody than STAMP (p < 0.05) and could not differentiate between healthy and allergic mouse skin. Errors bars indicate SD N = 4.
To assess the ability of STAMP to sample markers from diseased skin, a mouse model of allergic dermatitis was developed and sampling of allergy biomarkers (IgE antibodies) was evaluated. STAMP samples contained about 3-fold higher amount of IgE antibodies in allergic skin than healthy mouse skin (Fig. 3B, p < 0.05). Consistent with the pathology of allergic disease, no statistically significant difference was found between the amounts of IgG antibodies sampled from allergic and healthy mice skin (Fig. 3C). Cotton swabbing, performed as a control procedure, sampled significantly lower amounts of IgE and IgG antibodies than STAMP from both healthy and allergic skin (Fig. 3 B and C, p < 0.05). IgE sampled by cotton swabbing could not differentiate the pathology of allergic skin.
STAMP-Assisted Sampling of Lipids and Nucleic Acids.
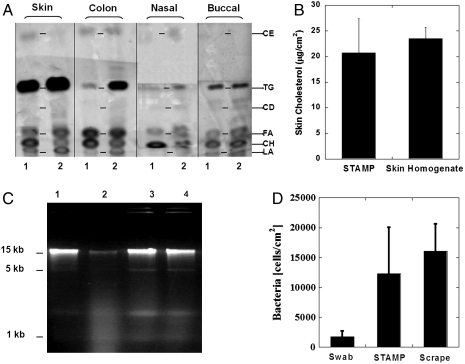
STAMP was able to retrieve all major types of nonpolar lipids from porcine skin and mucosal tissues in vitro, which compared well with the lipid profiles in tissue homogenate samples (Fig. 4A). STAMP sampled six types of lipids: cholesteryl esters (CE), triglycerides (TG), cholesteryl diesters (CD), free fatty acids (FA), cholesterol (CH) and lanosterol (LA). In vivo sampling of mouse skin showed that the amount of cholesterol sampled by STAMP from skin was representative of the actual amount of cholesterol natively present in skin (Fig. 4B).
Fig. 4.
STAMP-assisted sampling of lipids and nucleic acids. (A) Chromatographs of STAMP samples (lane 2) show that STAMP was able to retrieve a major type of nonpolar lipids from porcine skin and mucosal tissues in vitro. STAMP lipid profiles compared well with those in tissue homogenate samples (lane 1). (B) Sampling of mouse skin in vivo showed that the amount of cholesterol sampled by STAMP from skin was representative of the actual amount of cholesterol natively present in skin. (C) E. coli cell suspensions were subjected to STAMP procedure and integrity of solubilized DNA was evaluated with gel electrophoresis. Sonication significantly fragmented bacterial DNA for E. coli cells prepared in TBS (lane 2) compared to nonsonicated bacterial suspension in TBS (lane 1) or in DPS-B30 surfactant formulation (lane 3). DPS-B30 surfactant formulation (lane 4), when added to the bacterial suspension, provided outstanding protection to the DNA’s structural integrity from the sonication stress of STAMP procedure. (D) Amount of bacteria sampled by STAMP was measured with PCR by using specific primers for amplifying the conserved 16S bacterial gene in the samples. STAMP sampled at least a 7-fold higher amount of bacteria from porcine skin than the conventional cotton swabbing procedure and was equivalent to the amount sampled by aggressive scraping of skin. Errors bars indicate SD N = 4 (B and D). Representative chromatogram from three experiments (A and C).
The ability of STAMP to sample genomic DNA from excised porcine skin was also confirmed. For this purpose DNA from skin-resident bacteria was chosen as a target. DNA, which is highly susceptible to disintegration upon ultrasound exposure (Fig. 4C, lane 2 compared to lane 1), was protected when DPS-B30 was added during sonication (Fig. 4C, lane 4). The amount of DNA sampled by STAMP was measured with quantitative PCR by using specific primers for amplifying the conserved 16S bacterial gene in the samples. STAMP sampled at least a 7-fold higher amount of bacterial genome from skin compared to the conventional cotton swabbing procedure (Fig. 4D) and as much as that sampled by mechanical scraping of the epidermal skin. Amplification of genes sampled by STAMP showed that the functionality of genomic DNA was preserved during the sampling procedure.
Discussion
Efficient release of structurally intact biomolecules from tissues is an important first step in many diagnostic procedures (14). However, no standardized methods are presently available to safely and conveniently acquire tissues as well as rapidly solubilize their constituents, especially in vivo. This challenge is addressed here by STAMP. The leading design of STAMP, comprising a combination of DPS-B30 formulation with ultrasound, achieved high protein recovery and activity. Further, the results were applicable to a broad variety of tissues and biomolecules. Current strategies for preparing functional tissue samples from living tissues are invasive, laborious, and challenging. Additional issues such as degradation during handling and storage also pose limitations on their use. STAMP can acquire and solubilize living tissues and prepare a liquefied ready-to-analyze sample, all in one convenient quick step. These attributes facilitate the use of STAMP for “tissue-based” molecular diagnostics in a clinical setting (Fig. S3). Since local tissue microenvironment governs the pathophysiology of diseases manifested in tissues, STAMP-assisted molecular profiling of local biomarkers can lead to sensitive disease diagnosis such as skin IgE for allergy, skin cholesterol for atherosclerosis (15), and skin microbial DNA for infectious diseases.
The most critical enabling feature of STAMP is perhaps the preservation of the functionality and structural integrity of biomolecules during sonication. Ultrasound-induced shear and cavitation often lead to denaturation, proteolysis, and disintegration of biomolecules (16–19). This limitation is addressed by using the surfactant mixtures described here. Surfactants may potentially reduce cavitation (20, 21), but at the same time exhibit better penetration and dispersion into the skin due to ultrasound (21, 22). Indeed, increased presence of localized transport pathways on skin was found in the case of DPS-B30 compared to that seen for 1% (wt/vol) SLS (Fig. S4A), suggesting better access of DPS-B30 to the tissue. DPS-B30-ultrasound combination also yielded about a 3-fold higher proportion of soluble protein in the sample compared to that yielded by SLS-ultrasound combination (Fig. S4B). The benefits of using DPS-B30 in STAMP are further escalated by its ability to preserve the protein structure under denaturing stress of ultrasound exposure. Dynamic light scattering and Fourier-transform infrared spectroscopy studies confirmed that DPS-B30 stabilizes proteins against aggregation and other structure alterations (Fig. S5 A and B).
Ultrasound has been previously used to permeabilize the skin so as to enable transdermal drug delivery or extraction of interstitial fluid (23, 24). In that context, ultrasound exposure to skin at conditions used in this study has a history of clinical safety (25). In our experiments, no visual damage or inflammation in skin was observed for up to 48 h after the STAMP procedure. Further, both DPS and B30, the surfactants used in STAMP, have a history of use in topical products (26). However, additional safety studies focused on detailed evaluation of local tissue response as well as tolerance of the STAMP procedure by diseased tissues are warranted before the technique can be advanced to human studies. With these and other studies focused on detailed mechanistic understanding, STAMP may open additional applications for in vitro sample preparation as well as for tissue-based molecular diagnostics in vivo.
Methods
Surfactant Formulation Library.
A set of 19 representative surfactants was chosen and classified in four groups according to the surfactant’s head group chemistries (see SI Text for a list of surfactants). A library of binary surfactant formulations was prepared in 10 mM PBS (Sigma-Aldrich) by combining an anionic, cationic, or zwitterionic surfactant with a nonionic surfactant. Formulations that presented instability at room temperature were eliminated from the library design. For each pair of surfactant formulation, three different total concentrations of 0.5, 1, and 2% (wt/vol) were tested. The weight fractions of the two surfactants were kept equal in all of the formulations. A total of 153 binary surfactant formulations resulted and were studied.
Screening of Surfactant Formulations for Protein Bioactivity Retention and Tissue Solubilization.
The surfactant library was screened for identifying surfactant formulations that would retain protein bioactivity in solubilized tissues. Mouse monoclonal immunoglobulin E antibody (IgE; MCA2259, AbD Serotec) with functional binding specificity toward chicken albumin was used as a model protein for screening purposes. Activity of IgE was determined by ELISA (see details of the assay in SI Text). The surfactant formulations were ranked by calculating the percent of bioactivity retention in comparison with IgE dissolved in PBS as the positive control (100% bioactivity retention). Surfactant formulations that showed IgE bioactivity retention ≥90% were further screened for their ability to solubilize tissue proteins. Tissue solubilization was performed with surfactant dissolution in conjunction with a short 3-min sonication (2.4 W/cm2; see SI Methods for protocol). The solubilization ability for each surfactant formulation was quantified by the total amount of solubilized protein (mg) sampled per cm2 of skin exposed to surfactant formulations and ultrasound. Supernatants were isolated from the samples using a centrifuge operating at 10,000 × g and 4 °C for 15 min. The solubilized protein amount was measured in the sample supernatant by using a colorimetric detection kit (Micro BCA Protein Assay Kit; Pierce). The insoluble protein amount was determined from the centrifuged pellet. The pellet was washed in 1 mL of saline (3 times); 100 μL of saline was added to the washed pellet and homogenized in a sonication bath (Branson 1510) for 0.5 h before analysis. For each surfactant formulation, a calibration curve was obtained using freshly prepared bovine serum albumin standards.
Bioactivity Assessment of Proteins Subjected to STAMP.
The leading surfactant formulation identified after the two-step screening design—0.5% (wt/vol) DPS-B30—was tested for its ability to preserve functionality of various types of proteins under ultrasonic exposure (STAMP procedure). IgE, LDH, β-Gal, and lysozyme were tested as representative proteins. Ultrasound was applied at operating conditions as described above. In separate experiments, these proteins were added to 5 mL of surfactant formulation and transferred to a sonication chamber (centrifuge tube #430290, Corning Inc.). Proteins dissolved in PBS were prepared as comparative controls. Ultrasound was exposed by lowering the probe transducer to a distance of 5 mm from the bottom of the chamber. A maximum of two samples (100 μL each), amounting to 4% of total sonicated volume (5 mL), were periodically collected for analyzing protein bioactivity during ultrasound exposure. A higher formulation volume (5 mL) was used in these experiments to minimize sampling bias due to periodical retrieval of samples. IgE functionality was assessed using ELISA (see SI Methods for protocol) using chemiluminescent substrate (LumiGLO; KPL). LDH, β-Gal, and lysozyme enzymatic activity were measured using a colorimetric assay kit according to manufacturer’s guidelines (LDH: G1780, Promega Corp.; β-Gal: 72134, Anaspec Inc.; and lysozyme: E-22013, Invitrogen). Samples were prepared at an initial concentration of 100 ng/mL IgE (MCA2259, AbD Serotec), 1∶500 dilution of LDH stock provided in the assay kit, 10 μg/mL β-Gal (G5635; Sigma-Aldrich), and 300 U/mL lysozyme (L6876; Sigma-Aldrich).
Analysis of Sampled Biomolecules.
Lipids, proteins, and DNA collected by STAMP were analyzed using methods described in SI Text.
STAMP-Assisted Skin Sampling in Vivo.
The allergy mouse model was developed by an epicutaneous exposure protocol as described previously (27). STAMP sampling was performed with DPS-B30 formulation and ultrasound exposure at operating parameters described above (see details in SI Text). As a comparative control, samples were obtained by swabbing the skin with cotton swabs (B4320115, BD Diagnostics). Sampling was performed by swabbing the skin surfaces enclosed within a chamber (see SI Text for details). Swabs were soaked in PBS and gently rubbed against 4skin surface for 20 s. Each swab was extracted with 1 mL of PBS solution for 1 h at 4 °C.
Statistical Analysis.
Student’s t test was applied to test the significance of protein bioactivity and the amount of various biomolecules sampled by different types of surfactants, and among different sample sources including STAMP methodology and tissue homogenate.
Supplementary Material
Acknowledgments.
The authors acknowledge Dr. Raphael Simon and Dr. Anubhav Arora for helpful discussions and Casey Schmidt, Ilker Erbil, and Ari A. Pritchard-Bell for help with performing surfactant screening study. This work was funded in part by U.S. Army Medical Research and Materiel Command (W81XWH-060100400).
Footnotes
Conflict of interest statement: The authors are inventors on patent applications related to the technology described in the manuscript. University of California holds rights to the patents.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004302107/-/DCSupplemental.
References
- 1.Rifai N, Gillette M, Carr S. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–984. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Fernie A, Trethewey R, Krotzky A, Willmitzer L. Metabolite profiling: From diagnostics to systems biology. Nat Rev Mol Cell Bio. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 3.Weston A, Hood L. Systems biology, proteomics, and the future of health care: Toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 4.Hood L, Heath J, Phelps M, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 5.Thompson J, Schaeffer-Reiss C, Ueffing M. Functional Proteomics. New York: Humana Press; 2008. [DOI] [PubMed] [Google Scholar]
- 6.Scopes R. Protein Purification: Principles and Practice. New York: Springer; 1994. [Google Scholar]
- 7.Ornstein D, et al. Proteomic analysis of laser capture microdissected human prostate cancer and in vitro prostate cell lines. Electrophoresis. 2000;21:2235–2242. doi: 10.1002/1522-2683(20000601)21:11<2235::AID-ELPS2235>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 8.McDowall R. Sample preparation for biomedical analysis. J Chromatogr. 1989;492:3–5. doi: 10.1016/s0378-4347(00)84463-1. [DOI] [PubMed] [Google Scholar]
- 9.Taylor C. The total test approach to standardization of immunohistochemistry. Arch Pathol Lab Med. 2004;124:945–951. doi: 10.5858/2000-124-0945-TTTATS. [DOI] [PubMed] [Google Scholar]
- 10.Yu C, Cohen L. Tissue sample preparation: Not the same old grind. LC GC N Am. 2003;21:1038–1048. [Google Scholar]
- 11.Bodzon-Kulakowska A, et al. Methods for samples preparation in proteomic research. J Chromatogr B. 2007;849(1–2):1–31. doi: 10.1016/j.jchromb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 12.Jones MN. Surfactants in membrane solubilisation. Int J Pharm. 1999;177:137–159. doi: 10.1016/s0378-5173(98)00345-7. [DOI] [PubMed] [Google Scholar]
- 13.Rice R, Green H. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell. 1977;11:417–422. doi: 10.1016/0092-8674(77)90059-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectro Rev. 2005;24:413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 15.Bouissou H, et al. Skin cholesterol and skin apoprotein B in atherosclerosis. Biomed Pharmacother. 1982;36:159–162. [PubMed] [Google Scholar]
- 16.Hansen KC, et al. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol Cell Proteomics. 2009;8:1648–1657. doi: 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bermejo P, et al. Enzymatic digestion and ultrasonication: a powerful combination in analytical chemistry. Trac-Trend Anal Chem. 2004;23:654–663. [Google Scholar]
- 18.Elsner HI, Lindblad EB. Ultrasonic degradation of DNA. DNA. 1989;8:697–701. doi: 10.1089/dna.1989.8.697. [DOI] [PubMed] [Google Scholar]
- 19.Miller DL, Thomas RM. The role of cavitation in the induction of cellular DNA damage by ultrasound and lithotripter shock waves in vitro. Ultrasound Med Biol. 1996;22:681–687. doi: 10.1016/0301-5629(95)02077-2. [DOI] [PubMed] [Google Scholar]
- 20.Stottlemyer T, Apfel R. The effects of surfactant additives on the acoustic and light emissions from a single stable sonoluminescing bubble. J Acoust Soc Am. 1997;102:1418–1423. doi: 10.1121/1.420100. [DOI] [PubMed] [Google Scholar]
- 21.Mitragotri S, et al. Synergistic effect of low-frequency ultrasound and sodium lauryl sulfate on transdermal transport. J Pharm Sci. 2000;89:892–900. doi: 10.1002/1520-6017(200007)89:7<892::AID-JPS6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 22.Tezel A, Sens A, Tuchscherer J, Mitragotri S. Synergistic effect of low-frequency ultrasound and surfactants on skin permeability. J Pharm Sci. 2002;91:91–100. doi: 10.1002/jps.10000. [DOI] [PubMed] [Google Scholar]
- 23.Kost J, et al. Transdermal monitoring of glucose and other analytes using ultrasound. Nat Med. 2000;6:347–350. doi: 10.1038/73213. [DOI] [PubMed] [Google Scholar]
- 24.Mitragotri S, Coleman M, Kost J, Langer R. Transdermal extraction of analytes using low-frequency ultrasound. Pharm Res. 2000;17:466–470. doi: 10.1023/a:1007537222591. [DOI] [PubMed] [Google Scholar]
- 25.Ogura M, Paliwal S, Mitragotri S. Low-frequency sonophoresis: Current status and future prospects. Adv Drug Deliver Rev. 2008;60:1218–1223. doi: 10.1016/j.addr.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Skin deep: Cosmetics Safety Review Database. (Environmental Working Group) 2010. www.cosmeticsdatabase.com.
- 27.Spergel JM, et al. Epicutaneous sensitization with protein antigen induces localized allergic dermatitis and hyperresponsiveness to methacholine after single exposure to aerosolized antigen in mice. J Clin Invest. 1998;101:1614–1622. doi: 10.1172/JCI1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.